Whole Genome Analysis and Assessment of the Metabolic Potential of Gordonia rubripertincta Strain 112, a Degrader of Aromatic and Aliphatic Compounds
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain
2.2. Chemicals
2.3. Growth Media and Conditions
2.4. Determination of Hydrocarbon Concentration
2.5. Genome Sequencing and Analysis
3. Results and Discussion
3.1. Identification of Strain 112
3.2. The Plasmid of the Gordonia rubripertincta Strain 112
- Phenylacetate catabolism gene cluster;
- Styrene monooxygenase StyA, 3 beta-hydroxysteroid dehydrogenase/Delta 5-->4-isomerase;
- Genes involved in type IV secretory system Conjugative DNA transfer.
3.3. Pangenome Analysis of Gordonia rubripertincta Strains
3.4. Functional Annotation of the Genome of the Strain 112
3.4.1. Diversity of Aromatic Compound Catabolism Genes in the Genome of Gordonia rubripertincta 112
3.4.2. Diversity of Alkane Catabolism Genes in Strain Gordonia rubripertincta 112
3.5. Peculiarities of Alkane Catabolism by Strain G. rubripertincta 112
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cha, J.-H.; Cha, C.-J. Gordonia alkaliphila sp. nov., an actinomycete isolated from tidal flat sediment. Int. J. Syst. Evol. Microbiol. 2013, 63 Pt 1, 327–331. [Google Scholar] [CrossRef]
- He, Y.; Lyu, L.; Hu, Z.; Yu, Z.; Shao, Z. Gordonia tangerina sp. nov., isolated from seawater. Int. J. Syst. Evol. Microbiol. 2022, 72, 005632. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Dong, K.; Kim, J.; Lee, S. Characteristics of crude oil-degrading bacteria Gordonia iterans isolated from marine coastal in Taean sediment. Microbiologyopen 2019, 8, e00754. [Google Scholar] [CrossRef]
- Sangkanu, S.; Suriyachadkun, C.; Phongpaichit, S. Gordonia sediminis sp. nov., an actinomycete isolated from mangrove sediment. Int. J. Syst. Evol. Microbiol. 2019, 69, 1814–1820. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.-T.; Goodfellow, M.; Jones, A.L.; Chen, Y.-P.; Arun, A.B.; Lai, W.-A.; Rekha, P.D.; Young, C.-C. Gordonia soli sp. nov., a novel actinomycete isolated from soil. Int. J. Syst. Evol. Microbiol. 2006, 56, 2597–2601. [Google Scholar] [CrossRef] [PubMed]
- Brandão, P.F.B.; Maldonado, L.A.; Ward, A.C.; Bull, A.T.; Goodfellow, M. Gordonia namibiensis sp. nov., a Novel Nitrile MetabolisingActinomycete Recovered from an African Sand. Syst. Appl. Microbiol. 2001, 24, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Xu, M.; Liu, H.; Yu, T.; Guo, P.; Liu, W.; Jin, X. Antimicrobial compounds were isolated from the secondary metabolites of Gordonia, a resident of intestinal tract of Periplaneta americana. AMB Express 2021, 11, 111. [Google Scholar] [CrossRef]
- Mohammadipanah, F.; Wink, J. Actinobacteria from Arid and Desert Habitats: Diversity and Biological Activity. Front. Microbiol. 2016, 6, 1541. [Google Scholar] [CrossRef]
- Kepenek, E.S.; Severcan, M.; Gozen, A.G.; Severcan, F. Discrimination of heavy metal acclimated environmental strains by chemometric analysis of FTIR spectra. Ecotoxicol. Environ. Saf. 2020, 202, 110953. [Google Scholar] [CrossRef]
- Kayasth, M.; Kumar, V.; Gera, R. Gordonia sp.: A salt tolerant bacterial inoculant for growth promotion of pearl millet under saline soil conditions. 3 Biotech 2014, 4, 553–557. [Google Scholar] [CrossRef]
- Ehiosun, K.I.; Godin, S.; Urios, L.; Lobinski, R.; Grimaud, R. Degradation of long-chain alkanes through biofilm formation by bacteria isolated from oil-polluted soil. Int. Biodeterior. Biodegradation 2022, 175, 105508. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, J.; Liu, Y.; Wu, X. Biological Process of Alkane Degradation by Gordonia sihwaniensis. ACS Omega 2022, 7, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Lo Piccolo, L.; De Pasquale, C.; Fodale, R.; Puglia, A.M.; Quatrini, P. Involvement of an Alkane Hydroxylase System of Gordonia sp. Strain SoCg in Degradation of Solid n-Alkanes. Appl. Environ. Microbiol. 2011, 77, 1204–1213. [Google Scholar] [CrossRef] [PubMed]
- Silva, N.M.; de Oliveira, A.M.S.A.; Pegorin, S.; Giusti, C.E.; Ferrari, V.B.; Barbosa, D.; Martins, L.F.; Morais, C.; Setubal, J.C.; Vasconcellos, S.P.; et al. Characterization of novel hydrocarbon-degrading Gordonia paraffinivorans and Gordonia sihwensis strains isolated from composting. PLoS ONE 2019, 14, e0215396. [Google Scholar] [CrossRef] [PubMed]
- Frantsuzova, E.; Solomentsev, V.; Vetrova, A.; Travkin, V.; Solyanikova, I.; Delegan, Y. Complete Genome Sequence of Gordonia polyisoprenivorans 135, a Promising Degrader of Aromatic Compounds. Microbiol. Resour. Announc. 2023, 12, e0005823. [Google Scholar] [CrossRef]
- Lin, C.-L.; Shen, F.-T.; Tan, C.-C.; Huang, C.-C.; Chen, B.-Y.; Arun, A.; Young, C.-C. Characterization of Gordonia sp. strain CC-NAPH129-6 capable of naphthalene degradation. Microbiol. Res. 2012, 167, 395–404. [Google Scholar] [CrossRef]
- Silva, A.S.; de Oliveira Camargo, F.A.; Andreazza, R.; Jacques, R.J.S.; Baldoni, D.B.; Bento, F.M. Enzymatic activity of catechol 1,2-dioxygenase and catechol 2,3-dioxygenase produced by Gordonia polyisoprenivorans. Química Nova 2012, 35, 1587–1592. [Google Scholar] [CrossRef]
- Suzina, N.E.; Sorokin, V.V.; Polivtseva, V.N.; Klyueva, V.V.; Emelyanova, E.V.; Solyanikova, I.P. From Rest to Growth: Life Collisions of Gordonia polyisoprenivorans 135. Microorganisms 2022, 10, 465. [Google Scholar] [CrossRef]
- Akhtar, N.; Akhtar, K.; Ghauri, M.A. Biodesulfurization of Thiophenic Compounds by a 2-Hydroxybiphenyl-Resistant Gordonia sp. HS126-4N Carrying dszABC Genes. Curr. Microbiol. 2018, 75, 597–603. [Google Scholar] [CrossRef]
- Delegan, Y.; Kocharovskaya, Y.; Frantsuzova, E.; Streletskii, R.; Vetrova, A. Characterization and genomic analysis of Gordonia alkanivorans 135, a promising dibenzothiophene-degrading strain. Biotechnol. Rep. 2021, 29, e00591. [Google Scholar] [CrossRef]
- Wang, W.; Ma, T.; Lian, K.; Zhang, Y.; Tian, H.; Ji, K.; Li, G. Genetic Analysis of Benzothiophene Biodesulfurization Pathway of Gordonia terrae Strain C-6. PLoS ONE 2013, 8, e84386. [Google Scholar] [CrossRef]
- Hu, T.; Yang, C.; Hou, Z.; Liu, T.; Mei, X.; Zheng, L.; Zhong, W. Phthalate Esters Metabolic Strain Gordonia sp. GZ-YC7, a Potential Soil Degrader for High Concentration Di-(2-ethylhexyl) Phthalate. Microorganisms 2022, 10, 641. [Google Scholar] [CrossRef]
- Jin, D.; Kong, X.; Jia, M.; Yu, X.; Wang, X.; Zhuang, X.; Deng, Y.; Bai, Z. Gordonia phthalatica sp. nov., a di-n-butyl phthalate-degrading bacterium isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 2017, 67, 5128–5133. [Google Scholar] [CrossRef]
- Kanaujiya, D.K.; Sivashanmugam, S.; Pakshirajan, K. Biodegradation and toxicity removal of phthalate mixture by Gordonia sp. in a continuous stirred tank bioreactor system. Environ. Technol. Innov. 2022, 26, 102324. [Google Scholar] [CrossRef]
- Nahurira, R.; Ren, L.; Song, J.; Jia, Y.; Wang, J.; Fan, S.; Wang, H.; Yan, Y. Degradation of Di(2-Ethylhexyl) Phthalate by a Novel Gordonia alkanivorans Strain YC-RL2. Curr. Microbiol. 2017, 74, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, J.; Chowdhury, P.P.; Dutta, T.K. Complete degradation of di-n-octyl phthalate by Gordonia sp. strain Dop5. Chemosphere 2013, 90, 2571–2577. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.B.; Brown, R.; Oldfield, C.; Gilbert, S.C.; Iliarionov, S.; Goodfellow, M. Gordonia amicalis sp. nov., a novel dibenzothiophene-desulphurizing actinomycete. Int. J. Syst. Evol. Microbiol. 2000, 50, 2031–2036. [Google Scholar] [CrossRef]
- Kummer, C.; Schumann, P.; Stackebrandt, E. Gordonia alkanivorans sp. nov., isolated from tar-contaminated soil. Int. J. Syst. Evol. Microbiol. 1999, 49, 1513–1522. [Google Scholar] [CrossRef] [PubMed]
- Linos, A.; Steinbüchel, A.; Spröer, C.; Kroppenstedt, R.M. Gordonia polyisoprenivorans sp. nov., a rubber-degrading actinomycete isolated from an automobile tyre. Int. J. Syst. Evol. Microbiol. 1999, 49, 1785–1791. [Google Scholar] [CrossRef]
- Linos, A.; Berekaa, M.M.; Steinbüchel, A.; Kim, K.K.; Sproer, C.; Kroppenstedt, R.M. Gordonia westfalica sp. nov., a novel rubber-degrading actinomycete. Int. J. Syst. Evol. Microbiol. 2002, 52, 1133–1139. [Google Scholar] [CrossRef]
- Xue, Y.; Sun, X.; Zhou, P.; Liu, R.; Liang, F.; Ma, Y. Gordonia paraffinivorans sp. nov., a hydrocarbon-degrading actinomycete isolated from an oil-producing well. Int. J. Syst. Evol. Microbiol. 2003, 53, 1643–1646. [Google Scholar] [CrossRef] [PubMed]
- Esuola, C.O.; Babalola, O.O.; Heine, T.; Schwabe, R.; Schlömann, M.; Tischler, D. Identification and characterization of a FAD-dependent putrescine N-hydroxylase (GorA) from Gordonia rubripertincta CWB2. J. Mol. Catal. B Enzym. 2016, 134, 378–389. [Google Scholar] [CrossRef]
- Hofmann, M.; Martin del Campo, J.S.; Sobrado, P.; Tischler, D. Biosynthesis of desferrioxamine siderophores initiated by decarboxylases: A functional investigation of two lysine/ornithine-decarboxylases from Gordonia rubripertincta CWB2 and Pimelobacter simplex 3E. Arch. Biochem. Biophys. 2020, 689, 108429. [Google Scholar] [CrossRef] [PubMed]
- Schwabe, R.; Obst, B.; Mehnert, M.; Tischler, D.; Wiche, O. Gallium Mobilization in Soil by Bacterial Metallophores. Solid State Phenom. 2017, 262, 513–516. [Google Scholar] [CrossRef]
- Schwabe, R.; Senges, C.H.R.; Bandow, J.E.; Heine, T.; Lehmann, H.; Wiche, O.; Schlömann, M.; Levicán, G.; Tischler, D. Data on metal-chelating, -immobilisation and biosorption properties by Gordonia rubripertincta CWB2 in dependency on rare earth adaptation. Data Brief 2020, 31, 105739. [Google Scholar] [CrossRef] [PubMed]
- Schwabe, R.; Senges, C.H.R.; Bandow, J.E.; Heine, T.; Lehmann, H.; Wiche, O.; Schlömann, M.; Levicán, G.; Tischler, D. Cultivation dependent formation of siderophores by Gordonia rubripertincta CWB2. Microbiol. Res. 2020, 238, 126481. [Google Scholar] [CrossRef] [PubMed]
- Eggerichs, D.; Mügge, C.; Mayweg, J.; Apfel, U.-P.; Tischler, D. Enantioselective Epoxidation by Flavoprotein Monooxygenases Supported by Organic Solvents. Catalysts 2020, 10, 568. [Google Scholar] [CrossRef]
- Heine, T.; Zimmerling, J.; Ballmann, A.; Kleeberg, S.B.; Rückert, C.; Busche, T.; Winkler, A.; Kalinowski, J.; Poetsch, A.; Scholtissek, A.; et al. On the Enigma of Glutathione-Dependent Styrene Degradation in Gordonia rubripertincta CWB2. Appl. Environ. Microbiol. 2018, 84, e00154-18. [Google Scholar] [CrossRef] [PubMed]
- Zimmerling, J.; Oelschlägel, M.; Großmann, C.; Voitel, M.; Schlömann, M.; Tischler, D. Biochemical Characterization of Phenylacetaldehyde Dehydrogenases from Styrene-degrading Soil Bacteria. Appl. Biochem. Biotechnol. 2020, 193, 650–667. [Google Scholar] [CrossRef] [PubMed]
- Han, S.-S.; Kang, H.K.; Jo, B.Y.; Ryu, B.-G.; Jin, H.M.; Chung, E.J.; Jung, J.Y. Complete Genome Sequence of Gordonia rubripertincta SD5, a Soil Bacterium Isolated from a Di-(2-Ethylhexyl) Phthalate-Degrading Enrichment Culture. Genome Announc. 2020, 9, e01087-20. [Google Scholar] [CrossRef]
- Trögl, J.; Esuola, C.O.; Kříženecká, S.; Kuráň, P.; Seidlová, L.; Veronesi-Dáňová, P.; Popelka, J.; Babalola, O.O.; Hrabák, P.; Czinnerová, M.; et al. Biodegradation of High Concentrations of Aliphatic Hydrocarbons in Soil from a Petroleum Refinery: Implications for Applicability of New Actinobacterial Strains. Appl. Sci. 2018, 8, 1855. [Google Scholar] [CrossRef]
- Kotani, T.; Yamamoto, T.; Yurimoto, H.; Sakai, Y.; Kato, N. PropaneMonooxygenase and NAD + -Dependent Secondary AlcoholDehydrogenase in Propane Metabolism by Gordonia sp. Strain TY-5. J. Bacteriol. 2003, 185, 7120–7128. [Google Scholar] [CrossRef] [PubMed]
- Kotani, T.; Yurimoto, H.; Kato, N.; Sakai, Y. Novel Acetone Metabolism in a Propane-Utilizing Bacterium, Gordonia sp. Strain TY-5. J. Bacteriol. 2007, 189, 886–893. [Google Scholar] [CrossRef]
- van Beilen, J.B.; Funhoff, E.G.; van Loon, A.; Just, A.; Kaysser, L.; Bouza, M.; Holtackers, R.; Röthlisberger, M.; Li, Z.; Witholt, B. Cytochrome P450 Alkane Hydroxylases of the CYP153 Family Are Common in Alkane-Degrading Eubacteria Lacking Integral Membrane Alkane Hydroxylases. Appl. Environ. Microbiol. 2006, 72, 59–65. [Google Scholar] [CrossRef]
- Delegan, Y.A.; Valentovich, L.N.; Shafieva, S.M.; Ganbarov, K.G.; Filonov, A.E.; Vainstein, M.B. Characterization and genomic analysis of highly efficient thermotolerant oil-degrading bacterium Gordonia sp. 1D. Folia Microbiol. 2018, 64, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Liang, J.; Fang, H.; Tang, Y.-Q.; Wu, X.-L. Two Novel Alkane Hydroxylase-Rubredoxin Fusion Genes Isolated from a Dietzia Bacterium and the Functions of Fused Rubredoxin Domains in Long-Chain n-Alkane Degradation. Appl. Environ. Microbiol. 2011, 77, 7279–7288. [Google Scholar] [CrossRef]
- Nie, Y.; Liang, J.-L.; Fang, H.; Tang, Y.-Q.; Wu, X.-L. Characterization of a CYP153 alkane hydroxylase gene in a Gram-positive Dietzia sp. DQ12-45-1b and its “team role” with alkW1 in alkane degradation. Appl. Microbiol. Biotechnol. 2014, 98, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Bertani, G. STUDIES ON LYSOGENESIS I. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program: Table 1. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Reeve, W.; O’hara, G.; Chain, P.; Ardley, J.; Brau, L.; Nandesena, K.; Tiwari, R.; Malfatti, S.; Kiss, H.; Lapidus, A.; et al. Complete genome sequence of Rhizobium leguminosarum bv trifolii strain WSM2304, an effective microsymbiont of the South American clover Trifolium polymorphum. Stand. Genom. Sci. 2010, 2, 66–76. [Google Scholar] [CrossRef]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple Alignment of Conserved Genomic Sequence with Rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef]
- Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Ivshina, I.B.; Kevorkov, N.N.; Koblova, I.V.; Nesterenko, O.A.; Kvasnikov, E.I.; Kasumova, S.A. Identification of bacteria belonging to the genus Rhodococcus using the technique of immunodiffusion. Mikrobiologiya 1980, 51, 636–641. [Google Scholar]
- Iminova, L.; Delegan, Y.; Frantsuzova, E.; Bogun, A.; Zvonarev, A.; Suzina, N.; Anbumani, S.; Solyanikova, I. Physiological and biochemical characterization and genome analysis of Rhodococcus qingshengii strain 7B capable of crude oil degradation and plant stimulation. Biotechnol. Rep. 2022, 35, e00741. [Google Scholar] [CrossRef]
- Goral, A.M.; Tkaczuk, K.L.; Chruszcz, M.; Kagan, O.; Savchenko, A.; Minor, W. Crystal structure of a putative isochorismatase hydrolase from Oleispira antarctica. J. Struct. Funct. Genom. 2012, 13, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Drzyzga, O.; Fernández de las Heras, L.; Morales, V.; Navarro Llorens, J.M.; Perera, J. Cholesterol Degradation by Gordonia cholesterolivorans. Appl. Environ. Microbiol. 2011, 77, 4802–4810. [Google Scholar] [CrossRef]
- Ge, F.; Li, W.; Chen, G.; Liu, Y.; Zhang, G.; Yong, B.; Wang, Q.; Wang, N.; Huang, Z.; Li, W.; et al. Draft Genome Sequence of Gordonia neofelifaecis NRRL B-59395, a Cholesterol-Degrading Actinomycete. J. Bacteriol. 2011, 193, 5045–5046. [Google Scholar] [CrossRef]
- Zhang, Q.; Ren, Y.; He, J.; Cheng, S.; Yuan, J.; Ge, F.; Li, W.; Zhang, Y.; Xie, G. Multiplicity of 3-ketosteroid Δ1-dehydrogenase enzymes in Gordonia neofelifaecis NRRL B-59395 with preferences for different steroids. Ann. Microbiol. 2015, 65, 1961–1971. [Google Scholar] [CrossRef]
- Shen, F.-T.; Lin, J.-L.; Huang, C.-C.; Ho, Y.-N.; Arun, A.B.; Young, L.-S.; Young, C.-C. Molecular detection and phylogenetic analysis of the catechol 1,2-dioxygenase gene from Gordonia spp. Syst. Appl. Microbiol. 2009, 32, 291–300. [Google Scholar] [CrossRef]
- Solyanikova, I.P.; Emelyanova, E.V.; Shumkova, E.S.; Egorova, D.O.; Korsakova, E.S.; Plotnikova, E.G.; Golovleva, L.A. Peculiarities of the degradation of benzoate and its chloro- and hydroxy-substituted analogs by actinobacteria. Int. Biodeterior. Biodegradation 2015, 100, 155–164. [Google Scholar] [CrossRef]
- Macaya, C.C.; Durán, R.E.; Hernández, L.; Rodríguez-Castro, L.; Barra-Sanhueza, B.; Dorochesi, F.; Seeger, M. Bioremediation of Petroleum. In Reference Module in Life Sciences; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar] [CrossRef]

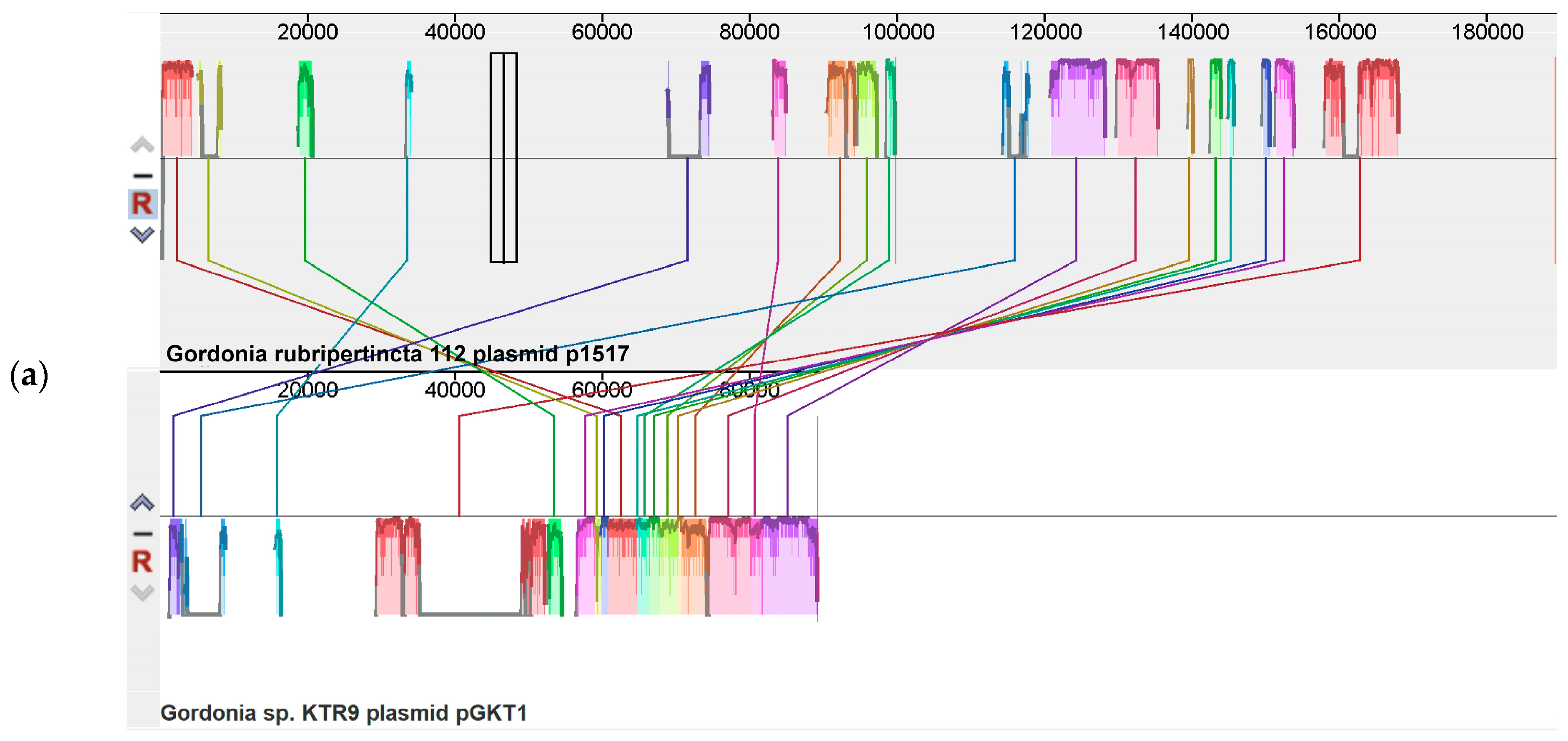
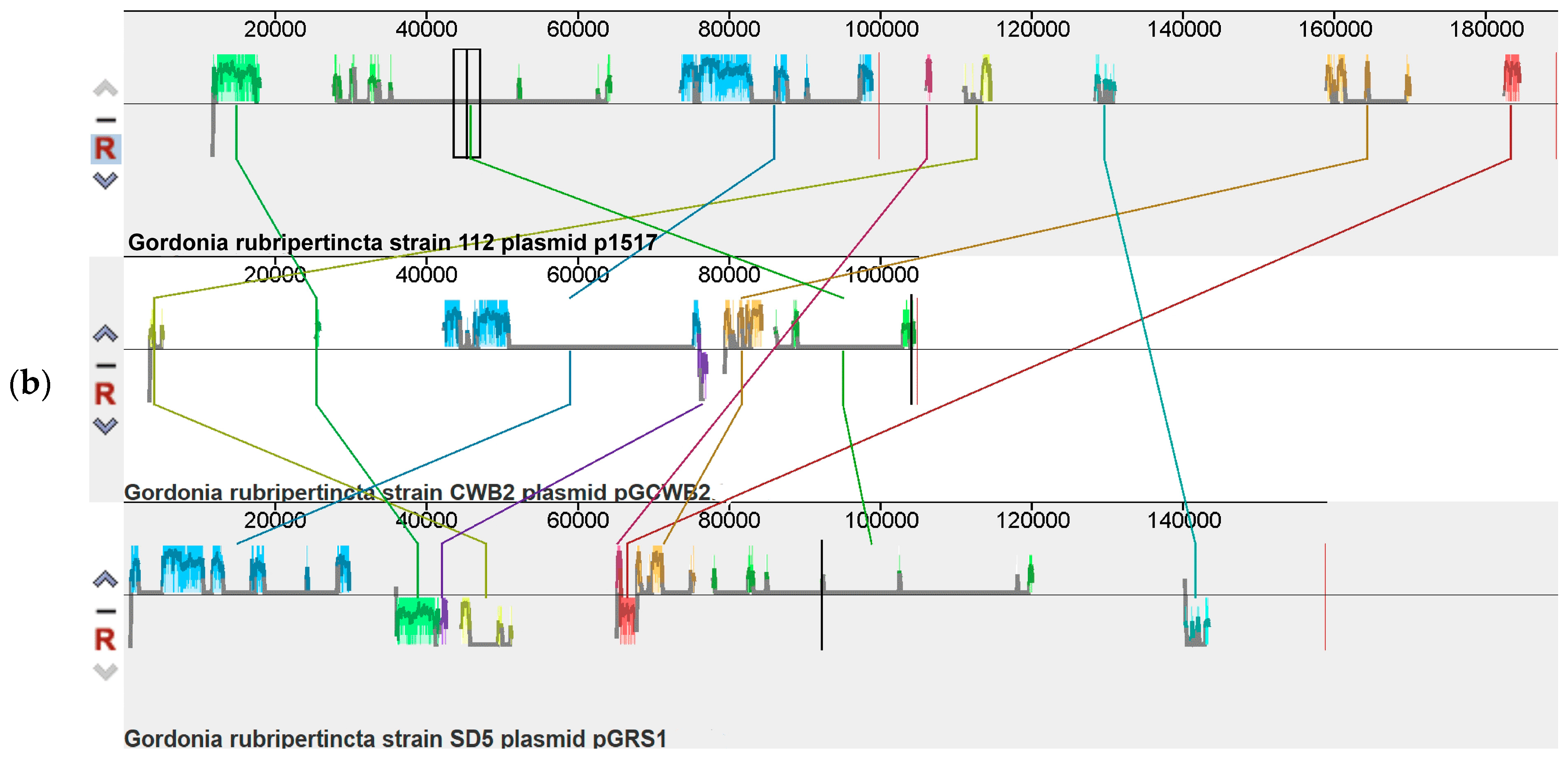
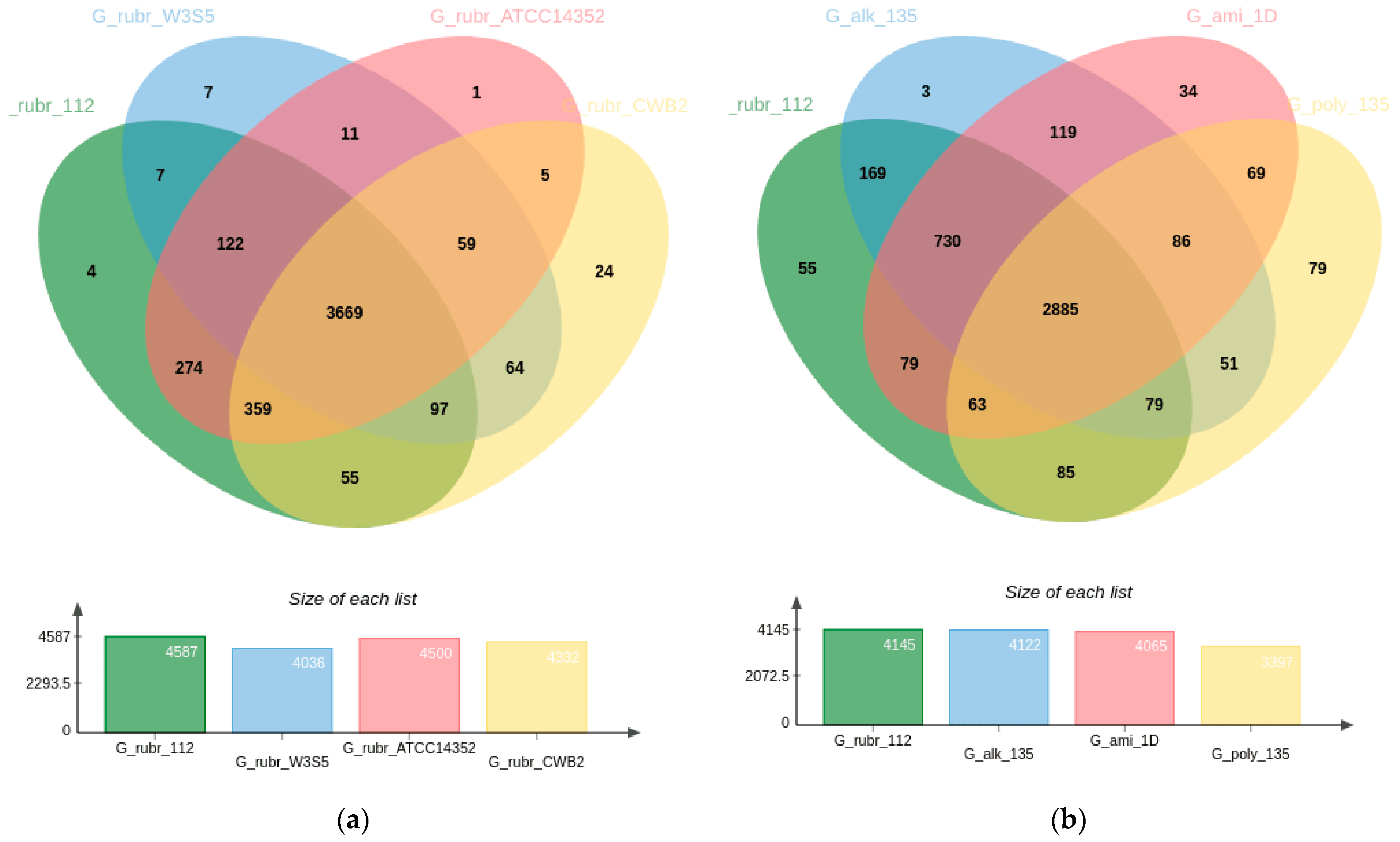

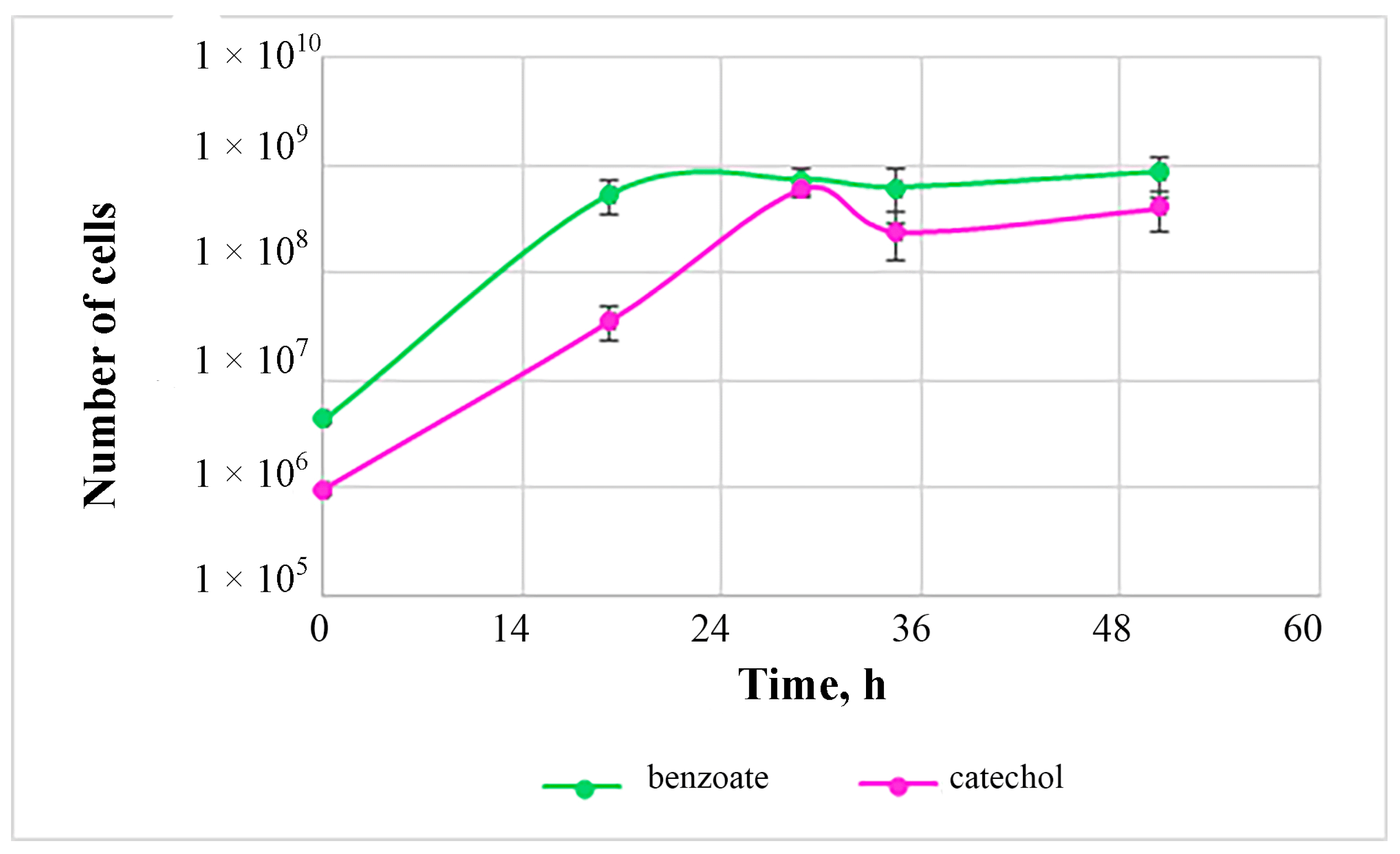
| Parameter | Length |
|---|---|
| Genome size, bp | 5,281,129 |
| The longest contig, bp | 845,133 |
| The shortest contig, bp | 502 |
| N50, bp | 233,583 |
| N75, bp | 130,856 |
| N90, bp | 59,209 |
| GenBank Accession Number | Strain Name | Chromosome Size, Mb | Plasmid Size, kb |
|---|---|---|---|
| JARUXG000000000 | Gordonia rubripertincta 112 | 5.09 | 189 |
| JAAXPB000000000.1 | Gordonia rubripertincta ATCC14352T | 5.70 | - |
| JAFFGU000000000.1 | Gordonia rubripertincta BP-295 | 5.15 | - |
| CP022580.1 | Gordonia rubripertincta CWB2 | 5.23 | 105 |
| Strain Name | ANI Value, % | DDH Value, % |
|---|---|---|
| Gordonia rubripertincta ATCC14352T | 99.98 | 98.90 |
| Gordonia rubripertincta BP-295 | 98.18 | 78.40 |
| Gordonia rubripertincta CWB2 | 98.08 | 86.40 |
| Gordonia rubripertincta SD5 | 98.43 | 77.70 |
| Gordonia rubripertincta W3S5 | 98.06 | 81.80 |
| Gene Onthology (GO) Category Identifier | Function | Gene Identifier According to the GenBank Annotation | |
|---|---|---|---|
| 112 | 135 | ||
| 0019439 | aromatic compound catabolic process | 01466 | 00188 |
| 0006707 | cholesterol catabolic process | 00126 | 03312 |
| 0008202 | steroid metabolic process | 00127 | 03311 |
| 0006694 | steroid biosynthetic process | 00124 00123 | 03314 03315 |
| 0009712 | catechol-containing compound metabolic process | 01163 | 01469 |
| Strain | Sequence Name | Query Cover, % | Per. Ident, % |
|---|---|---|---|
| G. alkanivorans 135 | isochorismatase family protein | 100 | 100 |
| G. rubripertincta SD5 | isochorismatase family protein | 100 | 97.29 |
| G. alkanivorans YC-RL2 | isochorismatase | 100 | 96.68 |
| G. alkanivorans GH-1 | isochorismatase | 100 | 96.68 |
| G. rubripertincta CWB2 | maleamate amidohydrolase | 69 | 96.98 |
| GenBank Accession Number | Strain Name | Total Number of Genes | Functionally Annotated Genes | Functionally Annotated Genes, % |
|---|---|---|---|---|
| JARUXG000000000 | Gordonia rubripertincta 112 | 4787 | 2212 | 46.2 |
| JAAXPB000000000.1 | Gordonia rubripertincta ATCC14352T | 5023 | 2352 | 46.8 |
| JAFFGU000000000.1 | Gordonia rubripertincta BP-295 | 4650 | 2086 | 44.9 |
| CP022580.1 | Gordonia rubripertincta CWB2 | 4707 | 2092 | 44.4 |
| CP059694.1 | Gordonia rubripertincta SD5 | 4670 | 2058 | 44.1 |
| VLNS00000000.1 | Gordonia rubripertincta W3S5 | 4252 | 1993 | 46.9 |
| Strain | C1,2DO | C2,3DO |
|---|---|---|
| ATCC14352 | 2 | 1 |
| W3S5 | 2 | 1 |
| BP-295 | 2 | - |
| SD5 | 2 | - |
| CWB2 | 2 | - |
| 112 | 2 | 1 |
| Strain | Genbank Acc Number | Query Cover, % | Percent Identity, % |
|---|---|---|---|
| Rhodococcus pseudokoreensis R79 | CP070619 | 93 | 74.96 |
| Rhodococcus sp. USK10 | CP076048 | 94 | 74.80 |
| Rhodococcus opacus B4 | AP011115 | 94 | 74.19 |
| Growth Substrate | Abiotic Loss, % | Degradation Degree, % |
|---|---|---|
| hexadecane | 1.21 ± 0.13 | 10.79 ± 1.17 |
| decane | 62.87 ± 0.41 | 16.14 ± 0.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frantsuzova, E.; Bogun, A.; Solomentsev, V.; Vetrova, A.; Streletskii, R.; Solyanikova, I.; Delegan, Y. Whole Genome Analysis and Assessment of the Metabolic Potential of Gordonia rubripertincta Strain 112, a Degrader of Aromatic and Aliphatic Compounds. Biology 2023, 12, 721. https://doi.org/10.3390/biology12050721
Frantsuzova E, Bogun A, Solomentsev V, Vetrova A, Streletskii R, Solyanikova I, Delegan Y. Whole Genome Analysis and Assessment of the Metabolic Potential of Gordonia rubripertincta Strain 112, a Degrader of Aromatic and Aliphatic Compounds. Biology. 2023; 12(5):721. https://doi.org/10.3390/biology12050721
Chicago/Turabian StyleFrantsuzova, Ekaterina, Alexander Bogun, Viktor Solomentsev, Anna Vetrova, Rostislav Streletskii, Inna Solyanikova, and Yanina Delegan. 2023. "Whole Genome Analysis and Assessment of the Metabolic Potential of Gordonia rubripertincta Strain 112, a Degrader of Aromatic and Aliphatic Compounds" Biology 12, no. 5: 721. https://doi.org/10.3390/biology12050721
APA StyleFrantsuzova, E., Bogun, A., Solomentsev, V., Vetrova, A., Streletskii, R., Solyanikova, I., & Delegan, Y. (2023). Whole Genome Analysis and Assessment of the Metabolic Potential of Gordonia rubripertincta Strain 112, a Degrader of Aromatic and Aliphatic Compounds. Biology, 12(5), 721. https://doi.org/10.3390/biology12050721







