Monitoring Snake Venom-Induced Extracellular Matrix Degradation and Identifying Proteolytically Active Venom Toxins Using Fluorescently Labeled Substrates
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Venoms
2.2. Chemicals and Reagents
2.3. Detection of Proteolytic Activity of Snake Venoms
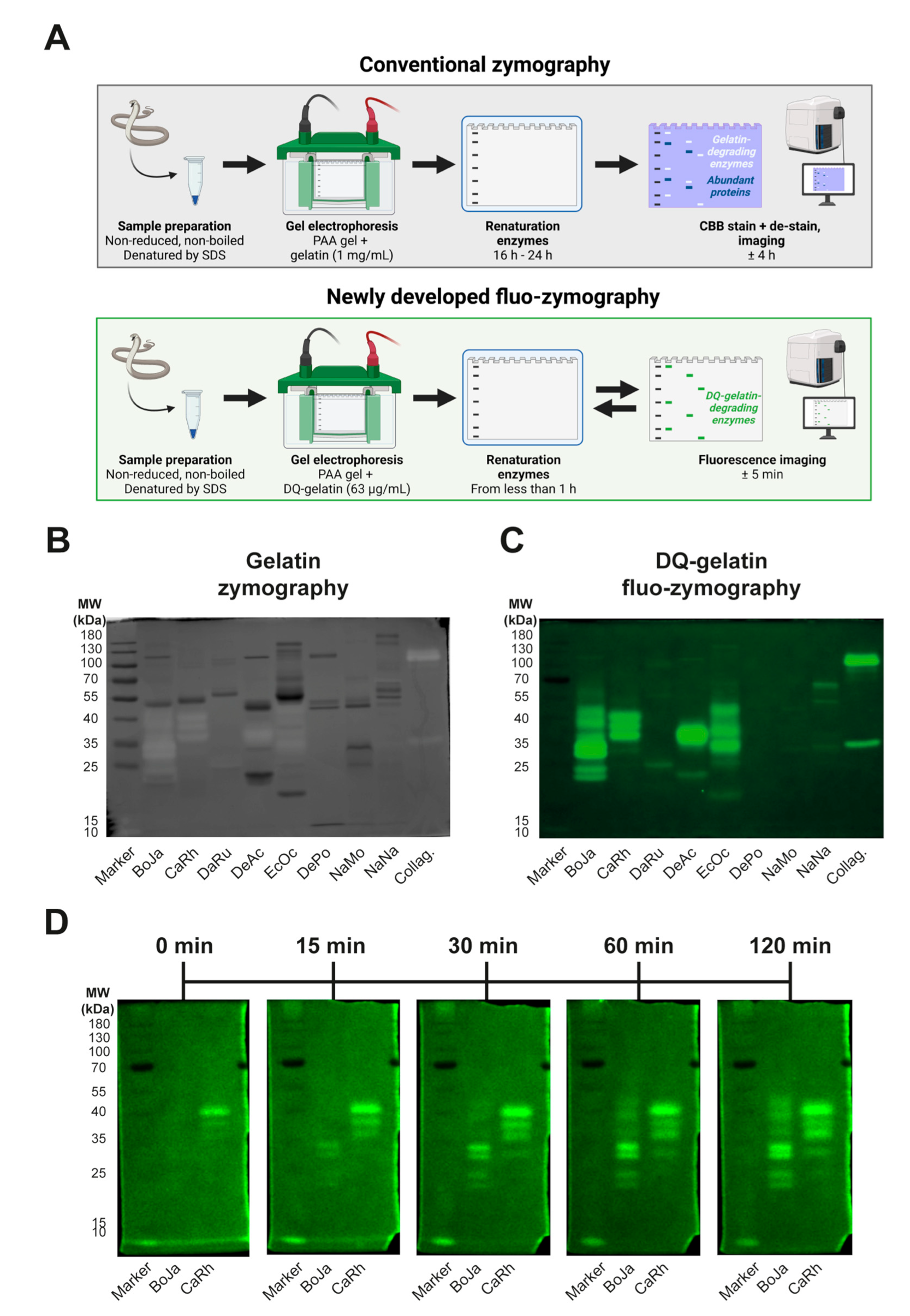
2.4. Identification of Proteolytically Active Proteins in Venoms Using In-Gel (Fluo-)Zymography
2.5. In-Gel Tryptic Digestion of Proteolytically Active Venom Components
2.6. Toxin Identification Using Proteomics
2.7. Statistical Analysis
3. Results and Discussion
3.1. Profiling of Proteolytic Degradation Using SDS-PAGE Gels
3.2. Assessing Potency and Kinetics for Degradation of Fluorescent ECM Components
3.3. Extending the Method to Monitor Substrate-Specific Inhibition of ECM Degradation
3.4. Identification of Proteolytic Components Using (Fluo-)Zymography
3.5. Validation of Proteolytically Active Proteins within Crude Snake Venoms Using Proteomics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gutiérrez, J.M.; Calvete, J.J.; Habib, A.G.; Harrison, R.A.; Williams, D.J.; Warrell, D.A. Snakebite envenoming. Nat. Rev. Dis. Prim. 2017, 3, 17063. [Google Scholar] [CrossRef] [PubMed]
- Kasturiratne, A.; Wickremasinghe, A.R.; De Silva, N.; Gunawardena, N.K.; Pathmeswaran, A.; Premaratna, R.; Savioli, L.; Lalloo, D.G.; De Silva, H.J. The global burden of snakebite: A literature analysis and modelling based on regional estimates of envenoming and deaths. PLoS Med. 2008, 5, e218. [Google Scholar] [CrossRef] [PubMed]
- Habib, A.G.; Kuznik, A.; Hamza, M.; Abdullahi, M.I.; Chedi, B.A.; Chippaux, J.-P.; Warrell, D.A. Snakebite is Under Appreciated: Appraisal of Burden from West Africa. PLoS Negl. Trop. Dis. 2015, 9, e0004088. [Google Scholar] [CrossRef]
- Warrell, D.A. Guidelines of Management of Snake bite. Lancet 2010, 375, 77–88. [Google Scholar] [CrossRef]
- Waheed, H.; Moin, S.F.; Choudhary, M.I. Snake Venom: From Deadly Toxins to Life-saving Therapeutics. Curr. Med. Chem. 2017, 24, 1874–1891. [Google Scholar] [CrossRef] [PubMed]
- Tasoulis, T.; Isbister, G.K. A review and database of snake venom proteomes. Toxins 2017, 9, 290. [Google Scholar] [CrossRef] [PubMed]
- Longbottom, J.; Shearer, F.M.; Devine, M.; Alcoba, G.; Chappuis, F.; Weiss, D.J.; Ray, S.E.; Ray, N.; Warrell, D.A.; de Castañeda, R.R.; et al. Vulnerability to snakebite envenoming: A global mapping of hotspots. Lancet 2018, 392, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Chippaux, J.P. Snakebite envenomation turns again into a neglected tropical disease! J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 1–2. [Google Scholar] [CrossRef]
- Alberts, B.; Hopkin, K.; Johnson, A.D.; Morgan, D.; Raff, M.; Roberts, K.; Walter, P. Essential Cell Biology: Fifth International Student Edition; WW Norton & Company: New York, NY, USA, 2018. [Google Scholar]
- Gutiérrez, J.M.; Escalante, T.; Rucavado, A.; Herrera, C.; Fox, J.W. A comprehensive view of the structural and functional alterations of extracellular matrix by snake venom metalloproteinases (SVMPs): Novel perspectives on the pathophysiology of envenoming. Toxins 2016, 8, 304. [Google Scholar] [CrossRef]
- Gutiérrez, J.M.; Escalante, T.; Rucavado, A.; Herrera, C. Hemorrhage caused by snake venom metalloproteinases: A journey of discovery and understanding. Toxins 2016, 8, 93. [Google Scholar] [CrossRef]
- Jayadev, R.; Sherwood, D.R. Basement membranes. Curr. Biol. 2017, 27, R207–R211. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M.; Rucavado, A.; Escalante, T.; Díaz, C. Hemorrhage induced by snake venom metalloproteinases: Biochemical and biophysical mechanisms involved in microvessel damage. Toxicon 2005, 45, 997–1011. [Google Scholar] [CrossRef]
- Takeda, S.; Takeya, H.; Iwanaga, S. Snake venom metalloproteinases: Structure, function and relevance to the mammalian ADAM/ADAMTS family proteins. Biochim. Biophys. Acta Proteins Proteom. 2012, 1824, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Escalante, T.; Ortiz, N.; Rucavado, A.; Sanchez, E.F.; Richardson, M.; Fox, J.W.; Gutiérrez, J.M. Role of collagens and perlecan in microvascular stability: Exploring the mechanism of capillary vessel damage by snake venom metalloproteinases. PLoS ONE 2011, 6, e028017. [Google Scholar] [CrossRef] [PubMed]
- Markland, F.S.; Swenson, S. Snake venom metalloproteinases. Toxicon 2013, 62, 3–18. [Google Scholar] [CrossRef]
- Girish, K.S.; Jagadeesha, D.K.; Rajeev, K.B.; Kemparaju, K. Snake venom hyaluronidase: An evidence for isoforms and extracellular matrix degradation. Mol. Cell. Biochem. 2002, 240, 105–110. [Google Scholar] [CrossRef]
- Bala, E.; Hazarika, R.; Singh, P.; Yasir, M.; Shrivastava, R. A biological overview of Hyaluronidase: A venom enzyme and its inhibition with plants materials. Mater. Today Proc. 2018, 5, 6406–6412. [Google Scholar] [CrossRef]
- Terra, R.M.S.; Pinto, A.F.M.; Guimarães, J.A.; Fox, J.W. Proteomic profiling of snake venom metalloproteinases (SVMPs): Insights into venom induced pathology. Toxicon 2009, 54, 836–844. [Google Scholar] [CrossRef]
- Mackessy, S.P.; Leroy, J.; Mociño-Deloya, E.; Setser, K.; Bryson, R.W.; Saviola, A.J. Venom ontogeny in the mexican lance-headed rattlesnake (Crotalus polystictus). Toxins 2018, 10, 271. [Google Scholar] [CrossRef]
- Roldán-Padrón, O.; Castro-Guillén, J.L.; García-Arredondo, J.A.; Cruz-Pérez, M.S.; Díaz-Peña, L.F.; Saldaña, C.; Blanco-Labra, A.; García-Gasca, T. Snake venom hemotoxic enzymes: Biochemical comparison between Crotalus species from central Mexico. Molecules 2019, 24, 1489. [Google Scholar] [CrossRef]
- Meléndez-Martínez, D.; Plenge-Tellechea, L.F.; Gatica-Colima, A.; Cruz-Pérez, M.S.; Aguilar-Yáñez, J.M.; Licona-Cassani, C. Functional Mining of the Crotalus Spp. Venom Protease Repertoire Reveals Potential for Chronic Wound Therapeutics. Molecules 2020, 25, 3401. [Google Scholar] [CrossRef] [PubMed]
- CBD Secretariat. Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from their Utilization to the convention on biological diversity—Text and annex. Nagoya Protoc. 2011, 12, 1–320. [Google Scholar]
- Slagboom, J.; Mladić, M.; Xie, C.; Kazandjian, T.D.; Vonk, F.; Somsen, G.W.; Casewell, N.R.; Kool, J. High throughput screening and identification of coagulopathic snake venom proteins and peptides using nanofractionation and proteomics approaches. PLoS Negl. Trop. Dis. 2020, 14, e0007802. [Google Scholar] [CrossRef] [PubMed]
- Vandooren, J.; Geurts, N.; Martens, E.; Van Den Steen, P.E.; Opdenakker, G. Zymography methods for visualizing hydrolytic enzymes. Nat. Methods 2013, 10, 211–220. [Google Scholar] [CrossRef]
- Bee, A.; Theakston, R.D.G.; Harrison, R.A.; Carter, S.D. Novel in vitro assays for assessing the haemorrhagic activity of snake venoms and for demonstration of venom metalloproteinase inhibitors. Toxicon 2001, 39, 1429–1434. [Google Scholar] [CrossRef]
- Tasoulis, T.; Pukala, T.L.; Isbister, G.K. Investigating Toxin Diversity and Abundance in Snake Venom Proteomes. Front. Pharmacol. 2022, 12, 768015. [Google Scholar] [CrossRef]
- Fry, B.G. Venomous Reptiles and Their Toxins: Evolution, Pathophysiology and Biodiscovery; Oxford University Press: Oxford, UK, 2015. [Google Scholar] [CrossRef]
- Gasanov, S.E.; Dagda, R.K.; Rael, E.D. Snake Venom Cytotoxins, Phospholipase A2 s, and Zn2+ -dependent Metalloproteinases: Mechanisms of Action and Pharmacological Relevance. J. Clin. Toxicol. 2014, 4. [Google Scholar] [CrossRef]
- Bjarnason, J.B.; Fox, J.W. Characterization of two hemorrhagic zinc proteinases, toxin c and toxin d, from western diamondback rattlesnake (Crotalus atrox) venom. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1987, 911, 356–363. [Google Scholar] [CrossRef]
- Antunes, T.C.; Yamashita, K.M.; Barbaro, K.C.; Saiki, M.; Santoro, M.L. Comparative analysis of newborn and adult Bothrops jararaca snake venoms. Toxicon 2010, 56, 1443–1458. [Google Scholar] [CrossRef]
- Serrano, S.M.T.; Shannon, J.D.; Wang, D.; Camargo, A.C.M.; Fox, J.W. A multifaceted analysis of viperid snake venoms by two-dimensional gel electrophoresis: An approach to understanding venom proteomics. Proteomics 2005, 5, 501–510. [Google Scholar] [CrossRef]
- Möller, C.; Vanderweit, N.; Bubis, J.; Mari, F. Comparative analysis of proteases in the injected and dissected venom of cone snail species. Toxicon 2013, 65, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M.; Rucavado, A. Snake venom metalloproteinases: Their role in the pathogenesis of local tissue damage. Biochimie 2000, 82, 841–850. [Google Scholar] [CrossRef]
- Teixeira, C.; Cury, Y.; Moreira, V.; Picolo, G.; Chaves, F. Inflammation induced by Bothrops asper venom. Toxicon 2009, 54, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Sitprija, V. Animal toxins and the kidney. Nat. Clin. Pract. Nephrol. 2008, 4, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Liu, S.; Yao, Y.; Zhang, Q.; Sun, M.Z. Past decade study of snake venom l-amino acid oxidase. Toxicon 2012, 60, 302–311. [Google Scholar] [CrossRef]
- Costal-Oliveira, F.; Stransky, S.; Guerra-Duarte, C.; de Souza, D.L.N.; Vivas-Ruiz, D.E.; Yarlequé, A.; Sanchez, E.F.; Chávez-Olórtegui, C.; Braga, V.M.M. L-amino acid oxidase from Bothrops atrox snake venom triggers autophagy, apoptosis and necrosis in normal human keratinocytes. Sci. Rep. 2019, 9, 781. [Google Scholar] [CrossRef]
- Izidoro, L.F.M.; Sobrinho, J.C.; Mendes, M.M.; Costa, T.R.; Grabner, A.N.; Rodrigues, V.M.; da Silva, S.L.; Zanchi, F.B.; Zuliani, J.P.; Fernandes, C.F.C.; et al. Snake venom L-amino acid oxidases: Trends in pharmacology and biochemistry. BioMed Res. Int. 2014, 2014, 196754. [Google Scholar] [CrossRef]
- Ande, S.R.; Kommoju, P.R.; Draxl, S.; Murkovic, M.; Macheroux, P.; Ghisla, S.; Ferrando-May, E. Mechanisms of cell death induction by L-amino acid oxidase, a major component of ophidian venom. Apoptosis 2006, 11, 1439–1451. [Google Scholar] [CrossRef]
- Serrano, S.M.T. The long road of research on snake venom serine proteinases. Toxicon 2013, 62, 19–26. [Google Scholar] [CrossRef]
- Montecucco, C.; Gutiérrez, J.M.; Lomonte, B. Cellular pathology induced by snake venom phospholipase A2 myotoxins and neurotoxins: Common aspects of their mechanisms of action. Cell. Mol. Life Sci. 2008, 65, 2897–2912. [Google Scholar] [CrossRef]
- Saikia, D.; Bordoloi, N.K.; Chattopadhyay, P.; Choklingam, S.; Ghosh, S.S.; Mukherjee, A.K. Differential mode of attack on membrane phospholipids by an acidic phospholipase A2 (RVVA-PLA2-I) from Daboia russelli venom. Biochim. Biophys. Acta Biomembr. 2012, 1818, 3149–3157. [Google Scholar] [CrossRef] [PubMed]
- Morita, T. Structures and functions of snake venom CLPs (C-type lectin-like proteins) with anticoagulant-, procoagulant-, and platelet-modulating activities. Toxicon 2005, 45, 1099–1114. [Google Scholar] [CrossRef] [PubMed]
- Clemetson, K.J. Snaclecs (snake C-type lectins) that inhibit or activate platelets by binding to receptors. Toxicon 2010, 56, 1236–1246. [Google Scholar] [CrossRef] [PubMed]
- Eble, J.A. Structurally robust and functionally highly versatile—C-type lectin (-related) proteins in snake venoms. Toxins 2019, 11, 136. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bittenbinder, M.A.; Bergkamp, N.D.; Slagboom, J.; Bebelman, J.P.M.; Casewell, N.R.; Siderius, M.H.; Smit, M.J.; Kool, J.; Vonk, F.J. Monitoring Snake Venom-Induced Extracellular Matrix Degradation and Identifying Proteolytically Active Venom Toxins Using Fluorescently Labeled Substrates. Biology 2023, 12, 765. https://doi.org/10.3390/biology12060765
Bittenbinder MA, Bergkamp ND, Slagboom J, Bebelman JPM, Casewell NR, Siderius MH, Smit MJ, Kool J, Vonk FJ. Monitoring Snake Venom-Induced Extracellular Matrix Degradation and Identifying Proteolytically Active Venom Toxins Using Fluorescently Labeled Substrates. Biology. 2023; 12(6):765. https://doi.org/10.3390/biology12060765
Chicago/Turabian StyleBittenbinder, Mátyás A., Nick D. Bergkamp, Julien Slagboom, Jan Paul M. Bebelman, Nicholas R. Casewell, Marco H. Siderius, Martine J. Smit, Jeroen Kool, and Freek J. Vonk. 2023. "Monitoring Snake Venom-Induced Extracellular Matrix Degradation and Identifying Proteolytically Active Venom Toxins Using Fluorescently Labeled Substrates" Biology 12, no. 6: 765. https://doi.org/10.3390/biology12060765
APA StyleBittenbinder, M. A., Bergkamp, N. D., Slagboom, J., Bebelman, J. P. M., Casewell, N. R., Siderius, M. H., Smit, M. J., Kool, J., & Vonk, F. J. (2023). Monitoring Snake Venom-Induced Extracellular Matrix Degradation and Identifying Proteolytically Active Venom Toxins Using Fluorescently Labeled Substrates. Biology, 12(6), 765. https://doi.org/10.3390/biology12060765






