Targeted Central Nervous System Irradiation with Proton Microbeam Induces Mitochondrial Changes in Caenorhabditis elegans
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. C. elegans Strain and Culture Conditions
2.2. Site Specific Microbeam Irradiation
2.3. Monitoring In Vivo Reactive Oxygen Species (ROS) Production Response to Ionizing Radiation in C. elegans
2.4. Mitochondrial Membrane Potential Imaging Using TMRE Staining
2.5. Measurement of Mitochondrial DNA Copy Number
- DNA extraction:
- by qPCR:
- by ddPCR
2.6. Single Worm Gene Expression Analysis
2.7. Analysis of Fluorescence Intensity
2.8. Oxygen Consumption Rate of C. elegans
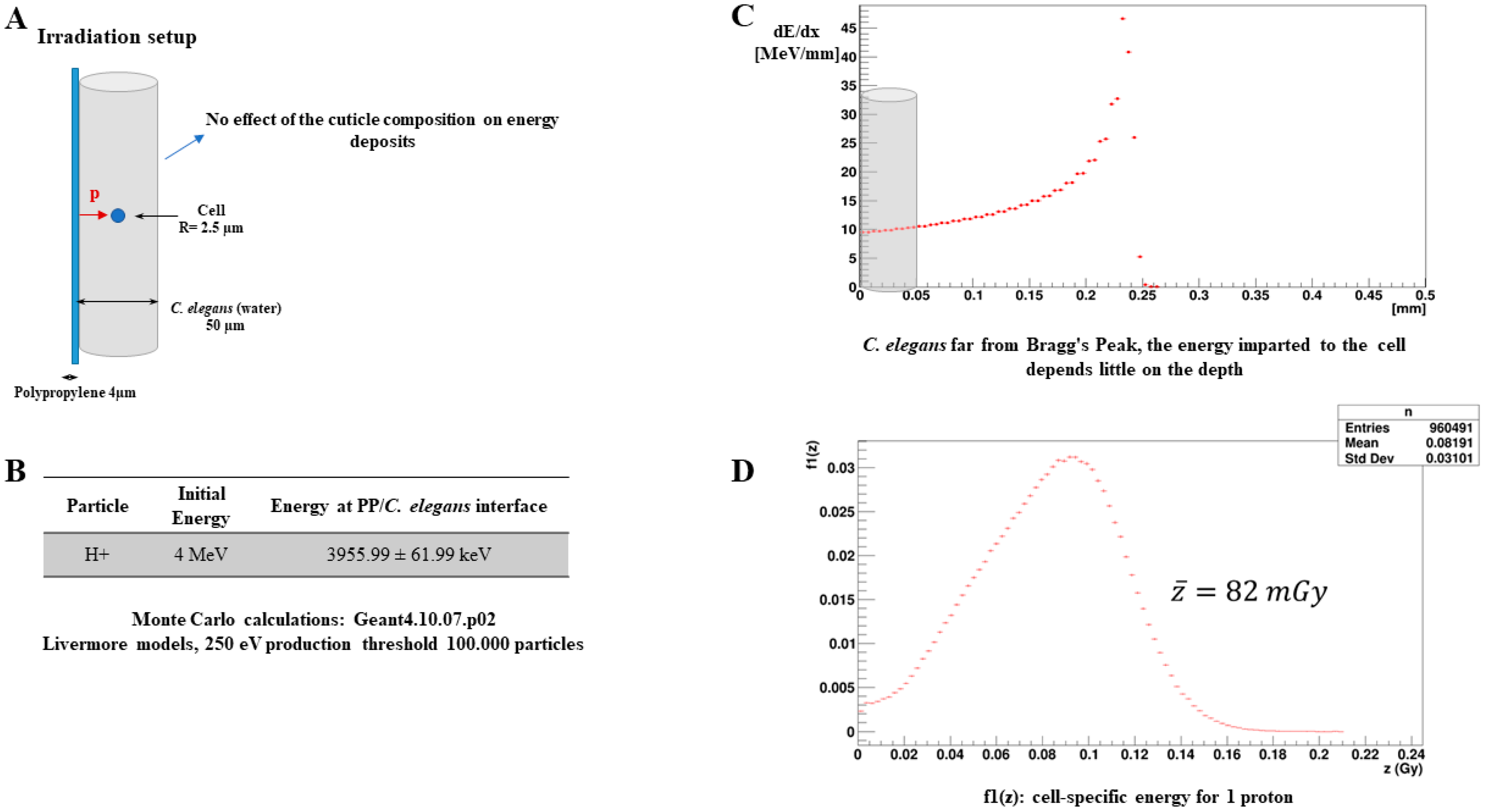
2.9. Microdosimetry Calculations
2.10. Statistical Analysis
3. Results
3.1. Monte Carlo Simulations for Dosimetry
3.2. Protons Induce Oxidative Stress
3.3. Increased Mitochondrial Copy Number
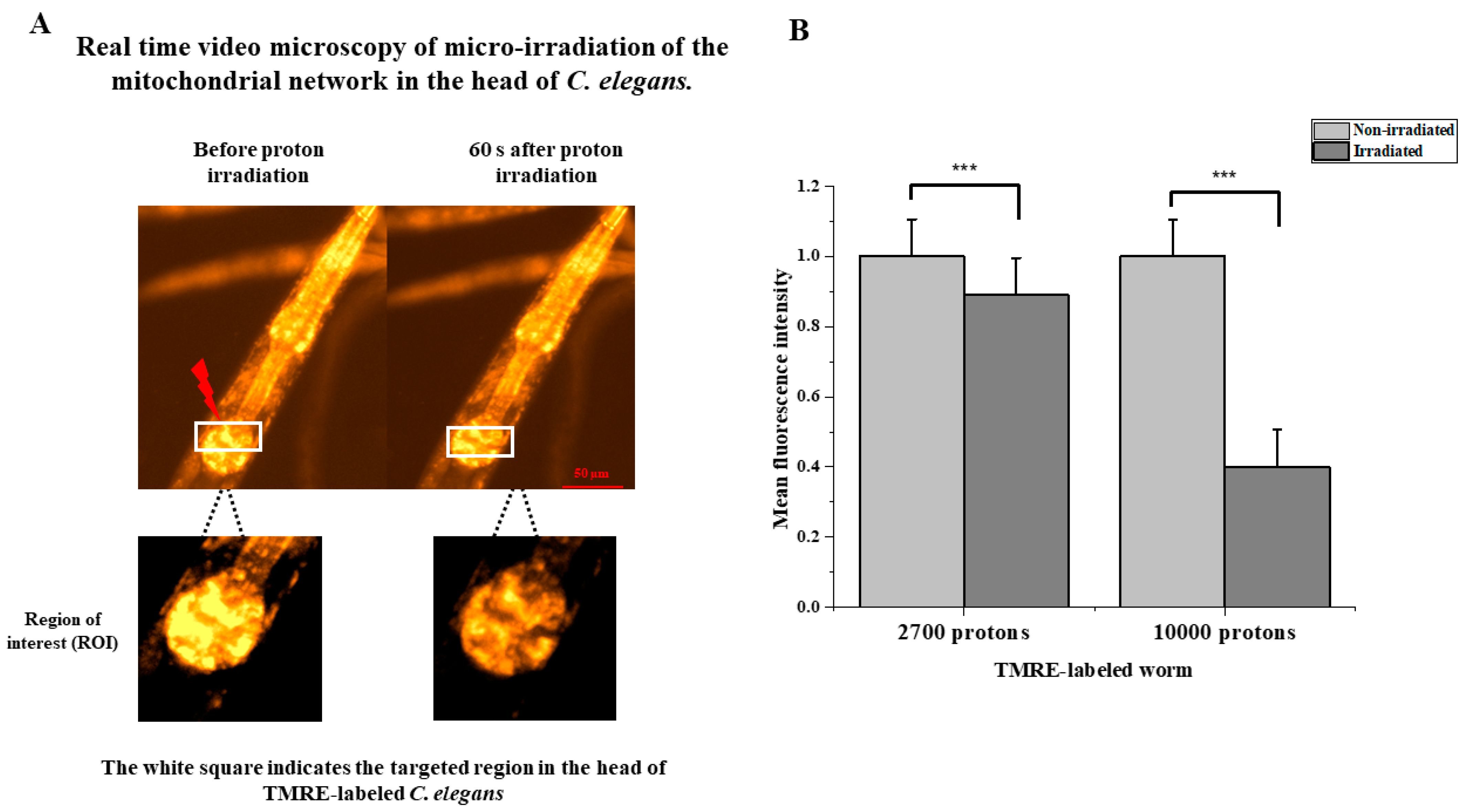
3.4. Protons Induce Immediate Loss of Mitochondrial Membrane Potential
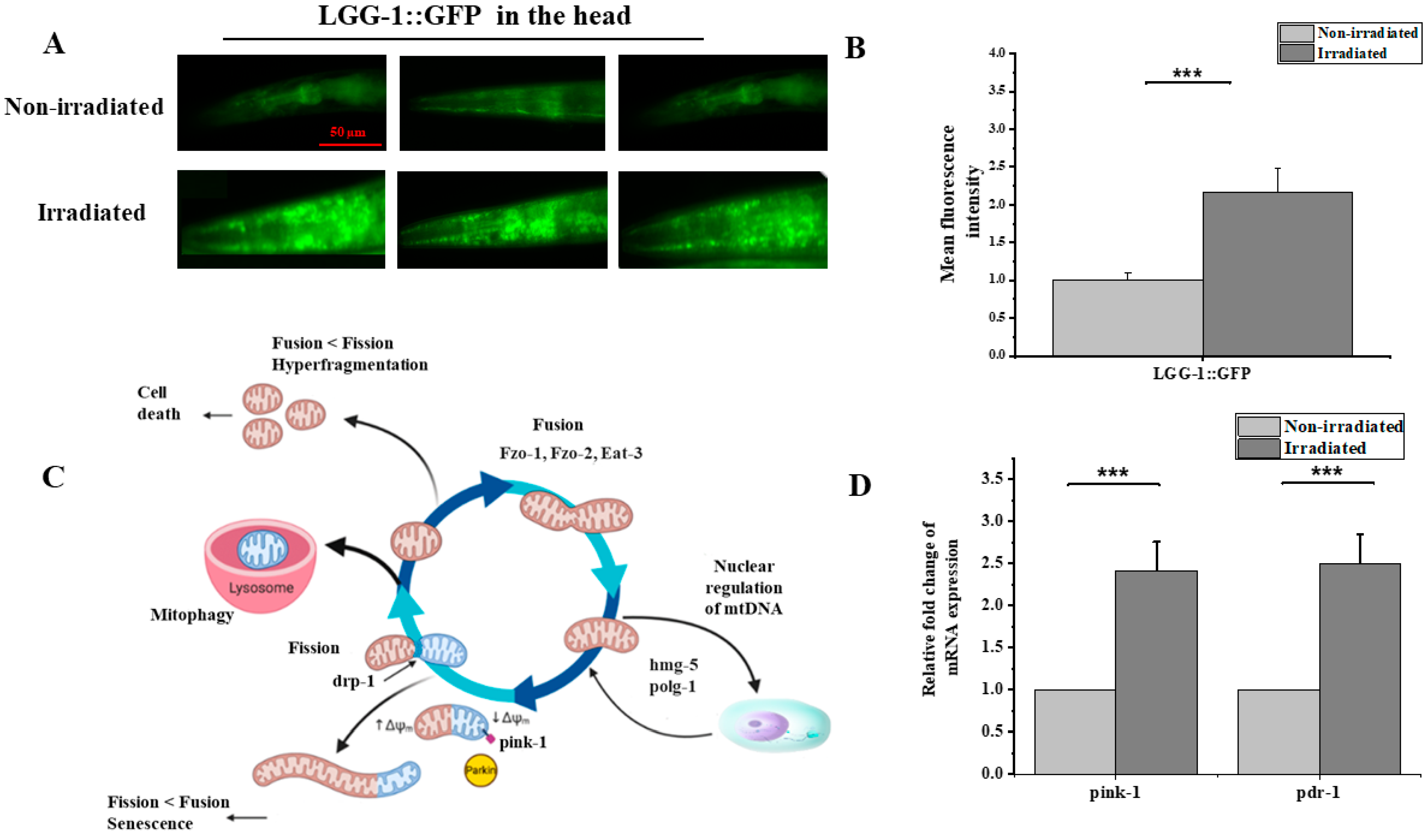
3.5. Proton Irradiation Induced Autophagy
3.6. Measurement of Oxygen Consumption Using SEAHORSE XFe96 Analyzer after Irradiation
4. Discussion
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baskar, R.; Lee, K.A.; Yeo, R.; Yeoh, K.-W. Cancer and Radiation Therapy: Current Advances and Future Directions. Int. J. Med. Sci. 2012, 9, 193–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majeed, H.; Gupta, V. Adverse Effects Of Radiation Therapy. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Mumtaz, S.; Rana, J.N.; Choi, E.H.; Han, I. Microwave Radiation and the Brain: Mechanisms, Current Status, and Future Prospects. Int. J. Mol. Sci. 2022, 23, 9288. [Google Scholar] [CrossRef] [PubMed]
- Turnquist, C.; Harris, B.T.; Harris, C.C. Radiation-induced brain injury: Current concepts and therapeutic strategies targeting neuroinflammation. Neuro-Oncol. Adv. 2020, 2, vdaa057. [Google Scholar] [CrossRef] [PubMed]
- Greene-Schloesser, D.; Robbins, M.E. Radiation-induced cognitive impairment-from bench to bedside. Neuro-Oncology 2012, 14, iv37–iv44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulz-Ertner, D.; Nikoghosyan, A.; Thilmann, C.; Haberer, T.; Jäkel, O.; Karger, C.; Kraft, G.; Wannenmacher, M.; Debus, J. Results of carbon ion radiotherapy in 152 patients. Int. J. Radiat. Oncol. Biol. Phys. 2004, 58, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Kahalley, L.S.; Peterson, R.; Ris, M.D.; Janzen, L.; Okcu, M.F.; Grosshans, D.R.; Ramaswamy, V.; Paulino, A.C.; Hodgson, D.; Mahajan, A.; et al. Superior Intellectual Outcomes after Proton Radiotherapy Compared with Photon Radiotherapy for Pediatric Medulloblastoma. J. Clin. Oncol. 2020, 38, 454–461. [Google Scholar] [CrossRef]
- Robbins, M.; Greene-Schloesser, D.; Peiffer, A.; Shaw, E.; Chan, M.; Wheeler, K. Radiation-induced brain injury: A review. Front. Oncol. 2012, 2, 73. [Google Scholar]
- Durante, M.; Loeffler, J.S. Charged particles in radiation oncology. Nat. Rev. Clin. Oncol. 2010, 7, 37–43. [Google Scholar] [CrossRef]
- Eaton, B.R.; Yock, T. The Use of Proton Therapy in the Treatment of Benign or Low-Grade Pediatric Brain Tumors. Cancer J. 2014, 20, 403. [Google Scholar] [CrossRef]
- Mohan, R. A review of proton therapy—Current status and future directions. Precis. Radiat. Oncol. 2022, 6, 164–176. [Google Scholar] [CrossRef]
- Niemierko, A.; Schuemann, J.; Niyazi, M.; Giantsoudi, D.; Maquilan, G.; Shih, H.A.; Paganetti, H. Brain necrosis in adult patients after proton therapy: Is there evidence for dependency on linear energy transfer (LET)? Int. J. Radiat. Oncol. Biol. Phys. 2021, 109, 109–119. [Google Scholar] [CrossRef]
- Parihar, V.K.; Pasha, J.; Tran, K.K.; Craver, B.M.; Acharya, M.M.; Limoli, C.L. Persistent changes in neuronal structure and synaptic plasticity caused by proton irradiation. Brain Struct. Funct. 2015, 220, 1161–1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudobeck, E.; Bellone, J.A.; Szücs, A.; Bonnick, K.; Mehrotra-Carter, S.; Badaut, J.; Nelson, G.A.; Hartman, R.E.; Vlkolinský, R. Low-dose proton radiation effects in a transgenic mouse model of Alzheimer’s disease—Implications for space travel. PLoS ONE 2017, 12, e0186168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suckert, T.; Beyreuther, E.; Müller, J.; Azadegan, B.; Meinhardt, M.; Raschke, F.; Bodenstein, E.; von Neubeck, C.; Lühr, A.; Krause, M.; et al. Late Side Effects in Normal Mouse Brain Tissue after Proton Irradiation. Front. Oncol. 2021, 10, 598360. [Google Scholar] [CrossRef] [PubMed]
- Akolawala, Q.; Rovituso, M.; Versteeg, H.H.; Rondon, A.M.R.; Accardo, A. Evaluation of Proton-Induced DNA Damage in 3D-Engineered Glioblastoma Microenvironments. ACS Appl. Mater. Interfaces 2022, 14, 20778–20789. [Google Scholar] [CrossRef] [PubMed]
- Kam, W.W.-Y.; Banati, R.B. Effects of ionizing radiation on mitochondria. Free Radic. Biol. Med. 2013, 65, 607–619. [Google Scholar] [CrossRef]
- Averbeck, D.; Rodriguez-Lafrasse, C. Role of Mitochondria in Radiation Responses: Epigenetic, Metabolic, and Signaling Impacts. Int. J. Mol. Sci. 2021, 22, 11047. [Google Scholar] [CrossRef] [PubMed]
- Vianna, F.; Gonon, G.; Lalanne, K.; Adam-Guillermin, C.; Bottollier-Depois, J.-F.; Daudin, L.; Dugué, D.; Moretto, P.; Petit, M.; Serani, L.; et al. Characterization of MIRCOM, IRSN’s new ion microbeam dedicated to targeted irradiation of living biological samples. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. Atoms 2022, 515, 20–30. [Google Scholar] [CrossRef]
- Sakashita, T.; Takanami, T.; Yanase, S.; Hamada, N.; Suzuki, M.; Kimura, T.; Kobayashi, Y.; Ishii, N.; Higashitani, A. Radiation biology of Caenorhabditis elegans: Germ cell response, aging and behavior. J. Radiat. Res. 2010, 51, 107–121. [Google Scholar] [CrossRef] [Green Version]
- Dhakal, R.; Yosofvand, M.; Yavari, M.; Abdulrahman, R.; Schurr, R.; Moustaid-Moussa, N.; Moussa, H. Review of Biological Effects of Acute and Chronic Radiation Exposure on Caenorhabditis elegans. Cells 2021, 10, 1966. [Google Scholar] [CrossRef]
- Quevarec, L.; Réale, D.; Dufourcq-Sekatcheff, E.; Car, C.; Armant, O.; Dubourg, N.; Adam-Guillermin, C.; Bonzom, J.-M. Male frequency in Caenorhabditis elegans increases in response to chronic irradiation. Evol. Appl. 2022, 15, 1331–1343. [Google Scholar] [CrossRef] [PubMed]
- Madhu, B.; Lakdawala, M.F.; Gumienny, T.L. Small-Scale Extraction of Caenorhabditis elegans Genomic DNA. J. Vis. Exp. 2022, 63716. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Maremonti, E.; Brede, D.A.; Olsen, A.-K.; Eide, D.M.; Berg, E.S. Ionizing radiation, genotoxic stress, and mitochondrial DNA copy-number variation in Caenorhabditis elegans: Droplet digital PCR analysis. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2020, 858–860, 503277. [Google Scholar] [CrossRef]
- SenGupta, T.; Palikaras, K.; Esbensen, Y.Q.; Konstantinidis, G.; Galindo, F.J.N.; Achanta, K.; Kassahun, H.; Stavgiannoudaki, I.; Bohr, V.A.; Akbari, M.; et al. Base excision repair causes age-dependent accumulation of single-stranded DNA breaks that contribute to Parkinson disease pathology. Cell Rep. 2021, 36, 109668. [Google Scholar] [CrossRef] [PubMed]
- Agostinelli, S.; Allison, J.; Amako, K.; Apostolakis, J.; Araujo, H.; Arce, P.; Asai, M.; Axen, D.; Banerjee, S.; Barrand, G.; et al. Geant4—A simulation toolkit. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrometers Detect. Assoc. Equip. 2003, 506, 250–303. [Google Scholar] [CrossRef] [Green Version]
- Allison, J.; Amako, K.; Apostolakis, J.; Arce, P.; Asai, M.; Aso, T.; Bagli, E.; Bagulya, A.; Banerjee, S.; Barrand, G.; et al. Recent developments in Geant4. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrometers Detect. Assoc. Equip. 2016, 835, 186–225. [Google Scholar] [CrossRef]
- Biersack, J.P.; Haggmark, L.G. A Monte Carlo computer program for the transport of energetic ions in amorphous targets. Nucl. Instrum. Methods 1980, 174, 257–269. [Google Scholar] [CrossRef]
- Dubois, C.; Pophillat, M.; Audebert, S.; Fourquet, P.; Lecomte, C.; Dubourg, N.; Galas, S.; Camoin, L.; Frelon, S. Differential modification of the C. elegans proteome in response to acute and chronic gamma radiation: Link with reproduction decline. Sci. Total Environ. 2019, 676, 767–781. [Google Scholar] [CrossRef] [Green Version]
- Cadenas, E.; Davies, K.J. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic. Biol. Med. 2000, 29, 222–230. [Google Scholar] [CrossRef]
- Kowalczyk, P.; Sulejczak, D.; Kleczkowska, P.; Bukowska-Ośko, I.; Kucia, M.; Popiel, M.; Wietrak, E.; Kramkowski, K.; Wrzosek, K.; Kaczyńska, K. Mitochondrial Oxidative Stress—A Causative Factor and Therapeutic Target in Many Diseases. Int. J. Mol. Sci. 2021, 22, 13384. [Google Scholar] [CrossRef] [PubMed]
- Osellame, L.D.; Blacker, T.S.; Duchen, M.R. Cellular and molecular mechanisms of mitochondrial function. Best Pract. Res. Clin. Endocrinol. Metab. 2012, 26, 711–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugimoto, T.; Mori, C.; Takanami, T.; Sasagawa, Y.; Saito, R.; Ichiishi, E.; Higashitani, A. Caenorhabditis elegans par2.1/mtssb-1 is essential for mitochondrial DNA replication and its defect causes comprehensive transcriptional alterations including a hypoxia response. Exp. Cell Res. 2008, 314, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Sumitani, M.; Kasashima, K.; Matsugi, J.; Endo, H. Biochemical properties of Caenorhabditis elegans HMG-5, a regulator of mitochondrial DNA. J. Biochem. 2011, 149, 581–589. [Google Scholar] [CrossRef] [Green Version]
- Addo, M.G.; Cossard, R.; Pichard, D.; Obiri-Danso, K.; Rötig, A.; Delahodde, A. Caenorhabditis elegans, a pluricellular model organism to screen new genes involved in mitochondrial genome maintenance. Biochim. Biophys. Acta BBA—Mol. Basis Dis. 2010, 1802, 765–773. [Google Scholar] [CrossRef] [Green Version]
- Haroon, S.; Li, A.; Weinert, J.L.; Fritsch, C.; Ericson, N.G.; Alexander-Floyd, J.; Braeckman, B.P.; Haynes, C.M.; Bielas, J.H.; Gidalevitz, T.; et al. Multiple Molecular Mechanisms Rescue mtDNA Disease in C. elegans. Cell Rep. 2018, 22, 3115–3125. [Google Scholar] [CrossRef] [Green Version]
- Walsh, D.W.M.; Siebenwirth, C.; Greubel, C.; Ilicic, K.; Reindl, J.; Girst, S.; Muggiolu, G.; Simon, M.; Barberet, P.; Seznec, H.; et al. Live cell imaging of mitochondria following targeted irradiation in situ reveals rapid and highly localized loss of membrane potential. Sci. Rep. 2017, 7, 46684. [Google Scholar] [CrossRef] [Green Version]
- Sarasija, S.; Norman, K.R. Analysis of Mitochondrial Structure in the Body Wall Muscle of Caenorhabditis elegans. Bio-Protocol 2018, 8, e2801. [Google Scholar] [CrossRef] [Green Version]
- Leboutet, R.; Chen, Y.; Legouis, R.; Culetto, E. Mitophagy during development and stress in C. elegans. Mech. Ageing Dev. 2020, 189, 111266. [Google Scholar] [CrossRef]
- Yamasaki, A.; Suzuki, M.; Funayama, T.; Moriwaki, T.; Sakashita, T.; Kobayashi, Y.; Zhang-Akiyama, Q.-M. High-Dose Irradiation Inhibits Motility and Induces Autophagy in Caenorhabditis elegans. Int. J. Mol. Sci. 2021, 22, 9810. [Google Scholar] [CrossRef]
- Garbern, J.C.; Lee, R.T. Mitochondria and metabolic transitions in cardiomyocytes: Lessons from development for stem cell-derived cardiomyocytes. Stem Cell Res. Ther. 2021, 12, 177. [Google Scholar] [CrossRef]
- Richaud, M. Modèles Intégrés de Mécanistique et de Résistance en Oncopharmacologie-Sénescence: Caenorhabditis elegans et Hypsibius Dujardini. Ph.D. Thesis, Université Montpellier, Montpellier, France, 2016. [Google Scholar]
- Wang, Q.-Q.; Yin, G.; Huang, J.-R.; Xi, S.-J.; Qian, F.; Lee, R.-X.; Peng, X.-C.; Tang, F.-R. Ionizing Radiation-Induced Brain Cell Aging and the Potential Underlying Molecular Mechanisms. Cells 2021, 10, 3570. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Soh, Z.; Yamashita, H.; Tsuji, T.; Funayama, T. Targeted Central Nervous System Irradiation of Caenorhabditis elegans Induces a Limited Effect on Motility. Biology 2020, 9, 289. [Google Scholar] [CrossRef]
- Loo, M.; Clavier, J.-B.; Attal Khalifa, J.; Moyal, E.; Khalifa, J. Dose-Response Effect and Dose-Toxicity in Stereotactic Radiotherapy for Brain Metastases: A Review. Cancers 2021, 13, 6086. [Google Scholar] [CrossRef] [PubMed]
- Gaidamakova, E.K.; Sharma, A.; Matrosova, V.Y.; Grichenko, O.; Volpe, R.P.; Tkavc, R.; Conze, I.H.; Klimenkova, P.; Balygina, I.; Horne, W.H.; et al. Small-Molecule Mn Antioxidants in Caenorhabditis elegans and Deinococcus radiodurans Supplant MnSOD Enzymes during Aging and Irradiation. mBio 2022, 13, e03394-21. [Google Scholar] [CrossRef] [PubMed]
- Maremonti, E.; Eide, D.M.; Rossbach, L.M.; Lind, O.C.; Salbu, B.; Brede, D.A. In Vivo assessment of reactive oxygen species production and oxidative stress effects induced by chronic exposure to gamma radiation in Caenorhabditis elegans. Free Radic. Biol. Med. 2020, 152, 583–596. [Google Scholar] [CrossRef]
- Yin, H.; Si, J.; Xu, H.; Dong, J.; Zheng, D.; Lu, X.; Li, X. Resveratrol-loaded nanoparticles reduce oxidative stress induced by radiation or amyloid-beta in transgenic Caenorhabditis elegans. J. Biomed. Nanotechnol. 2014, 10, 1536–1544. [Google Scholar] [CrossRef]
- Zhang, C.; Xing, X.; Zhang, F.; Shao, M.; Jin, S.; Yang, H.; Wang, G.; Cui, J.; Cai, L.; Li, W.; et al. Low-dose radiation induces renal SOD1 expression and activity in type 1 diabetic mice. Int. J. Radiat. Biol. 2014, 90, 224–230. [Google Scholar] [CrossRef]
- Smith, J.T.; Willey, N.J.; Hancock, J.T. Low dose ionizing radiation produces too few reactive oxygen species to directly affect antioxidant concentrations in cells. Biol. Lett. 2012, 8, 594–597. [Google Scholar] [CrossRef] [Green Version]
- Krisko, A.; Leroy, M.; Radman, M.; Meselson, M. Extreme anti-oxidant protection against ionizing radiation in bdelloid rotifers. Proc. Natl. Acad. Sci. USA 2012, 109, 2354–2357. [Google Scholar] [CrossRef] [Green Version]
- Finkel, T.; Holbrook, N.J. Oxidants, oxidative stress and the biology of ageing. Nature 2000, 408, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.T.; Beal, M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 2006, 443, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Abdullaev, S.; Gubina, N.; Bulanova, T.; Gaziev, A. Assessment of Nuclear and Mitochondrial DNA, Expression of Mitochondria-Related Genes in Different Brain Regions in Rats after Whole-Body X-ray Irradiation. Int. J. Mol. Sci. 2020, 21, 1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, Z.; Yang, G.; Deng, S.; Wang, Q. Oxidative stress levels and dynamic changes in mitochondrial gene expression in a radiation-induced lung injury model. J. Radiat. Res. 2019, 60, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Kesäniemi, J.; Lavrinienko, A.; Tukalenko, E.; Moutinho, A.F.; Mappes, T.; Møller, A.P.; Mousseau, T.A.; Watts, P.C. Exposure to environmental radionuclides alters mitochondrial DNA maintenance in a wild rodent. Evol. Ecol. 2020, 34, 163–174. [Google Scholar] [CrossRef] [Green Version]
- Yi, S.; Kim, S.; Song, J. Analysis of the Effect of Space Radiations on the Nematode, Caenorhabditis elegans, through the Simulated Space Radiation. Int. J. Astron. Astrophys. 2013, 3, 291–302. [Google Scholar] [CrossRef] [Green Version]
- Bess, A.S.; Crocker, T.L.; Ryde, I.T.; Meyer, J.N. Mitochondrial dynamics and autophagy aid in removal of persistent mitochondrial DNA damage in Caenorhabditis elegans. Nucleic Acids Res. 2012, 40, 7916–7931. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.-H.; Lin, Y.-W. Bioenergetic Health Assessment of a Single Caenorhabditis elegans from Postembryonic Development to Aging Stages via Monitoring Changes in the Oxygen Consumption Rate within a Microfluidic Device. Sensors 2018, 18, 2453. [Google Scholar] [CrossRef] [Green Version]
- Stackley, K.D.; Beeson, C.C.; Rahn, J.J.; Chan, S.S.L. Bioenergetic profiling of zebrafish embryonic development. PLoS ONE 2011, 6, e25652. [Google Scholar] [CrossRef] [Green Version]
- Suda, H.; Shouyama, T.; Yasuda, K.; Ishii, N. Direct measurement of oxygen consumption rate on the nematode Caenorhabditis elegans by using an optical technique. Biochem. Biophys. Res. Commun. 2005, 330, 839–843. [Google Scholar] [CrossRef]
- Crokart, N.; Jordan, B.F.; Baudelet, C.; Cron, G.O.; Hotton, J.; Radermacher, K.; Grégoire, V.; Beghein, N.; Martinive, P.; Bouzin, C.; et al. Glucocorticoids Modulate Tumor Radiation Response through a Decrease in Tumor Oxygen Consumption. Clin. Cancer Res. 2007, 13, 630–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salem, A.; Little, R.A.; Latif, A.; Featherstone, A.K.; Babur, M.; Peset, I.; Cheung, S.; Watson, Y.; Tessyman, V.; Mistry, H.; et al. Oxygen-enhanced MRI Is Feasible, Repeatable, and Detects Radiotherapy-induced Change in Hypoxia in Xenograft Models and in Patients with Non–small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 3818–3829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamori, T.; Yasui, H.; Yamazumi, M.; Wada, Y.; Nakamura, Y.; Nakamura, H.; Inanami, O. Ionizing radiation induces mitochondrial reactive oxygen species production accompanied by upregulation of mitochondrial electron transport chain function and mitochondrial content under control of the cell cycle checkpoint. Free Radic. Biol. Med. 2012, 53, 260–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sleiman, A.; Lalanne, K.; Vianna, F.; Perrot, Y.; Richaud, M.; SenGupta, T.; Cardot-Martin, M.; Pedini, P.; Picard, C.; Nilsen, H.; et al. Targeted Central Nervous System Irradiation with Proton Microbeam Induces Mitochondrial Changes in Caenorhabditis elegans. Biology 2023, 12, 839. https://doi.org/10.3390/biology12060839
Sleiman A, Lalanne K, Vianna F, Perrot Y, Richaud M, SenGupta T, Cardot-Martin M, Pedini P, Picard C, Nilsen H, et al. Targeted Central Nervous System Irradiation with Proton Microbeam Induces Mitochondrial Changes in Caenorhabditis elegans. Biology. 2023; 12(6):839. https://doi.org/10.3390/biology12060839
Chicago/Turabian StyleSleiman, Ahmad, Kévin Lalanne, François Vianna, Yann Perrot, Myriam Richaud, Tanima SenGupta, Mikaël Cardot-Martin, Pascal Pedini, Christophe Picard, Hilde Nilsen, and et al. 2023. "Targeted Central Nervous System Irradiation with Proton Microbeam Induces Mitochondrial Changes in Caenorhabditis elegans" Biology 12, no. 6: 839. https://doi.org/10.3390/biology12060839




