Multiple Instances of Adaptive Evolution in Aquaporins of Amphibious Fishes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Mining and Phylogenetic Reconstruction
2.2. Estimation of Gene Family Evolutionary Histories
2.3. Analyses of Adaptive Evolution
2.4. AQP 3D Structure Modelling
3. Results and Discussion
3.1. Diversity of Amphibious Fish Aquaporins
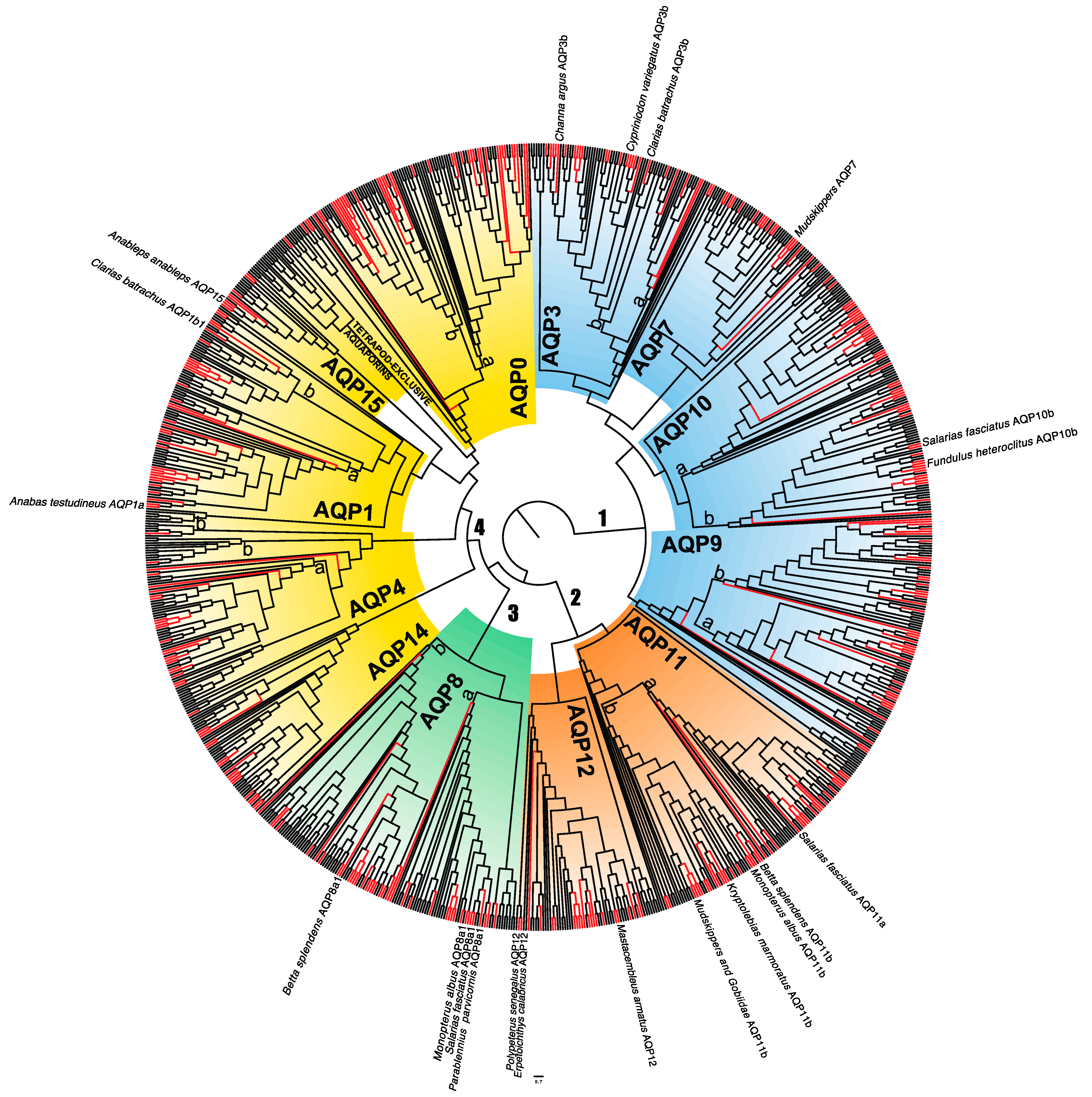

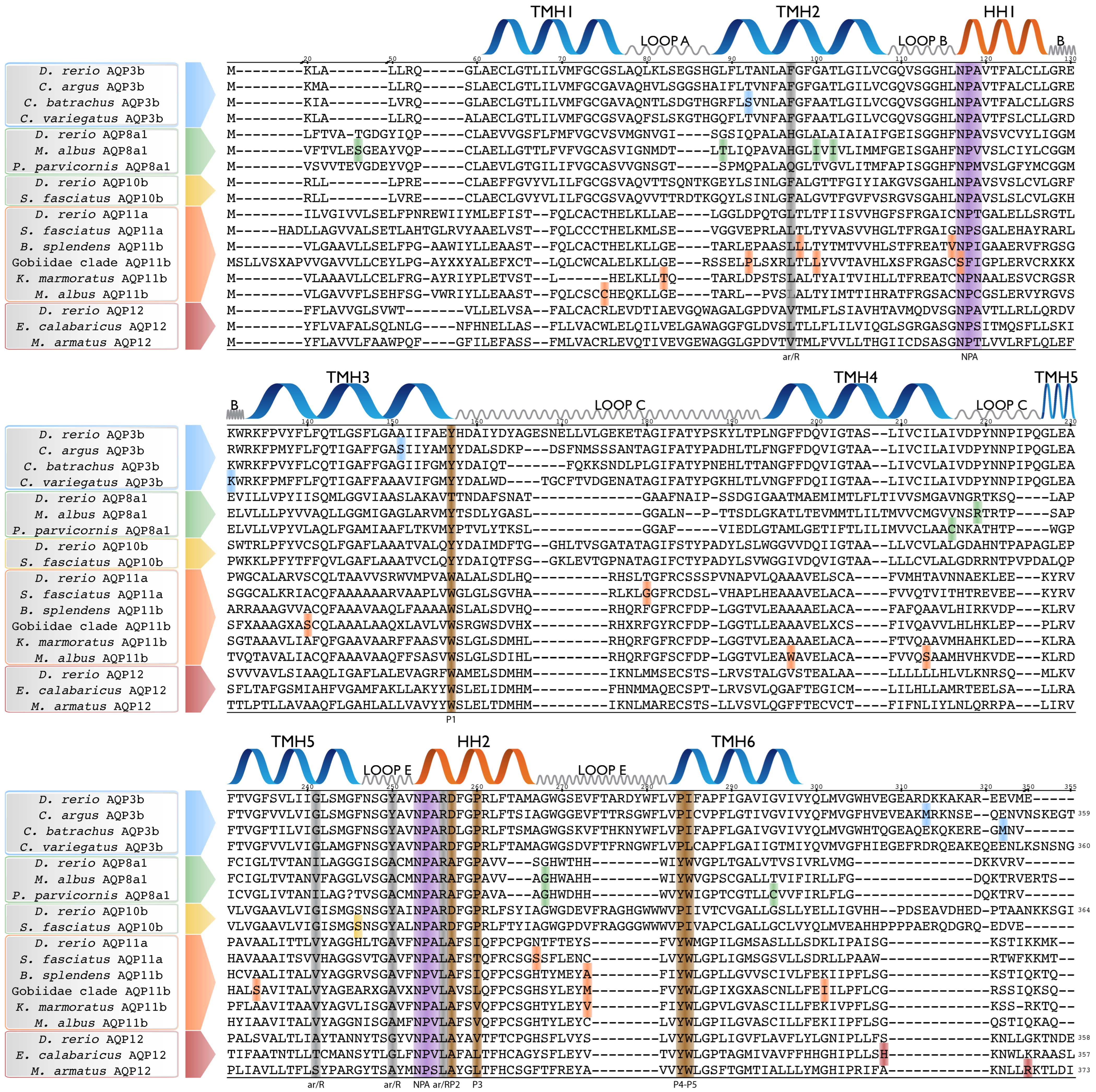
3.2. Adaptive Evolution in Amphibious Fish AQPs
3.3. Adaptive Evolution in Gobioidei AQP11b
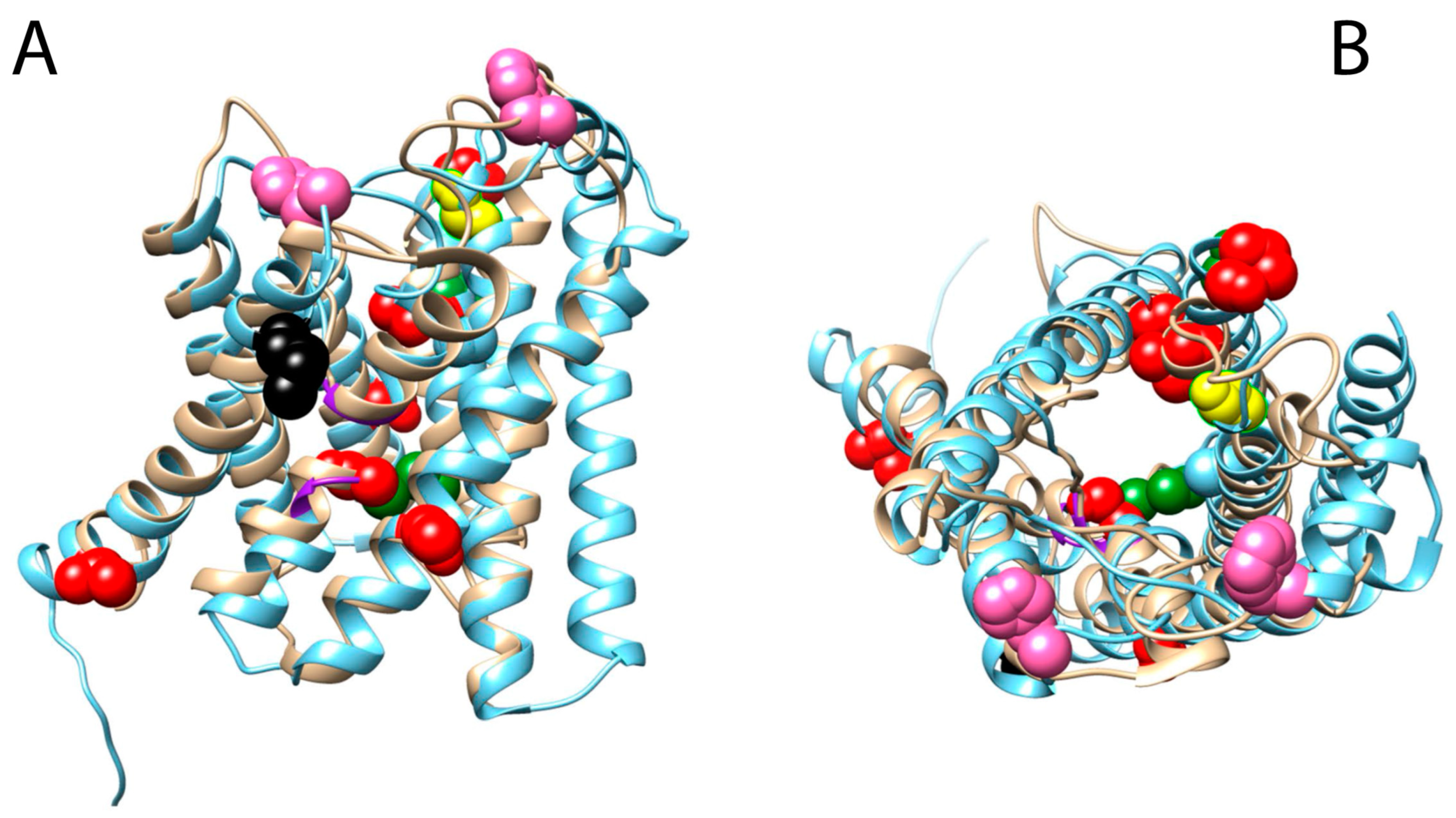
3.4. Mapping of Positively Selected Sites onto the 3D Structure
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Graham, J.B. Air-Breathing Fishes: Evolution, Diversity and Adaptation; Academic Press: San Diego, CA, USA, 1997. [Google Scholar]
- Schluter, D. The Ecology of Adaptive Radiation; OUP Oxford: Oxford, UK, 2000. [Google Scholar]
- Nosil, P. Ecological Speciation; Oxford University Press: Oxford, UK, 2012. [Google Scholar]
- Long, J.A.; Gordon, M.S. The Greatest Step in Vertebrate History: A Paleobiological Review of the Fish-Tetrapod Transition. Physiol. Biochem. Zool. 2004, 77, 700–719. [Google Scholar] [CrossRef] [Green Version]
- Laurin, M. How Vertebrates Left the Water; University of California Press: Berkeley, CA, USA, 2010. [Google Scholar]
- Sayer, M.D.J. Adaptations of Amphibious Fish for Surviving Life out of Water. Fish Fish. 2005, 6, 186–211. [Google Scholar] [CrossRef]
- Ord, T.J.; Cooke, G.M. Repeated Evolution of Amphibious Behavior in Fish and Its Implications for the Colonization of Novel Environments. Evolution 2016, 70, 1747–1759. [Google Scholar] [CrossRef]
- Wright, P.A.; Turko, A.J. Amphibious Fishes: Evolution and Phenotypic Plasticity. J. Exp. Biol. 2016, 219, 2245–2259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bridges, C.R. Respiratory Adaptations in Intertidal Fish. Am. Zool. 2015, 28, 79–96. [Google Scholar] [CrossRef] [Green Version]
- Graham, J.; Lee, H.L.; Wegner, N. Transition from Water to Land in an Extant Group of Fishes: Air Breathing and the Acquisition Sequence of Adaptations for Amphibious Life in Oxudercine Gobies. In Fish Respiration and Environment; Fernandes, M.N., Rantin, F.T., Glass, M.L., Kapoor, B., Eds.; Science Publishers: Enfield, NH, USA, 2007; pp. 255–288. [Google Scholar]
- LeBlanc, D.M.; Wood, C.M.; Fudge, D.S.; Wright, P.A. A Fish Out of Water: Gill and Skin Remodeling Promotes Osmo—and Ionoregulation in the Mangrove Killifish Kryptolebias Marmoratus. Physiol. Biochem. Zool. 2010, 83, 932–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frick, N.T.; Wright, P.A. Nitrogen Metabolism and Excretion in the Mangrove Killifish Rivulus Marmoratus I. The Influence of Environmental Salinity and External Ammonia. J. Exp. Biol. 2002, 205, 79–89. [Google Scholar]
- Frick, N.T.; Wright, P.A. Nitrogen Metabolism and Excretion in the Mangrove Killifish Rivulus Marmoratus II. Significant Ammonia Volatilization in a Teleost during Air-Exposure. J. Exp. Biol. 2002, 205, 91–100. [Google Scholar] [CrossRef]
- Chew, S.F.; Gan, J.; Ip, Y.K. Nitrogen Metabolism and Excretion in the Swamp Eel, Monopterus Albus, during 6 or 40 Days of Estivation in Mud. Physiol. Biochem. Zool. 2005, 78, 620–629. [Google Scholar] [CrossRef]
- Randall, D.J.; Ip, Y.K.; Chew, S.F.; Wilson, J.M. Air Breathing and Ammonia Excretion in the Giant Mudskipper, Periophthalmodon Schlosseri. Physiol. Biochem. Zool. 2015, 77, 783–788. [Google Scholar] [CrossRef]
- Ip, Y.K.; Chew, S.F.; Randall, D.J. Five Tropical Air-Breathing Fishes, Six Different Strategies to Defend against Ammonia Toxicity on Land. Physiol. Biochem. Zool. 2004, 7, 768–782. [Google Scholar] [CrossRef]
- Ip, Y.K.; Randall, D.J.; Kok, T.K.T.; Barzaghi, C.; Wright, P.A.; Ballantyne, J.S.; Wilson, J.M.; Chew, S.F. The Giant Mudskipper Periophthalmodon Schlosseri Facilitates Active NH 4+ Excretion by Increasing Acid Excretion and Decreasing NH3 Permeability in the Skin. J. Exp. Biol. 2004, 207, 787–801. [Google Scholar] [CrossRef] [Green Version]
- Davenport, J.; Matin, A.K.M.A. Terrestrial Locomotion in the Climbing Perch, Anabas Testudineus (Bloch) (Anabantidea, Pisces). J. Fish Biol. 1990, 37, 175–184. [Google Scholar] [CrossRef]
- Pace, C.M.; Gibb, A.C. Sustained Periodic Terrestrial Locomotion in Air-Breathing Fishes. J. Fish Biol. 2014, 84, 639–660. [Google Scholar] [CrossRef] [PubMed]
- Agre, P.; Kozono, D. Aquaporin Water Channels: Molecular Mechanisms for Human Diseases. FEBS Lett. 2003, 555, 72–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agre, P.; Preston, G.M.; Smith, B.L.; Jung, J.S.; Raina, S.; Moon, C.; Guggino, W.B.; Nielsen, S. Aquaporin CHIP: The Archetypal Molecular Water Channel. Am. J. Physiol. 1993, 265, F463–F476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Connolly, D.L.; Shanahan, C.M.; Weissberg, P.L. The Aquaporins. A Family of Water Channel Proteins. Int. J. Biochem. Cell Biol. 1998, 30, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Walz, T.; Hirai, T.; Murata, K.; Heymann, J.B.; Mitsuoka, K.; Fujiyoshi, Y.; Smith, B.L.; Agre, P.; Engel, A. The Three-Dimensional Structure of Aquaporin-1. Nature 1997, 387, 624–627. [Google Scholar] [CrossRef] [PubMed]
- Heymann, J.B.; Engel, A. Aquaporins: Phylogeny, Structure, and Physiology of Water Channels. News Physiol. Sci. 1999, 14, 187–193. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, M.; Andoo, A.; Shimono, M.; Takamatsu, N.; Taki, A.; Muta, K.; Matsushita, W.; Uechi, T.; Matsuzaki, T.; Kenmochi, N.; et al. The NPC Motif of Aquaporin-11, Unlike the NPA Motif of Known Aquaporins, Is Essential for Full Expression of Molecular Function. J. Biol. Chem. 2011, 286, 3342–3350. [Google Scholar] [CrossRef] [Green Version]
- Abascal, F.; Irisarri, I.; Zardoya, R. Diversity and Evolution of Membrane Intrinsic Proteins. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 1468–1481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heymann, J.B.; Engel, A. Structural Clues in the Sequences of the Aquaporins. J. Mol. Biol. 2000, 295, 1039–1053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sui, H.; Han, B.G.; Lee, J.K.; Walian, P.; Jap, B.K. Structural Basis of Water-Specific Transport through the AQP1 Water Channel. Nature 2001, 414, 872–878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Törnroth-Horsefield, S.; Hedfalk, K.; Fischer, G.; Lindkvist-Petersson, K.; Neutze, R. Structural Insights into Eukaryotic Aquaporin Regulation. FEBS Lett. 2010, 584, 2580–2588. [Google Scholar] [CrossRef] [Green Version]
- Bestetti, S.; Galli, M.; Sorrentino, I.; Pinton, P.; Rimessi, A.; Sitia, R.; Medraño-Fernandez, I. Human Aquaporin-11 Guarantees Efficient Transport of H2O2 across the Endoplasmic Reticulum Membrane. Redox Biol. 2020, 28, 101326. [Google Scholar] [CrossRef]
- Finn, R.N.; Cerdà, J. Evolution and Functional Diversity of Aquaporins. Biol. Bull. 2015, 229, 6–23. [Google Scholar] [CrossRef]
- Zardoya, R. Phylogeny and Evolution of the Major Intrinsic Protein Family. Biol. Cell 2005, 97, 397–414. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.N.; Chauvigné, F.; Hlidberg, J.B.; Cutler, C.P.; Cerdà, J. The Lineage—Specific Evolution of Aquaporin Gene Clusters Facilitated Tetrapod Terrestrial Adaptation. PLoS ONE 2014, 9, e113686. [Google Scholar] [CrossRef] [Green Version]
- Madsen, S.S.; Engelund, M.B.; Cutler, C.P. Water Transport and Functional Dynamics of Aquaporins in Osmoregulatory Organs of Fishes. Biol. Bull. 2015, 229, 70–92. [Google Scholar] [CrossRef]
- Ip, Y.K.; Soh, M.M.L.; Chen, X.L.; Ong, J.L.Y.; Chng, Y.R.; Ching, B.; Wong, W.P.; Lam, S.H.; Chew, S.F. Molecular Characterization of Branchial Aquaporin 1aa and Effects of Seawater Acclimation, Emersion or Ammonia Exposure on Its MRNA Expression in the Gills, Gut, Kidney and Skin of the Freshwater Climbing Perch, Anabas Testudineus. PLoS ONE 2013, 8, e61163. [Google Scholar] [CrossRef] [Green Version]
- Lorente-Martínez, H.; Agorreta, A.; Torres-Sánchez, M.; San Mauro, D. Evidence of Positive Selection Suggests Possible Role of Aquaporins in the Water-to-Land Transition of Mudskippers. Org. Divers. Evol. 2018, 18, 499–514. [Google Scholar] [CrossRef]
- Lorente-Martínez, H.; Agorreta, A.; San Mauro, D. Genomic Fishing and Data Processing for Molecular Evolution Research. Methods Protoc. 2022, 5, 26. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The Rapid Generation of Mutation Data Matrices from Protein Sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. Maximum Likelihood Phylogenetic Estimation from DNA Sequences with Variable Rates over Sites: Approximate Methods. J. Mol. Evol. 1994, 39, 306–314. [Google Scholar] [CrossRef] [Green Version]
- Reeves, J.H. Heterogeneity in the Substitution Process of Amino Acid Sites of Proteins Coded for by Mitochondrial DNA. J. Mol. Evol. 1992, 35, 17–31. [Google Scholar] [CrossRef]
- Abascal, F.; Zardoya, R.; Posada, D. ProtTest: Selection of Best-Fit Models of Protein Evolution What Can I Use ProtTest for?—Introduction The Program: Using ProtTest. Bioinformatics 2005, 21, 2104–2105. [Google Scholar] [CrossRef] [Green Version]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef]
- Hughes, L.C.; Ortí, G.; Huang, Y.; Sun, Y.; Baldwin, C.C.; Thompson, A.W.; Arcila, D.; Betancur-R, R.; Li, C.; Becker, L.; et al. Comprehensive Phylogeny of Ray-Finned Fishes (Actinopterygii) Based on Transcriptomic and Genomic Data. Proc. Natl. Acad. Sci. USA 2018, 115, 6249–6254. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Yu, L.; Kalavacharla, V.; Liu, Z. A Bayesian Model for Gene Family Evolution. BMC Bioinform. 2011, 12, 426. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [Green Version]
- Abascal, F.; Zardoya, R.; Telford, M.J. TranslatorX: Multiple Alignment of Nucleotide Sequences Guided by Amino Acid Translations. Nucleic Acids Res. 2010, 38, 7–13. [Google Scholar] [CrossRef] [Green Version]
- Tavaré, S. Some Probabilistic and Statistical Problems in the Analysis of DNA Sequences. Am. Math. Soc. Lect. Math. Life Sci. 1986, 17, 57–86. [Google Scholar]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Nielsen, R.; Yang, Z. Evaluation of an Improved Branch-Site Likelihood Method for Detecting Positive Selection at the Molecular Level. Mol. Biol. Evol. 2005, 22, 2472–2479. [Google Scholar] [CrossRef] [Green Version]
- Álvarez-Carretero, S.; Kapli, P.; Yang, Z. Beginner’s Guide on the Use of PAML to Detect Positive Selection. Mol. Biol. Evol. 2023, 40, msad041. [Google Scholar] [CrossRef] [PubMed]
- Self, S.G.; Liang, K.-Y. Asymptotic Properties of Maximum Likelihood Estimators and Likelihood Ratio Tests under Nonstandard Conditions. J. Am. Stat. Assoc. 1987, 82, 605–610. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Yang, Z.; Wong, W.S.W.; Nielsen, R. Bayes Empirical Bayes Inference of Amino Acid Sites Under Positive Selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosakovsky Pond, S.L.; Posada, D.; Gravenor, M.B.; Woelk, C.H.; Frost, S.D.W. GARD: A Genetic Algorithm for Recombination Detection. Bioinformatics 2006, 22, 3096–3098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weaver, S.; Shank, S.D.; Spielman, S.J.; Li, M.; Muse, S.V.; Kosakovsky Pond, S.L. Datamonkey 2.0: A Modern Web Application for Characterizing Selective and Other Evolutionary Processes. Mol. Biol. Evol. 2018, 35, 773–777. [Google Scholar] [CrossRef] [Green Version]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold—Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ittisoponpisan, S.; Islam, S.A.; Khanna, T.; Alhuzimi, E.; David, A.; Sternberg, M.J.E. Can Predicted Protein 3D Structures Provide Reliable Insights into Whether Missense Variants Are Disease Associated? J. Mol. Biol. 2019, 431, 2197–2212. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, O.; Chauvigné, F.; Ferré, A.; Nilsen, F.; Fjelldal, P.G.; Cerdà, J.; Finn, R.N. Unravelling the Complex Duplication History of Deuterostome Glycerol Transporters. Cells 2020, 9, 1663. [Google Scholar] [CrossRef]
- Cerdà, J.; Finn, R.N. Piscine Aquaporins: An Overview of Recent Advances. J. Exp. Zool. A Ecol. Genet. Physiol. 2010, 313A, 623–650. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.S.; van de Peer, Y.; Braasch, I.; Meyer, A. Comparative Genomics Provides Evidence for an Ancient Genome Duplication Event in Fish. Philos. Trans. R Soc. B Biol. Sci. 2001, 356, 1661–1679. [Google Scholar] [CrossRef] [Green Version]
- Xu, P.; Xu, J.; Liu, G.; Chen, L.; Zhou, Z.; Peng, W.; Jiang, Y.; Zhao, Z.; Jia, Z.; Sun, Y.; et al. The Allotetraploid Origin and Asymmetrical Genome Evolution of the Common Carp Cyprinus Carpio. Nat. Commun. 2019, 10, 4625. [Google Scholar] [CrossRef] [Green Version]
- Allendorf, F.W.; Thorgaard, G.H. Tetraploidy and the Evolution of Salmonid Fishes. In Evolutionary Genetics of Fishes; Turner, B.J., Ed.; Springer US: Boston, MA, USA, 1984; pp. 1–53. [Google Scholar]
- Ferré, A.; Chauvigné, F.; Vlasova, A.; Norberg, B.; Bargelloni, L.; Guigó, R.; Finn, R.N.; Cerdà, J. Functional Evolution of Clustered Aquaporin Genes Reveals Insights into the Oceanic Success of Teleost Eggs. Mol. Biol. Evol. 2023, 40, msad071. [Google Scholar] [CrossRef] [PubMed]
- Kosakovsky Pond, S.L.; Posada, D.; Gravenor, M.B.; Woelk, C.H.; Frost, S.D.W. Automated Phylogenetic Detection of Recombination Using a Genetic Algorithm. Mol. Biol. Evol. 2006, 23, 1891–1901. [Google Scholar] [CrossRef] [Green Version]
- Wolf, Y.I.; Koonin, E.V. Genome Reduction as the Dominant Mode of Evolution. BioEssays 2013, 35, 829–837. [Google Scholar] [CrossRef] [Green Version]
- Card, D.C.; Van Camp, A.G.; Santonastaso, T.; Jensen-Seaman, M.I.; Anthony, N.M.; Edwards, S.V. Structure and Evolution of the Squamate Major Histocompatibility Complex as Revealed by Two Anolis Lizard Genomes. Front. Genet. 2022, 13, 979746. [Google Scholar] [CrossRef]
- Martínez-Redondo, G.I.; Simón Guerrero, C.; Aristide, L.; Balart-García, P.; Tonzo, V.; Fernández, R. Parallel Duplication and Loss of Aquaporin-Coding Genes during the “out of the Sea” Transition as Potential Key Drivers of Animal Terrestrialization. Mol. Ecol. 2023, 32, 2022–2040. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. Adaptive Molecular Evolution. In Handbook of Statistical Genetics: Third Edition; Balding, D.J., Bishop, M.J., Cannings, C., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2008; Volume 1, pp. 375–406. ISBN 9780470058305. [Google Scholar]
- Zang, Y.; Chen, J.; Zhong, H.; Ren, J.; Zhao, W.; Man, Q.; Shang, S.; Tang, X. Genome-Wide Analysis of the Aquaporin Gene Family in Reptiles. Int. J. Biol. Macromol. 2019, 126, 1093–1098. [Google Scholar] [CrossRef] [PubMed]
- São Pedro, S.L.; Alves, J.M.P.; Barreto, A.S.; de Souza Lima, A.O. Evidence of Positive Selection of Aquaporins Genes from Pontoporia Blainvillei during the Evolutionary Process of Cetaceans. PLoS ONE 2015, 10, e0134516. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Lee, S.Y.; Kim, B.S.; Kim, D.S.; Nam, Y.K. Isolation and MRNA Expression Analysis of Aquaporin Isoforms in Marine Medaka Oryzias Dancena, a Euryhaline Teleost. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2014, 171, 1–8. [Google Scholar] [CrossRef]
- Ma, Q.; Liu, X.; Li, A.; Liu, S.; Zhuang, Z. Effects of Osmotic Stress on the Expression Profiling of Aquaporin Genes in the Roughskin Sculpin (Trachidermus Fasciatus). Acta Oceanol. Sin. 2020, 39, 19–25. [Google Scholar] [CrossRef]
- Ishibashi, K.; Tanaka, Y.; Morishita, Y. The Role of Mammalian Superaquaporins inside the Cell: An Update. Biochim. Et. Biophys. Acta BBA Biomembr. 2021, 1863, 183617. [Google Scholar] [CrossRef]
- Yakata, K.; Tani, K.; Fujiyoshi, Y. Water Permeability and Characterization of Aquaporin-11. J. Struct. Biol. 2011, 174, 315–320. [Google Scholar] [CrossRef]
- Bertolotti, M.; Bestetti, S.; García-Manteiga, J.M.; Medraño-Fernandez, I.; Dal Mas, A.; Malosio, M.L.; Sitia, R. Tyrosine Kinase Signal Modulation: A Matter of H2O2 Membrane Permeability? Antioxid. Redox Signal. 2013, 19, 1447–1451. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, S.; Moniaga, C.S.; Nielsen, S.; Hara-Chikuma, M. Aquaporin-9 Facilitates Membrane Transport of Hydrogen Peroxide in Mammalian Cells. Biochem. Biophys. Res. Commun. 2016, 471, 191–197. [Google Scholar] [CrossRef]
- Miller, E.W.; Dickinson, B.C.; Chang, C.J. Aquaporin-3 Mediates Hydrogen Peroxide Uptake to Regulate Downstream Intracellular Signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 15681–15686. [Google Scholar] [CrossRef] [Green Version]
- Tingaud-Sequeira, A.; Calusinska, M.; Finn, R.N.; Chauvigné, F.; Lozano, J.; Cerdà, J. The Zebrafish Genome Encodes the Largest Vertebrate Repertoire of Functional Aquaporins with Dual Paralogy and Substrate Specificities Similar to Mammals. BioMed Cent. Evol. Biol. 2010, 10, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chew, S.F.; Sim, M.Y.; Phua, Z.C.; Wong, W.P.; Ip, Y.K. Active Ammonia Excretion in the Giant Mudskipper, Periophthalmodon Schlosseri (Pallas), during Emersion. J. Exp. Zool. A Ecol. Genet. Physiol. 2007, 307, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Tay, A.S.L.; Chew, S.F.; Ip, Y.K. The Swamp Eel Monopterus Albus Reduces Endogenous Ammonia Production and Detoxifies Ammonia to Glutamine during 144 h of Aerial Exposure. J. Exp. Biol. 2003, 206, 2473–2486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ip, Y.K.; Tay, A.S.L.; Lee, K.H.; Chew, S.F. Strategies for Surviving High Concentrations of Environmental Ammonia in the Swamp Eel Monopterus Albus. Physiol. Biochem. Zool. 2004, 77, 390–405. [Google Scholar] [CrossRef]
- Tingaud-Sequeira, A.; Zapater, C.; Chauvigné, F.; Otero, D.; Cerdà, J. Adaptive Plasticity of Killifish (Fundulus Heteroclitus) Embryos: Dehydration-Stimulated Development and Differential Aquaporin-3 Expression. Am. J. Physiol Regul. Integr. Comp. Physiol. 2009, 296, 1041–1052. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.-C.; Hastings, P.A. Phylogeny and Biogeography of a Shallow Water Fish Clade (Teleostei: Blenniiformes). BMC Evol. Biol. 2013, 13, 210. [Google Scholar] [CrossRef] [Green Version]
- Rozemeije, M.J.C.; Plaut, I. Regulation of Nitrogen Excretion of the Amphibious Blenniidae Alticus Kirki (Guenther, 1868) during Emersion and Immersion. Comp. Biochem. Physiol. A Physiol. 1993, 104, 57–62. [Google Scholar] [CrossRef]
- Davenport, J.; Sayer, M.D.J. Ammonia and Urea Excretion in the Amphibious Teleost Blennius Pholis (L.) in Sea-Water and in Air. Comp. Biochem. Physiol. A Physiol. 1986, 84, 189–194. [Google Scholar] [CrossRef]
- Santos, C.R.A.; Estêvão; Fuentes, J.; Cardoso, J.C.R.; Fabra, M.; Passos, A.L.; Detmers, F.J.; Deen, P.M.T.; Cerdà, J.; Power, D.M. Isolation of a Novel Aquaglyceroporin from a Marine Teleost (Sparus Auratus): Function and Tissue Distribution. J. Exp. Biol. 2004, 207, 1217–1227. [Google Scholar] [CrossRef] [Green Version]
- Venkat, A.; Hahn, M.W.; Thornton, J.W. Multinucleotide Mutations Cause False Inferences of Lineage-Specific Positive Selection. Nat. Ecol. Evol. 2018, 2, 1280–1288. [Google Scholar] [CrossRef]
- Belinky, F.; Bykova, A.; Yurchenko, V.; Rogozin, I.B. No Evidence for Widespread Positive Selection on Double Substitutions within Codons in Primates and Yeasts. Front. Genet. 2022, 13, 991249. [Google Scholar] [CrossRef] [PubMed]
- Anisimova, M.; Nielsen, R.; Yang, Z. Effect of Recombination on the Accuracy of the Likelihood Method for Detecting Positive Selection at Amino Acid Sites. Genetics 2003, 164, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Gould, S.J.; Vrba, E.S. Exaptation—A Missing Term in the Science of Form. Paleobiology 1982, 8, 4–15. [Google Scholar] [CrossRef]
- Thacker, C.E. Phylogeny of Gobioidei and Placement within Acanthomorpha, with a New Classification and Investigation of Diversification and Character Evolution. Copeia 2009, 2009, 93–104. [Google Scholar] [CrossRef] [Green Version]
- Fricke, R.; Eschmeyer, W.N.; Van der Laan, R. Catalog of Fishes. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 23 April 2023).
- Kreida, S.; Törnroth-Horsefield, S. Structural Insights into Aquaporin Selectivity and Regulation. Curr. Opin. Struct. Biol. 2015, 33, 126–134. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [Green Version]
- King, M.-C.; Wilson, A.C. Evolution at Two Levels in Humans and Chimpanzees. Science 1975, 188, 107–116. [Google Scholar] [CrossRef] [Green Version]
| AQP | Foreground Branch | LRT | p-Value a | q-Value b | ω c | Prop. 2a d | Prop. 2b e | Selec. Sites |
|---|---|---|---|---|---|---|---|---|
| 1a | A. anableps | 7.711 | 0.003 | 0.0298 | 999 | 0.006 | 0.001 | 0 |
| 1b1 | C. batrachus * | 21.708 | 1.59 × 10−6 | 1.126 × 10−4 | 136.799 | 0.071 | 0.007 | 1 |
| 3b | C. argus | 16.551 | 2.367 × 10−5 | 2.603 × 10−4 | 31.599 | 0.032 | 0.008 | 2 |
| 3b | C. batrachus | 19.478 | 5.087 × 10−6 | 1.119 × 10−4 | 49.655 | 0.037 | 0.009 | 3 |
| 3b | C. variegatus | 9.906 | 8.233 × 10−4 | 0.006 | 67.061 | 0.013 | 0.003 | 1 |
| 7 | Mudskipper clade stem | 8.639 | 0.002 | 0.026 | 999 | 0.015 | 0.004 | 0 |
| 8a1 | M. albus | 12.334 | 2.217 × 10−4 | 0.007 | 12.806 | 0.068 | 0.013 | 6 |
| 8a1 | Mudskipper clade stem | 7.295 | 0.003 | 0.027 | 36.256 | 0.019 | 0.004 | 0 |
| 8a1 | P. parvicornis | 7.997 | 0.002 | 0.028 | 23.468 | 0.085 | 0.017 | 3 |
| 8b1 | B. splendens | 7.460 | 0.003 | 0.028 | 8.669 | 0.039 | 0.008 | 0 |
| 10b | F. heteroclitus * | 47.896 | 2.247 × 10−12 | 8.76 × 10−11 | 67.772 | 0.005 | 0.001 | 1 |
| 10b | S. pavo | 13.001 | 1.557 × 10−4 | 0.003 | 59.219 | 0.019 | 0.003 | 1 |
| 11b | Betta | 15.126 | 5.029 × 10−5 | 8.298 × 10−4 | 43.954 | 0.030 | 0.007 | 3 |
| 11b | Kryptolebias | 6.510 | 0.005 | 0.029 | 38.602 | 0.022 | 0.005 | 2 |
| 11b | Monopterus | 13.291 | 1.334 × 10−4 | 0.001 | 18.128 | 0.038 | 0.009 | 3 |
| 11b | Mudskipper clade stem | 9.607 | 9.690 × 10−4 | 0.006 | 999 | 0.029 | 0.006 | 0 |
| 11a | S. fasciatus | 12.613 | 1.916 × 10−4 | 0.002 | 16.503 | 0.047 | 0.011 | 2 |
| 12 | E. calabaricus | 10.325 | 6.560 × 10−4 | 0.004 | 708.322 | 0.021 | 0.005 | 1 |
| 12 | M. armatus | 10.907 | 4.790 × 10−4 | 0.004 | 998.999 | 0.014 | 0.003 | 1 |
| 12 | P. senegalus | 11.961 | 2.717 × 10−4 | 0.004 | 999 | 0.016 | 0.004 | 0 |
| 15 | A. anableps | 9.594 | 9.761 × 10−4 | 0.008 | 1 | 0.057 | 0.017 | 0 |
| 11b | Gobiidae stem branch ** | 17.813 | 1.218 × 10−5 | 4.021 × 10−4 | 41.228 | 0.110 | 0.025 | 7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lorente-Martínez, H.; Agorreta, A.; Irisarri, I.; Zardoya, R.; Edwards, S.V.; San Mauro, D. Multiple Instances of Adaptive Evolution in Aquaporins of Amphibious Fishes. Biology 2023, 12, 846. https://doi.org/10.3390/biology12060846
Lorente-Martínez H, Agorreta A, Irisarri I, Zardoya R, Edwards SV, San Mauro D. Multiple Instances of Adaptive Evolution in Aquaporins of Amphibious Fishes. Biology. 2023; 12(6):846. https://doi.org/10.3390/biology12060846
Chicago/Turabian StyleLorente-Martínez, Héctor, Ainhoa Agorreta, Iker Irisarri, Rafael Zardoya, Scott V. Edwards, and Diego San Mauro. 2023. "Multiple Instances of Adaptive Evolution in Aquaporins of Amphibious Fishes" Biology 12, no. 6: 846. https://doi.org/10.3390/biology12060846
APA StyleLorente-Martínez, H., Agorreta, A., Irisarri, I., Zardoya, R., Edwards, S. V., & San Mauro, D. (2023). Multiple Instances of Adaptive Evolution in Aquaporins of Amphibious Fishes. Biology, 12(6), 846. https://doi.org/10.3390/biology12060846





