MicroRNA miR-263b-5p Regulates Developmental Growth and Cell Association by Suppressing Laminin A in Drosophila
Abstract
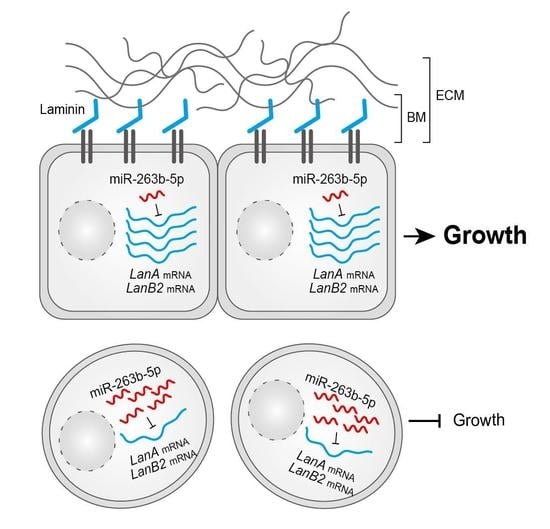
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Drosophila
2.2. Determination of mRNA and miRNA Expression
2.3. The Images of Larvae/Flies and Measurement of Body Weight
2.4. Analysis of Eclosion Rate
2.5. Wing Measurement
2.6. Analysis of the Larval Fat Body
2.7. The Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis
2.8. Luciferase Reporter Assay
3. Results
3.1. Overexpression of miR-263b in the Fat Body Leads to Defects in the Normal Growth of Drosophila
3.2. Overexpression of miR-263b Results in Cell Dissociation in the Fat Body
3.3. miR-263b-5p Suppresses the Expression of Drosophila Laminin A
3.4. Depletion of LanA in the fat Body Results in Defects in the Normal Growth
3.5. LanA Is Involved in Cell Association in the Fat Body
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yurchenco, P.D.; Amenta, P.S.; Patton, B.L. Basement membrane assembly, stability and activities observed through a developmental lens. Matrix Biol. 2004, 22, 521–538. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Estrada, B.; Jacobs, S.; Sanchez-Sanchez, B.J.; Tang, J.; Ma, M.; Magadan-Corpas, P.; Pastor-Pareja, J.C.; Martin-Bermudo, M.D. Dissection of Nidogen function in Drosophila reveals tissue-specific mechanisms of basement membrane assembly. PLoS Genet. 2018, 14, e1007483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uechi, G.; Sun, Z.; Schreiber, E.M.; Halfter, W.; Balasubramani, M. Proteomic View of Basement Membranes from Human Retinal Blood Vessels, Inner Limiting Membranes, and Lens Capsules. J. Proteome Res. 2014, 13, 3693–3705. [Google Scholar] [CrossRef]
- Jayadev, R.; Sherwood, D.R. Basement membranes. Curr. Biol. 2017, 27, R207–R211. [Google Scholar] [CrossRef] [Green Version]
- Yurchenco, P.D. Basement membranes: Cell scaffoldings and signaling platforms. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef] [Green Version]
- Hynes, R.O. The evolution of metazoan extracellular matrix. J. Cell Biol. 2012, 196, 671–679. [Google Scholar] [CrossRef] [Green Version]
- Timpl, R.; Rohde, H.; Robey, P.G.; Rennard, S.I.; Foidart, J.M.; Martin, G.R. Laminin—A glycoprotein from basement membranes. J. Biol. Chem. 1979, 254, 9933–9937. [Google Scholar] [CrossRef]
- Urbano, J.M.; Torgler, C.N.; Molnar, C.; Tepass, U.; Lopez-Varea, A.; Brown, N.H.; de Celis, J.F.; Martin-Bermudo, M.D. Drosophila laminins act as key regulators of basement membrane assembly and morphogenesis. Development 2009, 136, 4165–4176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henchcliffe, C.; Garcia-Alonso, L.; Tang, J.; Goodman, C.S. Genetic analysis of laminin A reveals diverse functions during morphogenesis in Drosophila. Development 1993, 118, 325–337. [Google Scholar] [CrossRef]
- Yarnitzky, T.; Volk, T. Laminin is required for heart, somatic muscles, and gut development in the Drosophila embryo. Dev. Biol. 1995, 169, 609–618. [Google Scholar] [CrossRef] [Green Version]
- Rowe, R.G.; Weiss, S.J. Breaching the basement membrane: Who, when and how? Trends. Cell Biol. 2008, 18, 560–574. [Google Scholar] [CrossRef] [PubMed]
- Jia, Q.; Liu, Y.; Liu, H.; Li, S. Mmp1 and Mmp2 cooperatively induce Drosophila fat body cell dissociation with distinct roles. Sci. Rep. 2014, 4, 7535. [Google Scholar] [CrossRef] [Green Version]
- Nelliot, A.; Bond, N.; Hoshizaki, D.K. Fat-body remodeling in Drosophila melanogaster. Genesis 2006, 44, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef]
- Lee, Y.S.; Nakahara, K.; Pham, J.W.; Kim, K.; He, Z.; Sontheimer, E.J.; Carthew, R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 2004, 117, 69–81. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Iwasaki, S.; Kawamata, T.; Tomari, Y. Drosophila argonaute1 and argonaute2 employ distinct mechanisms for translational repression. Mol. Cell. 2009, 34, 58–67. [Google Scholar] [CrossRef] [Green Version]
- Okamura, K.; Ishizuka, A.; Siomi, H.; Siomi, M.C. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004, 18, 1655–1666. [Google Scholar] [CrossRef] [Green Version]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [Green Version]
- Nian, X.; Chen, W.; Bai, W.; Zhao, Z.; Zhang, Y. miR-263b Controls Circadian Behavior and the Structural Plasticity of Pacemaker Neurons by Regulating the LIM-Only Protein Beadex. Cells 2019, 8. [Google Scholar] [CrossRef] [Green Version]
- Hilgers, V.; Bushati, N.; Cohen, S.M. Drosophila microRNAs 263a/b confer robustness during development by protecting nascent sense organs from apoptosis. PLoS Biol. 2010, 8, e1000396. [Google Scholar] [CrossRef] [Green Version]
- Lim, D.H.; Oh, C.T.; Lee, L.; Hong, J.S.; Noh, S.H.; Hwang, S.; Kim, S.; Han, S.J.; Lee, Y.S. The endogenous siRNA pathway in Drosophila impacts stress resistance and lifespan by regulating metabolic homeostasis. FEBS Lett. 2011, 585, 3079–3085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, J.H.; Kim, D.J.; Lee, D.E.; Han, J.Y.; Chung, J.H.; Ahn, H.K.; Lee, S.W.; Lim, D.H.; Lee, Y.S.; Park, S.Y.; et al. Genome-wide microRNA expression profiling in placentas of fetuses with Down syndrome. Placenta 2015, 36, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- McCabe, J.; French, V.; Partridge, L. Joint regulation of cell size and cell number in the wing blade of Drosophila melanogaster. Genet. Res. 1997, 69, 61–68. [Google Scholar] [CrossRef]
- Pesevski, M.; Dworkin, I. Genetic and environmental canalization are not associated among altitudinally varying populations of Drosophila melanogaster. Evolution 2020, 74, 1755–1771. [Google Scholar] [CrossRef]
- Agarwal, V.; Subtelny, A.O.; Thiru, P.; Ulitsky, I.; Bartel, D.P. Predicting microRNA targeting efficacy in Drosophila. Genome Biol. 2018, 19, 152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruby, J.G.; Stark, A.; Johnston, W.K.; Kellis, M.; Bartel, D.P.; Lai, E.C. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007, 17, 1850–1864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Edelheit, O.; Hanukoglu, A.; Hanukoglu, I. Simple and efficient site-directed mutagenesis using two single-primer reactions in parallel to generate mutants for protein structure-function studies. BMC Biotechnol. 2009, 9, 61. [Google Scholar] [CrossRef] [Green Version]
- Lim, D.H.; Lee, S.; Choi, M.S.; Han, J.Y.; Seong, Y.; Na, D.; Kwon, Y.S.; Lee, Y.S. The conserved microRNA miR-8-3p coordinates the expression of V-ATPase subunits to regulate ecdysone biosynthesis for Drosophila metamorphosis. FASEB J. 2020, 34, 6449–6465. [Google Scholar] [CrossRef] [Green Version]
- Asha, H.; Nagy, I.; Kovacs, G.; Stetson, D.; Ando, I.; Dearolf, C.R. Analysis of Ras-induced overproliferation in Drosophila hemocytes. Genetics 2003, 163, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Edgar, B.A. How flies get their size: Genetics meets physiology. Nat. Rev. Genet. 2006, 7, 907–916. [Google Scholar] [CrossRef]
- Llano, E.; Adam, G.; Pendas, A.M.; Quesada, V.; Sanchez, L.M.; Santamaria, I.; Noselli, S.; Lopez-Otin, C. Structural and enzymatic characterization of Drosophila Dm2-MMP, a membrane-bound matrix metalloproteinase with tissue-specific expression. J. Biol. Chem. 2002, 277, 23321–23329. [Google Scholar] [CrossRef] [Green Version]
- Heng, Y.W.; Koh, C.G. Actin cytoskeleton dynamics and the cell division cycle. Int. J. Biochem. Cell Biol. 2010, 42, 1622–1633. [Google Scholar] [CrossRef]
- Wessels, H.H.; Lebedeva, S.; Hirsekorn, A.; Wurmus, R.; Akalin, A.; Mukherjee, N.; Ohler, U. Global identification of functional microRNA-mRNA interactions in Drosophila. Nat. Commun. 2019, 10, 1626. [Google Scholar] [CrossRef] [Green Version]
- Yin, V.P.; Thummel, C.S. Mechanisms of steroid-triggered programmed cell death in Drosophila. Semin. Cell. Dev. Biol. 2005, 16, 237–243. [Google Scholar] [CrossRef]
- Jia, Q.; Liu, S.; Wen, D.; Cheng, Y.; Bendena, W.G.; Wang, J.; Li, S. Juvenile hormone and 20-hydroxyecdysone coordinately control the developmental timing of matrix metalloproteinase-induced fat body cell dissociation. J. Biol. Chem. 2017, 292, 21504–21516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bond, N.D.; Nelliot, A.; Bernardo, M.K.; Ayerh, M.A.; Gorski, K.A.; Hoshizaki, D.K.; Woodard, C.T. ssFTZ-F1 and Matrix metalloproteinase 2 are required for fat-body remodeling in Drosophila. Dev. Biol. 2011, 360, 286–296. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.W.; Song, S.; Weng, R.; Verma, P.; Kugler, J.M.; Buescher, M.; Rouam, S.; Cohen, S.M. Systematic study of Drosophila microRNA functions using a collection of targeted knockout mutations. Dev. Cell 2014, 31, 784–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilgus, T.A. Growth Factor-Extracellular Matrix Interactions Regulate Wound Repair. Adv. Wound. Care. 2012, 1, 249–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, C.J.; Kim, H.H.; Kim, H.K.; Lee, S.; Jang, D.; Kim, C.; Lim, D.-H. MicroRNA miR-263b-5p Regulates Developmental Growth and Cell Association by Suppressing Laminin A in Drosophila. Biology 2023, 12, 1096. https://doi.org/10.3390/biology12081096
Kim CJ, Kim HH, Kim HK, Lee S, Jang D, Kim C, Lim D-H. MicroRNA miR-263b-5p Regulates Developmental Growth and Cell Association by Suppressing Laminin A in Drosophila. Biology. 2023; 12(8):1096. https://doi.org/10.3390/biology12081096
Chicago/Turabian StyleKim, Chae Jeong, Hyun Ho Kim, Hee Kyung Kim, Sojeong Lee, Daegyu Jang, Chanhyeok Kim, and Do-Hwan Lim. 2023. "MicroRNA miR-263b-5p Regulates Developmental Growth and Cell Association by Suppressing Laminin A in Drosophila" Biology 12, no. 8: 1096. https://doi.org/10.3390/biology12081096
APA StyleKim, C. J., Kim, H. H., Kim, H. K., Lee, S., Jang, D., Kim, C., & Lim, D. -H. (2023). MicroRNA miR-263b-5p Regulates Developmental Growth and Cell Association by Suppressing Laminin A in Drosophila. Biology, 12(8), 1096. https://doi.org/10.3390/biology12081096