Droplet Digital PCR: A New Molecular Method to Detect G1105S/V Mutations in Plasmopara viticola CesA3 Gene
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Sample Collection
2.2. P. viticola Strains Isolation
2.3. DNA Extraction
2.4. Sequencing
2.5. Equipment and ddPCR Reaction Setup
2.6. Optimization of ddPCR Conditions
2.7. Sensitivity and Accuracy of the Probes
2.8. Specificity and False-Positive Rate Estimation
2.9. Multiplexing
2.10. Determination of Allele Frequencies in Field Samples
3. Results
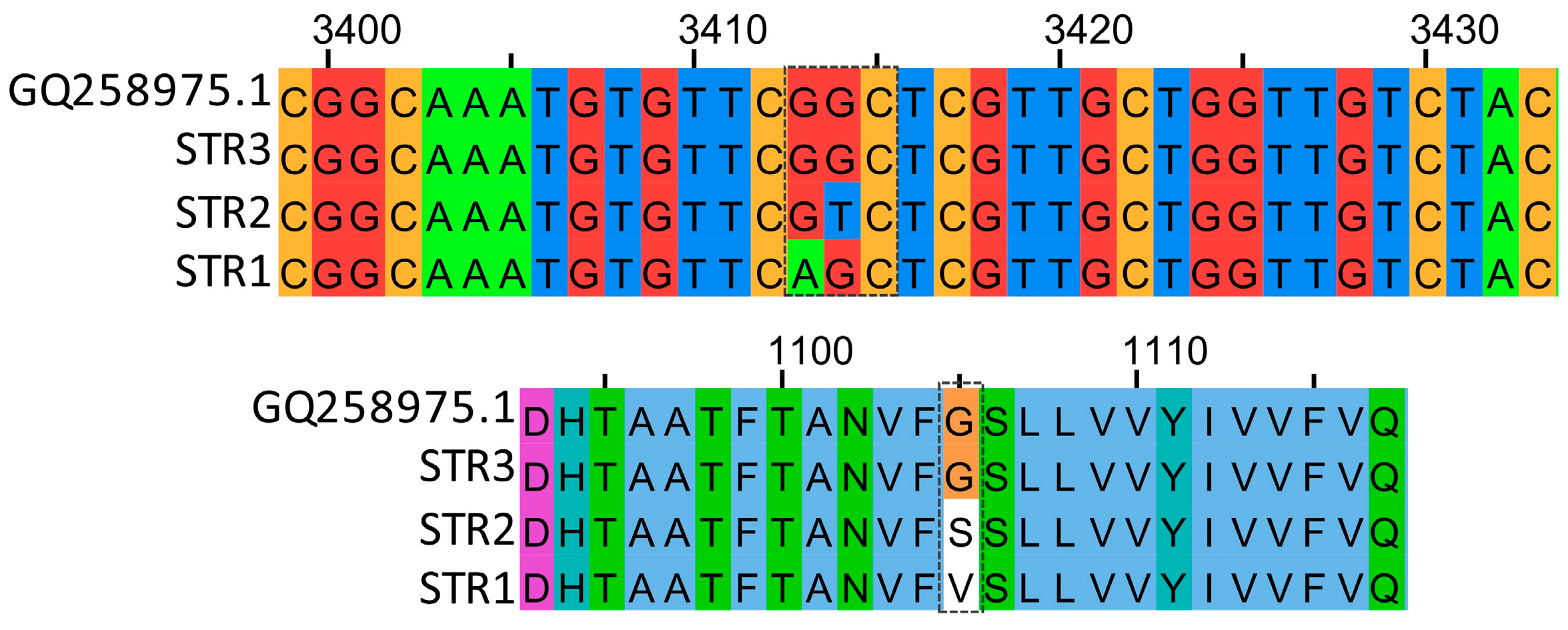
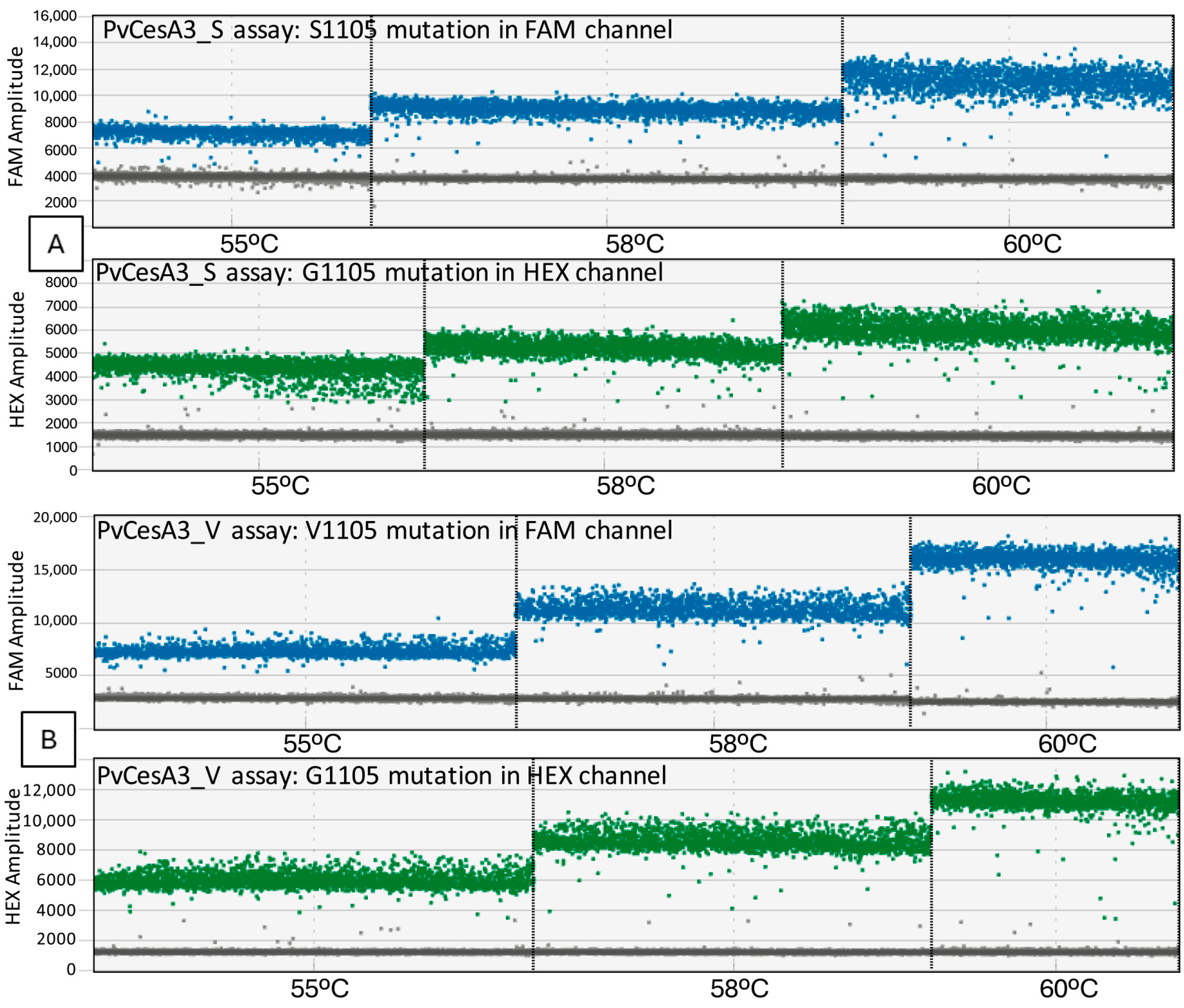
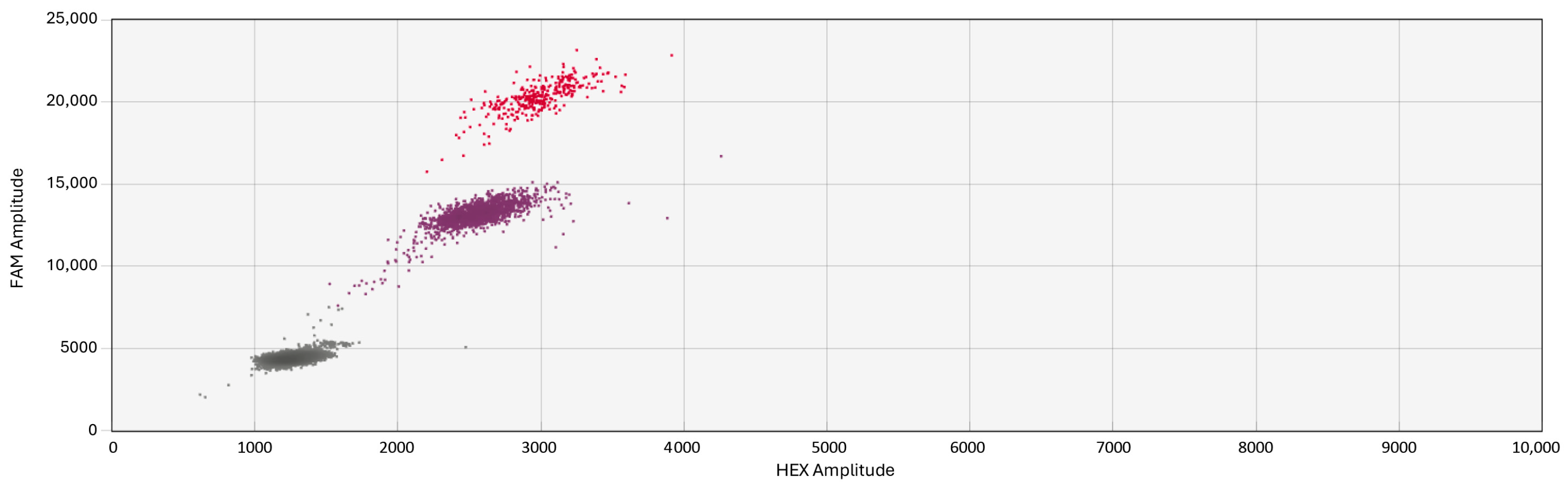
3.1. Optimization of ddPCR Conditions
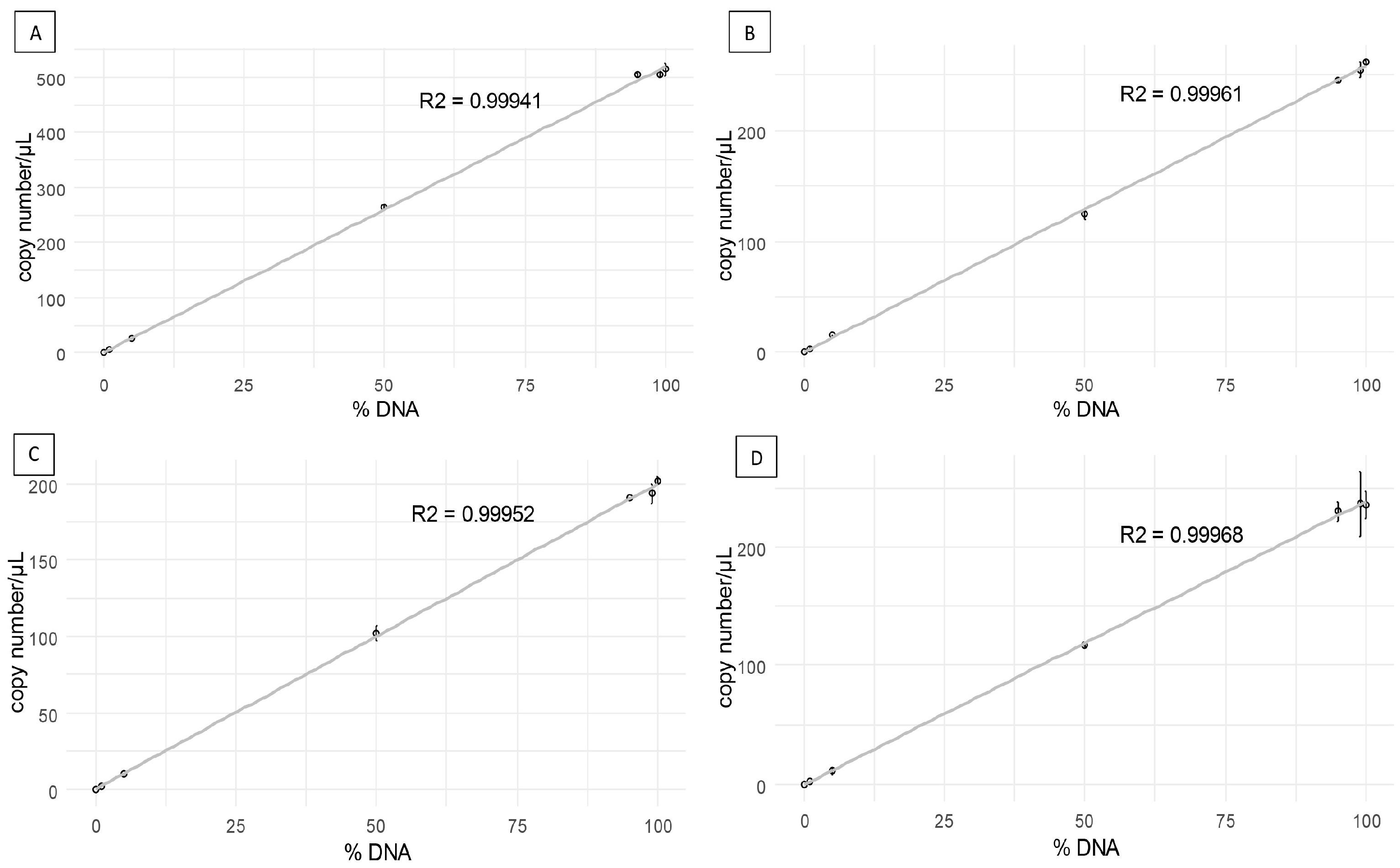
3.2. Sensitivity and Accuracy of the Probes
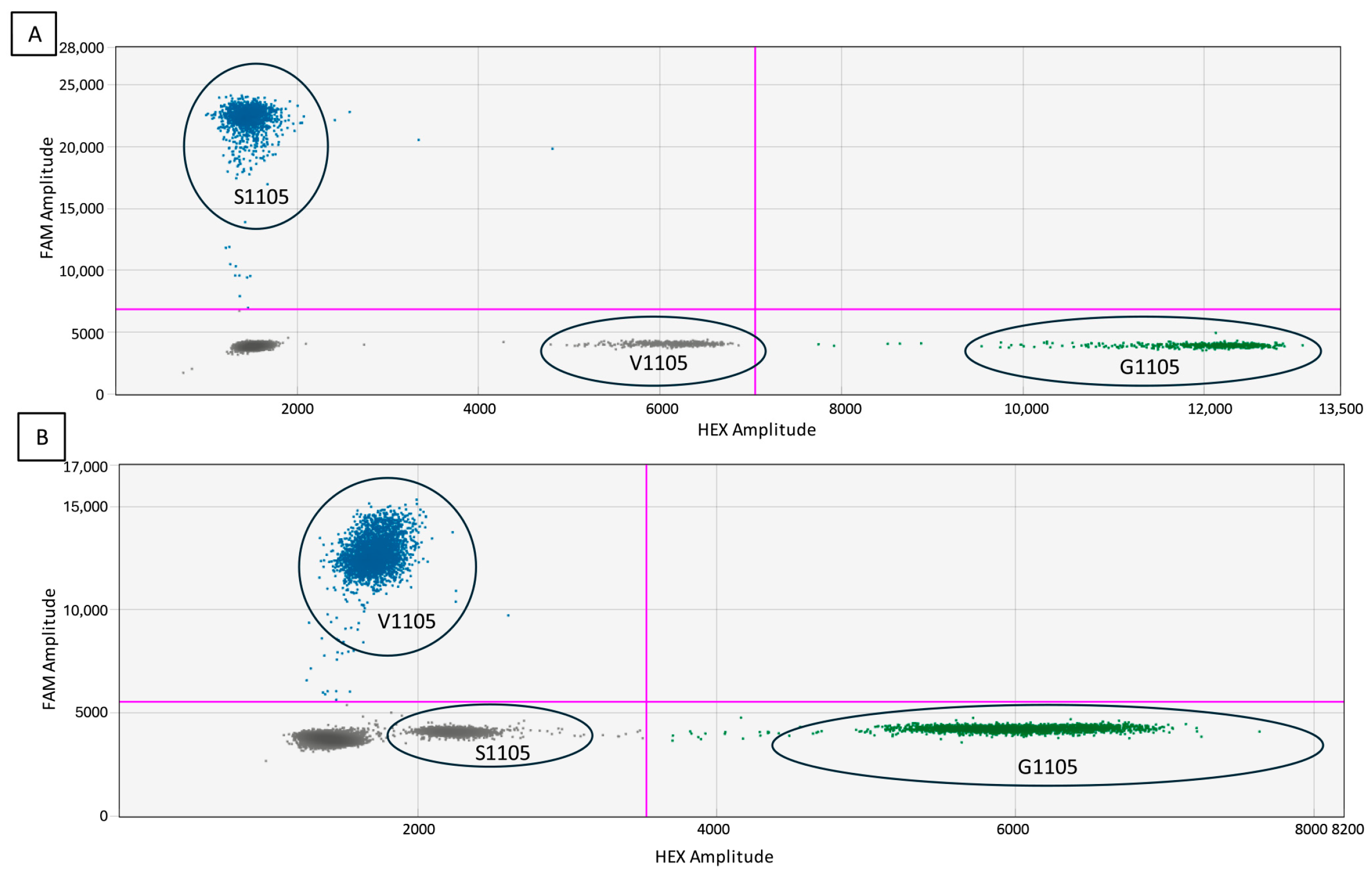
3.3. Specificity and False-Positive Rate Estimation
3.4. Multiplexing
3.5. Determination of Allele Frequencies in Field Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Massi, F.; Torriani, S.F.F.; Borghi, L.; Toffolatti, S.L. Fungicide Resistance Evolution and Detection in Plant Pathogens: Plasmopara viticola as a Case Study. Microorganisms 2021, 9, 119. [Google Scholar] [CrossRef] [PubMed]
- Fungicide Resistance Action Committee (FRAC). FRAC Recommendations for CAA Fungicides. Available online: https://www.frac.info/frac-teams/working-groups/caa-fungicides/recommendations-for-caa (accessed on 26 October 2024).
- Ministerio de Agricultura, Pesca y Alimentación. Productos Fitosanitarios: Registro de Productos Fitosanitarios. Gobierno de España. Available online: https://www.mapa.gob.es/es/agricultura/temas/sanidad-vegetal/productos-fitosanitarios/registro-productos/ (accessed on 29 August 2024).
- Gisi, U.; Waldner, M.; Kraus, N.; Dubuis, P.H.; Sierotzki, H. Inheritance of Resistance to Carboxylic Acid Amide (CAA) Fungicides in Plasmopara viticola. Plant Pathol. 2007, 56, 199–208. [Google Scholar] [CrossRef]
- Campbell, S.E.; Brannen, P.M.; Scherm, H.; Brewer, M.T. Fungicide Sensitivity Survey of Plasmopara viticola Populations in Georgia Vineyards. Plant Health Prog. 2020, 21, 256–261. [Google Scholar] [CrossRef]
- Nanni, I.M.; Pirondi, A.; Contaldo, N.; Collina, M. Screening of Sensitivity to Mandipropamid of Plasmopara viticola Populations from Italian Vineyards by Molecular and Biological Methods. Lett. Appl. Microbiol. 2016, 63, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.; Waldner, M.; Gisi, U. A Single Point Mutation in the Novel PvCesA3 Gene Confers Resistance to the Carboxylic Acid Amide Fungicide Mandipropamid in Plasmopara viticola. Fungal Genet. Biol. 2010, 47, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Sierotzki, H.; Blum, M.; Olaya, G.; Waldner-Zulauf, M.; Buitrago, C.; Wullschleger, J.; Cohen, Y.; Gisi, U. Sensitivity to CAA Fungicides and Frequency of Mutations in Cellulose Synthase (CesA3) Gene of Oomycete Pathogen Populations. In Modern Fungicides and Antifungal Compounds VI; Dehne, H.W., Gisi, U., Kuck, K.H., Russel, P.E., Lyr, H., Eds.; BCPC: Alton, UK, 2011; pp. 103–110. [Google Scholar]
- Zhang, H.; Kong, F.; Wang, X.; Liang, L.; Schoen, C.D.; Feng, J.; Wang, Z. Tetra-Primer ARMS PCR for Rapid Detection and Characterisation of Plasmopara viticola Phenotypes Resistant to Carboxylic Acid Amide Fungicides. Pest Manag. Sci. 2017, 73, 1655–1660. [Google Scholar] [CrossRef]
- Delmas, C.E.L.; Dussert, Y.; Delière, L.; Couture, C.; Mazet, I.D.; Richart Cervera, S.; Delmotte, F. Soft Selective Sweeps in Fungicide Resistance Evolution: Recurrent Mutations without Fitness Costs in Grapevine Downy Mildew. Mol. Ecol. 2017, 26, 1936–1951. [Google Scholar] [CrossRef]
- Toffolatti, S.L.; Russo, G.; Campia, P.; Bianco, P.A.; Borsa, P.; Coatti, M.; Torriani, S.F.F.; Sierotzki, H. A Time-Course Investigation of Resistance to the Carboxylic Acid Amide Mandipropamid in Field Populations of Plasmopara viticola Treated with Anti-Resistance Strategies. Pest Manag. Sci. 2018, 74, 2822–2834. [Google Scholar] [CrossRef]
- Rosenzweig, N.; Hanson, L.E.; Clark, G.; Franc, G.D.; Stump, W.L.; Jiang, Q.W.; Stewart, J.; Kirk, W.W. Use of PCR-RFLP Analysis to Monitor Fungicide Resistance in Cercospora Beticola Populations from Sugarbeet (Beta vulgaris) in Michigan, United States. Plant Dis. 2015, 99, 355–362. [Google Scholar] [CrossRef]
- Fontaine, S.; Remuson, F.; Caddoux, L.; Barrès, B. Investigation of the Sensitivity of Plasmopara viticola to Amisulbrom and Ametoctradin in French Vineyards Using Bioassays and Molecular Tools. Pest Manag. Sci. 2019, 75, 2115–2123. [Google Scholar] [CrossRef]
- Duan, Y.B.; Ge, C.Y.; Zhang, X.K.; Wang, J.X.; Zhou, M.G. Development and Evaluation of a Novel and Rapid Detection Assay for Botrytis cinerea based on Loop-Mediated Isothermal Amplification. PLoS ONE 2014, 9, e0111094. [Google Scholar] [CrossRef] [PubMed]
- Turan, C.; Nanni, I.M.; Landi, L.; Pirondi, A.; Collina, M. Fine Tuning of Real Time PCR as a First Tool for the Detection of G143A Substitution in Venturia inaequalis Samples. Am. J. Plant Sci. 2021, 12, 960–974. [Google Scholar] [CrossRef]
- Alzohairy, S.A.; Gillett, J.; Saito, S.; Naegele, R.N.; Xiao, C.L.; Miles, T.D. Fungicide Resistance Profiles of Botrytis cinerea Isolates from Michigan Vineyards and Development of a Taqman Assay for Detection of Fenhexamid Resistance. Plant Dis. 2021, 105, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Morley, A.A. Digital PCR: A Brief History. Biomol. Detect. Quantif. 2014, 1, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-Throughput Droplet Digital PCR System for Absolute Quantitation of DNA Copy Number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef]
- Hu, B.; Tao, Y.; Shao, Z.; Zheng, Y.; Zhang, R.; Yang, X.; Liu, J.; Li, X.; Sun, R. A Comparison of Blood Pathogen Detection Among Droplet Digital PCR, Metagenomic Next-Generation Sequencing, and Blood Culture in Critically Ill Patients With Suspected Bloodstream Infections. Front. Microbiol. 2021, 12, 641202. [Google Scholar] [CrossRef]
- Liu, X.; Feng, J.; Zhang, Q.; Guo, D.; Zhang, L.; Suo, T.; Hu, W.; Guo, M.; Wang, X.; Huang, Z.; et al. Analytical Comparisons of SARS-CoV-2 Detection by QRT-PCR and DdPCR with Multiple Primer/Probe Sets. Emerg. Microbes Infect. 2020, 9, 1175–1179. [Google Scholar] [CrossRef]
- Alcaide, M.; Yu, S.; Bushell, K.; Fornika, D.; Nielsen, J.S.; Nelson, B.H.; Mann, K.K.; Assouline, S.; Johnson, N.A.; Morin, R.D. Multiplex Droplet Digital PCR Quantification of Recurrent Somatic Mutations in Diffuse Large B-Cell and Follicular Lymphoma. Clin. Chem. 2016, 62, 1238–1247. [Google Scholar] [CrossRef]
- Miles, T.D.; Neill, T.M.; Colle, M.; Warneke, B.; Robinson, G.; Stergiopoulos, I.; Mahaffee, W.F. Allele-Specific Detection Methods for QoI Fungicide-Resistant Erysiphe necator in Vineyards. Plant Dis. 2021, 105, 175–182. [Google Scholar] [CrossRef]
- Mavridis, K.; Papapostolou, K.M.; Riga, M.; Ilias, A.; Michaelidou, K.; Bass, C.; Van Leeuwen, T.; Tsagkarakou, A.; Vontas, J. Multiple TaqMan QPCR and Droplet Digital PCR (DdPCR) Diagnostics for Pesticide Resistance Monitoring and Management, in the Major Agricultural Pest Tetranychus urticae. Pest Manag. Sci. 2022, 78, 263–273. [Google Scholar] [CrossRef]
- Battistini, G.; Gazzetti, K.; Collina, M. A New Approach: Determining Cyt b G143A Allele Frequency in Zymoseptoria tritici by Digital Droplet PCR. Biology 2022, 11, 240. [Google Scholar] [CrossRef] [PubMed]
- Gobbin, D.; Pertot, I.; Gessler, C. Genetic Structure of a Plasmopara viticola Population in an Isolated Italian Mountain Vineyard. J. Phytopathol. 2003, 151, 636–646. [Google Scholar] [CrossRef]
- Aoki, Y.; Furuya, S.; Suzuki, S. Method for Rapid Detection of the PvCesA3 Gene Allele Conferring Resistance to Mandipropamid, a Carboxylic Acid Amide Fungicide, in Plasmopara viticola Populations. Pest Manag. Sci. 2011, 67, 1557–1561. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2-A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Huggett, J.F.; Foy, C.A.; Benes, V.; Emslie, K.; Garson, J.A.; Haynes, R.; Hellemans, J.; Kubista, M.; Mueller, R.D.; Nolan, T.; et al. The Digital MIQE Guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments. Clin. Chem. 2013, 59, 892–902. [Google Scholar] [CrossRef]
- Whale, A.S.; De Spiegelaere, W.; Trypsteen, W.; Nour, A.A.; Bae, Y.K.; Benes, V.; Burke, D.; Cleveland, M.; Corbisier, P.; Devonshire, A.S.; et al. The Digital MIQE Guidelines Update: Minimum Information for Publication of Quantitative Digital PCR Experiments for 2020. Clin. Chem. 2020, 66, 1012–1029. [Google Scholar] [CrossRef]
- Hou, Y.; Chen, S.; Zheng, Y.; Zheng, X.; Lin, J.M. Droplet-Based Digital PCR (DdPCR) and Its Applications. TrAC—Trends Anal. Chem. 2023, 158, 116897. [Google Scholar] [CrossRef]
- Bio-Rad Laboratories. Droplet Digital TM PCR Applications Guide. Available online: https://www.bio-rad.com/es-es/literature-library (accessed on 2 November 2024).
- JCGM. International Vocabulary of Metrology Basic and General Concepts and Associated Terms, 3rd ed.; BIPM: Sèvres, France, 2012; Available online: http://www.bipm.org/utils/common/documents/jcgm/JCGM_200_2012.pdf (accessed on 7 October 2024).
- Venbrux, M.; Crauwels, S.; Rediers, H. Current and Emerging Trends in Techniques for Plant Pathogen Detection. Front. Plant Sci. 2023, 14, 1120968. [Google Scholar] [CrossRef]
- Dingle, T.C.; Sedlak, R.H.; Cook, L.; Jerome, K.R. Tolerance of Droplet-Digital PCR vs Real-Time Quantitative PCR to Inhibitory Substances. Clin. Chem. 2013, 59, 1670–1672. [Google Scholar] [CrossRef]
- Maheshwari, Y.; Selvaraj, V.; Godfrey, K.; Hajeri, S.; Yokomi, R. Multiplex Detection of “Candidatus Liberibacter Asiaticus” and Spiroplasma Citri by QPCR and Droplet Digital PCR. PLoS ONE 2021, 16, e0242392. [Google Scholar] [CrossRef]
- Van Heetvelde, M.; Van Loocke, W.; Trypsteen, W.; Baert, A.; Vanderheyden, K.; Crombez, B.; Vandesompele, J.; De Leeneer, K.; Claes, K.B.M. Evaluation of Relative Quantification of Alternatively Spliced Transcripts Using Droplet Digital PCR. Biomol. Detect. Quantif. 2017, 13, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Morcia, C.; Ghizzoni, R.; Delogu, C.; Andreani, L.; Carnevali, P.; Terzi, V. Digital Pcr: What Relevance to Plant Studies? Biology 2020, 9, 433. [Google Scholar] [CrossRef] [PubMed]
- Rowlands, V.; Rutkowski, A.J.; Meuser, E.; Carr, T.H.; Harrington, E.A.; Barrett, J.C. Optimisation of Robust Singleplex and Multiplex Droplet Digital PCR Assays for High Confidence Mutation Detection in Circulating Tumour DNA. Sci. Rep. 2019, 9, 12620. [Google Scholar] [CrossRef] [PubMed]
- Witte, A.K.; Mester, P.; Fister, S.; Witte, M.; Schoder, D.; Rossmanith, P. A Systematic Investigation of Parameters Influencing Droplet Rain in the Listeria Monocytogenes PrfA Assay—Reduction of Ambiguous Results in DdPCR. PLoS ONE 2016, 11, e0168179. [Google Scholar] [CrossRef] [PubMed]
- Nyaruaba, R.; Li, C.; Mwaliko, C.; Mwau, M.; Odiwuor, N.; Muturi, E.; Muema, C.; Xiong, J.; Li, J.; Yu, J.; et al. Developing Multiplex DdPCR Assays for SARS-CoV-2 Detection Based on Probe Mix and Amplitude Based Multiplexing. Expert. Rev. Mol. Diagn. 2021, 21, 119–129. [Google Scholar] [CrossRef]
- Huang, X.; Wang, X.; Kong, F.; van der Lee, T.; Wang, Z.; Zhang, H. Detection and Characterization of Carboxylic Acid Amide-Resistant Plasmopara viticola in China Using a Taqman-Mgb Real-Time PCR. Plant Dis. 2020, 104, 2338–2345. [Google Scholar] [CrossRef]

| Population | Village | PDO 1 | Cultivar | Fungicide Applications in 2023 |
|---|---|---|---|---|
| A | Olaberria | Getariko Txakolina | Hondarrabi Zuri | Never exposed to CAA fungicides |
| B | Muxika | Bizkaiko Txakolina | Hondarrabi Zuri | Ampexio (mandipropamid + zoxamide) Java (valifenalate + folpet) |
| C | Izurtza | Bizkaiko Txakolina | Hondarrabi Zuri | No CAA fungicide applications |
| D | Buradon Gatzaga | Rioja | Tempranillo | No CAA fungicide applications |
| Name | Sequence (5′ to 3′) | Probe Dye |
|---|---|---|
| Forward primer | ACGGCTGCTACCTTTAC | |
| Reverse primer | ACAAACACGACAATGTAGAC | |
| G1105 probe | AATGTGTTCGGCTCGTT | HEX |
| V1105 probe | TTCGTCTCGTTGCTGG | FAM |
| S1105 probe | AATGTGTTCAGCTCGTTG | FAM |
| Mix 1 | Mix 2 | Mix 3 | Mix 4 | Mix 5 | Mix 6 | Mix 7 | ||
|---|---|---|---|---|---|---|---|---|
| PvCesA3_S | S1105 | 0% | 1% | 5% | 50% | 95% | 99% | 100% |
| G1105 | 100% | 99% | 95% | 50% | 5% | 1% | 0% | |
| PvCesA3_V | V1105 | 0% | 1% | 5% | 50% | 95% | 99% | 100% |
| G1105 | 100% | 99% | 95% | 50% | 5% | 1% | 0% | |
| Populations | Duplex | Multiplex | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S1105 | V1105 | G1105 | Fractional Abundance (%) | SD 1 | S1105 | V1105 | G1105 | Fractional Abundance (%) | SD 1 | |
| A | 35.30 | 3.55 | 602.84 | 6.62 | 0.36 | 23.85 | 2.18 | 325.00 | 7.42 | 0.04 |
| B | 424.5 | 73.83 | 0.00 | 100 | 0.00 | 354.17 | 66.50 | 0.00 | 100 | 0.00 |
| C | 280.67 | 87.77 | 3.79 | 98.98 | 1.42 | 261.67 | 88.77 | 2.70 | 99.23 | 0.20 |
| D | 144.7 | 37.03 | 116.15 | 61.00 | 0.66 | 216.67 | 64.00 | 201.17 | 59.16 | 1.74 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Zelaia, H.; Nanni, I.M.; Oggiano, I.; Hernández, M.; Díez-Navajas, A.M.; Collina, M. Droplet Digital PCR: A New Molecular Method to Detect G1105S/V Mutations in Plasmopara viticola CesA3 Gene. Biology 2024, 13, 919. https://doi.org/10.3390/biology13110919
Sánchez-Zelaia H, Nanni IM, Oggiano I, Hernández M, Díez-Navajas AM, Collina M. Droplet Digital PCR: A New Molecular Method to Detect G1105S/V Mutations in Plasmopara viticola CesA3 Gene. Biology. 2024; 13(11):919. https://doi.org/10.3390/biology13110919
Chicago/Turabian StyleSánchez-Zelaia, Helene, Irene Maja Nanni, Ivano Oggiano, Mónica Hernández, Ana María Díez-Navajas, and Marina Collina. 2024. "Droplet Digital PCR: A New Molecular Method to Detect G1105S/V Mutations in Plasmopara viticola CesA3 Gene" Biology 13, no. 11: 919. https://doi.org/10.3390/biology13110919
APA StyleSánchez-Zelaia, H., Nanni, I. M., Oggiano, I., Hernández, M., Díez-Navajas, A. M., & Collina, M. (2024). Droplet Digital PCR: A New Molecular Method to Detect G1105S/V Mutations in Plasmopara viticola CesA3 Gene. Biology, 13(11), 919. https://doi.org/10.3390/biology13110919





