Enterobacteriaceae in the Human Gut: Dynamics and Ecological Roles in Health and Disease
Abstract
:Simple Summary
Abstract
1. Proteobacteria/Enterobacteriaceae in the Healthy Human Gut
1.1. Taxonomy, Diversity and Abundance
1.2. Strain Residency in the Gut
1.3. Functional Roles of Enterobacteriaceae in the Human Gut
- Maintenance of an efficient anaerobic environment in the gut
- Production of vitamins
- Protection against gut pathogen infections
2. Proteobacteria/Enterobacteriaceae Expansion in Dysbiosis Associated with Human Disease
2.1. Proteobacteria Bloom: A Marker of Gut Microbiota in Disease
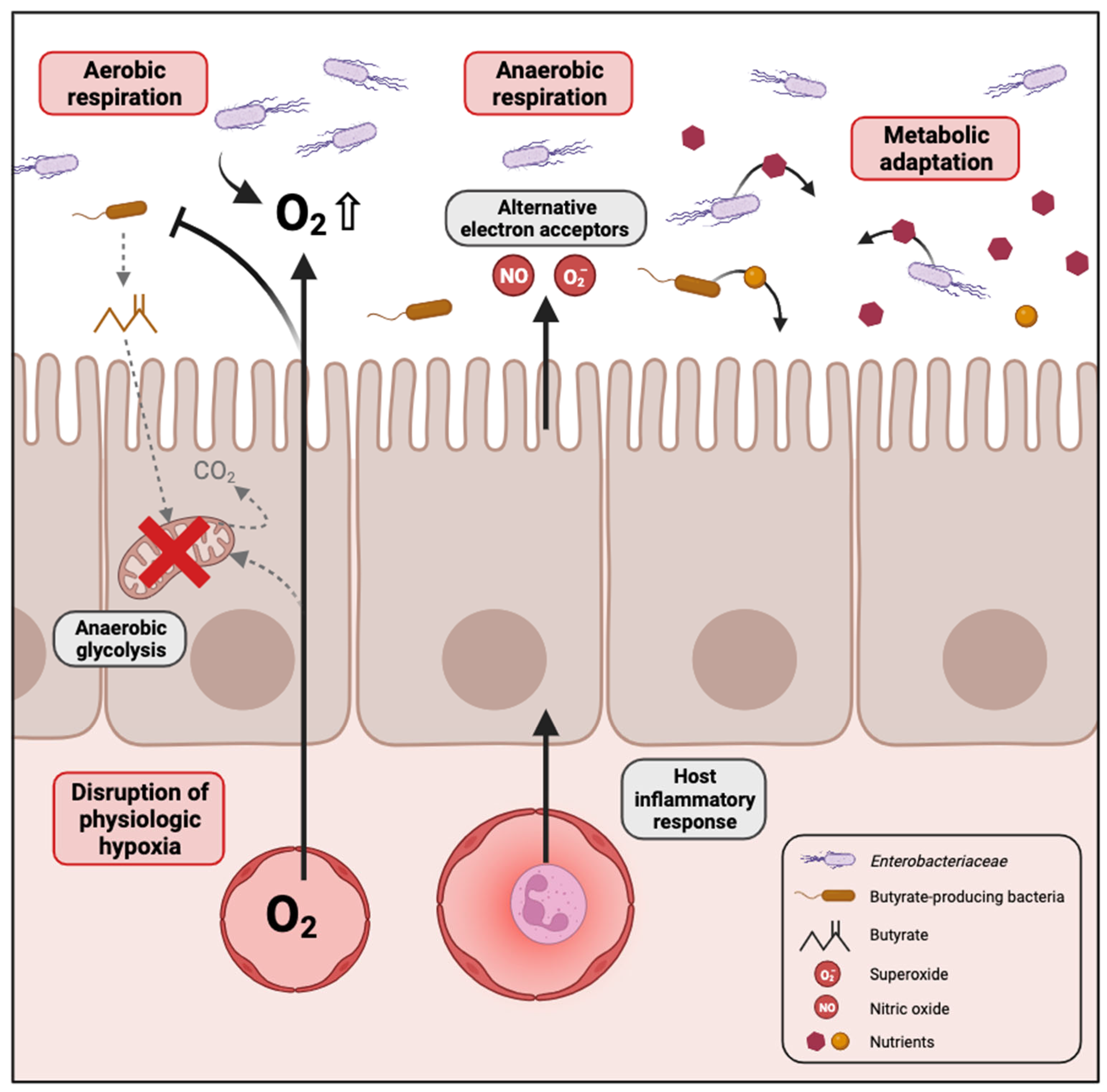
2.2. Causes of Enterobacteriaceae Proliferation in Dysbiotic Conditions
2.2.1. Disruption of Physiologic Hypoxia
2.2.2. Inflammation Generates Electron Acceptors for Anaerobic Respiration
2.2.3. Metabolic Adaptation to Dysbiotic Conditions
- Glycerol in cystic fibrosis patients
- Intestinal mucosa-derived substrates
- Diet-derived substrates
2.3. Consequences of Enterobacteriaceae Proliferation in Disease Progression
- Crohn’s disease
- Obesity-associated diseases
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sakkas, H.; Bozidis, P.; Touzios, C.; Kolios, D.; Athanasiou, G.; Athanasopoulou, E.; Gerou, I.; Gartzonika, C. Nutritional Status and the Influence of the Vegan Diet on the Gut Microbiota and Human Health. Medicina 2020, 56, 88. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What Is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; Fitzgerald, M.G.; Fulton, R.S.; et al. Structure, Function and Diversity of the Healthy Human Microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Martinson, J.N.V.; Pinkham, N.V.; Peters, G.W.; Cho, H.; Heng, J.; Rauch, M.; Broadaway, S.C.; Walk, S.T. Rethinking Gut Microbiome Residency and the Enterobacteriaceae in Healthy Human Adults. ISME J. 2019, 13, 2306–2318. [Google Scholar] [CrossRef]
- Tenaillon, O.; Skurnik, D.; Picard, B.; Denamur, E. The Population Genetics of Commensal Escherichia coli. Nat. Rev. Microbiol. 2010, 8, 207–217. [Google Scholar] [CrossRef]
- Alm, E.W.; Walk, S.T.; Gordon, D.M. The Niche of Escherichia coli. In Population Genetics of Bacteria; ASM Press: Washington, DC, USA, 2014; Available online: https://onlinelibrary.wiley.com/doi/abs/10.1128/9781555817114.ch6 (accessed on 20 February 2024).
- Marin, J.; Clermont, O.; Royer, G.; Mercier-Darty, M.; Decousser, J.W.; Tenaillon, O.; Denamur, E.; Blanquart, F. The Population Genomics of Increased Virulence and Antibiotic Resistance in Human Commensal Escherichia coli over 30 Years in France. Appl. Environ. Microbiol. 2022, 88, e0066422. [Google Scholar] [CrossRef]
- Gordon, D.M.; O’Brien, C.L.; Pavli, P. Escherichia coli Diversity in the Lower Intestinal Tract of Humans. Environ. Microbiol. Rep. 2015, 7, 642–648. [Google Scholar] [CrossRef] [PubMed]
- Calderón, D.; Cárdenas, P.A.; Prado-Vivar, B.; Graham, J.P.; Trueba, G. A Longitudinal Study of Dominant E. coli Lineages and Antimicrobial Resistance in the Gut of Children Living in an Upper Middle-Income Country. J. Glob. Antimicrob. Resist. 2022, 29, 136–140. [Google Scholar] [CrossRef]
- Priya, S.; Blekhman, R. Population Dynamics of the Human Gut Microbiome: Change Is the Only Constant. Genome Biol. 2019, 20, 150. [Google Scholar] [CrossRef]
- Martinson, J.N.V.; Walk, S.T. Escherichia coli Residency in the Gut of Healthy Human Adults. EcoSal Plus 2020, 9, 10–1128. [Google Scholar] [CrossRef]
- Tomas, J.; Reygner, J.; Mayeur, C.; Ducroc, R.; Bouet, S.; Bridonneau, C.; Cavin, J.B.; Thomas, M.; Langella, P.; Cherbuy, C. Early Colonizing Escherichia coli Elicits Remodeling of Rat Colonic Epithelium Shifting toward a New Homeostatic State. ISME J. 2015, 9, 46–58. [Google Scholar] [CrossRef]
- Mueller, N.T.; Bakacs, E.; Combellick, J.; Grigoryan, Z.; Dominguez-Bello, M.G. The Infant Microbiome Development: Mom Matters. Trends Mol. Med. 2015, 21, 109–117. [Google Scholar] [CrossRef]
- Jiang, M.; Cao, Y.; Guo, Z.F.; Chen, M.; Chen, X.; Guo, Z. Menaquinone Biosynthesis in Escherichia coli: Identification of 2-Succinyl-5-Enolpyruvyl-6-Hydroxy-3-Cyclohexene-1-Carboxylate as a Novel Intermediate and Re-Evaluation of MenD Activity. Biochemistry 2007, 46, 10979–10989. [Google Scholar] [CrossRef]
- Karl, J.P.; Meydani, M.; Barnett, J.B.; Vanegas, S.M.; Barger, K.; Fu, X.; Goldin, B.; Kane, A.; Rasmussen, H.; Vangay, P.; et al. Fecal Concentrations of Bacterially Derived Vitamin K Forms Are Associated with Gut Microbiota Composition but Not Plasma or Fecal Cytokine Concentrations in Healthy Adults. Am. J. Clin. Nutr. 2017, 106, 1052–1061. [Google Scholar] [CrossRef]
- Fang, H.; Kang, J.; Zhang, D. Microbial Production of Vitamin B12: A Review and Future Perspectives. Microb. Cell Fact. 2017, 16, 15. [Google Scholar] [CrossRef]
- Degnan, P.H.; Taga, M.E.; Goodman, A.L. Vitamin B12 as a Modulator of Gut Microbial Ecology. Cell Metab. 2014, 20, 769–778. [Google Scholar] [CrossRef]
- Magnúsdóttir, S.; Ravcheev, D.; De Crécy-Lagard, V.; Thiele, I. Systematic Genome Assessment of B-Vitamin Biosynthesis Suggests Cooperation among Gut Microbes. Front. Genet. 2015, 6, 148. [Google Scholar] [CrossRef] [PubMed]
- Ducarmon, Q.R.; Zwittink, R.D.; Hornung, B.V.H.; van Schaik, W.; Young, V.B.; Kuijper, E.J. Gut Microbiota and Colonization Resistance against Bacterial Enteric Infection. Microbiol. Mol. Biol. Rev. 2019, 83, e00007-19. [Google Scholar] [CrossRef] [PubMed]
- Hudault, S.; Guignot, J.; Servin, A.L. Escherichia coli Strains Colonising the Gastrointestinal Tract Protect Germfree Mice against Salmonella Typhimurium Infection. Gut 2001, 49, 47–55. [Google Scholar] [CrossRef]
- Chang, D.E.; Smalley, D.J.; Tucker, D.L.; Leatham, M.P.; Norris, W.E.; Stevenson, S.J.; Anderson, A.B.; Grissom, J.E.; Laux, D.C.; Cohen, P.S.; et al. Carbon Nutrition of Escherichia coli in the Mouse Intestine. Proc. Natl. Acad. Sci. USA 2004, 101, 7427–7432. [Google Scholar] [CrossRef] [PubMed]
- Fabich, A.J.; Jones, S.A.; Chowdhury, F.Z.; Cernosek, A.; Anderson, A.; Smalley, D.; McHargue, J.W.; Hightower, G.A.; Smith, J.T.; Autieri, S.M.; et al. Comparison of Carbon Nutrition for Pathogenic and Commensal Escherichia coli Strains in the Mouse Intestine. Infect. Immun. 2008, 76, 1143–1152. [Google Scholar] [CrossRef] [PubMed]
- Leatham, M.P.; Banerjee, S.; Autieri, S.M.; Mercado-Lubo, R.; Conway, T.; Cohen, P.S. Precolonized Human Commensal Escherichia coli Strains Serve as a Barrier to E. coli O157:H7 Growth in the Streptomycin-Treated Mouse Intestine. Infect. Immun. 2009, 77, 2876–2886. [Google Scholar] [CrossRef]
- Maltby, R.; Leatham-Jensen, M.P.; Gibson, T.; Cohen, P.S.; Conway, T. Nutritional Basis for Colonization Resistance by Human Commensal Escherichia coli Strains HS and Nissle 1917 against E. coli O157:H7 in the Mouse Intestine. PLoS ONE 2013, 8, e53957. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, A.M.; Shanahan, F. The Gut Flora as a Forgotten Organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The Impact of the Gut Microbiota on Human Health: An Integrative View. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef] [PubMed]
- Shin, N.R.; Whon, T.W.; Bae, J.W. Proteobacteria: Microbial Signature of Dysbiosis in Gut Microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A Common Factor in Human Diseases. BioMed. Res. Int. 2017, 2017, 9351507. [Google Scholar] [CrossRef]
- Winter, S.E.; Bäumler, A.J. Why Related Bacterial Species Bloom Simultaneously in the Gut: Principles Underlying the “like Will to like” Concept. Cell. Microbiol. 2014, 16, 179–184. [Google Scholar] [CrossRef]
- Seksik, P.; Rigottier-Gois, L.; Gramet, G.; Sutren, M.; Pochart, P.; Marteau, P.; Jian, R.; Doré, J. Alterations of the Dominant Faecal Bacterial Groups in Patients with Crohn’s Disease of the Colon. Gut 2003, 52, 237–242. [Google Scholar] [CrossRef]
- Wang, T.; Cai, G.; Qiu, Y.; Fei, N.; Zhang, M.; Pang, X.; Jia, W.; Cai, S.; Zhao, L. Structural Segregation of Gut Microbiota between Colorectal Cancer Patients and Healthy Volunteers. ISME J. 2012, 6, 320–329. [Google Scholar] [CrossRef]
- Krogius-Kurikka, L.; Lyra, A.; Malinen, E.; Aarnikunnas, J.; Tuimala, J.; Paulin, L.; Mäkivuokko, H.; Kajander, K.; Palva, A. Microbial Community Analysis Reveals High Level Phylogenetic Alterations in the Overall Gastrointestinal Microbiota of Diarrhoea-Predominant Irritable Bowel Syndrome Sufferers. BMC Gastroenterol. 2009, 9, 95. [Google Scholar] [CrossRef]
- Fei, N.; Zhao, L. An Opportunistic Pathogen Isolated from the Gut of an Obese Human Causes Obesity in Germfree Mice. ISME J. 2013, 7, 880–884. [Google Scholar] [CrossRef]
- Espey, M.G. Role of Oxygen Gradients in Shaping Redox Relationships between the Human Intestine and Its Microbiota. Free Radic. Biol. Med. 2013, 55, 130–140. [Google Scholar] [CrossRef]
- Albenberg, L.; Esipova, T.V.; Judge, C.P.; Bittinger, K.; Chen, J.; Laughlin, A.; Grunberg, S.; Baldassano, R.N.; Lewis, J.D.; Li, H.; et al. Correlation between Intraluminal Oxygen Gradient and Radial Partitioning of Intestinal Microbiota. Gastroenterology 2014, 147, 1055–1063.e8. [Google Scholar] [CrossRef] [PubMed]
- Henson, M.A.; Phalak, P. Microbiota Dysbiosis in Inflammatory Bowel Diseases: In Silico Investigation of the Oxygen Hypothesis. BMC Syst. Biol. 2017, 11, 145. [Google Scholar] [CrossRef] [PubMed]
- Rigottier-Gois, L. Dysbiosis in Inflammatory Bowel Diseases: The Oxygen Hypothesis. ISME J. 2013, 7, 1256–1261. [Google Scholar] [CrossRef] [PubMed]
- Hughes, E.R.; Winter, M.G.; Duerkop, B.A.; Spiga, L.; Furtado de Carvalho, T.; Zhu, W.; Gillis, C.C.; Büttner, L.; Smoot, M.P.; Behrendt, C.L.; et al. Microbial Respiration and Formate Oxidation as Metabolic Signatures of Inflammation-Associated Dysbiosis. Cell Host Microbe 2017, 21, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Stecher, B. The Roles of Inflammation, Nutrient Availability and the Commensal Microbiota in Enteric Pathogen Infection. Microbiol. Spectr. 2015, 3, 297–320. [Google Scholar] [CrossRef] [PubMed]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the Intestinal Microbiome in Inflammatory Bowel Disease and Treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef]
- Carroll, I.M.; Ringel-Kulka, T.; Siddle, J.P.; Ringel, Y. Alterations in Composition and Diversity of the Intestinal Microbiota in Patients with Diarrhea-Predominant Irritable Bowel Syndrome. Neurogastroenterol. Motil. 2012, 24, 521-e248. [Google Scholar] [CrossRef]
- Larsen, N.; Vogensen, F.K.; Van Den Berg, F.W.J.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sørensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut Microbiota in Human Adults with Type 2 Diabetes Differs from Non-Diabetic Adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef]
- Zhu, L.; Baker, S.S.; Gill, C.; Liu, W.; Alkhouri, R.; Baker, R.D.; Gill, S.R. Characterization of Gut Microbiomes in Nonalcoholic Steatohepatitis (NASH) Patients: A Connection between Endogenous Alcohol and NASH. Hepatology 2013, 57, 601–609. [Google Scholar] [CrossRef]
- Litvak, Y.; Byndloss, M.X.; Tsolis, R.M.; Bäumler, A.J. Dysbiotic Proteobacteria Expansion: A Microbial Signature of Epithelial Dysfunction. Curr. Opin. Microbiol. 2017, 39, 1–6. [Google Scholar] [CrossRef]
- Litvak, Y.; Byndloss, M.X.; Bäumler, A.J. Colonocyte Metabolism Shapes the Gut Microbiota. Science 2018, 362, eaat9076. [Google Scholar] [CrossRef]
- Andoh, A.; Nishida, A. Alteration of the Gut Microbiome in Inflammatory Bowel Disease. Digestion 2023, 104, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Winter, S.E.; Thiennimitr, P.; Winter, M.G.; Butler, B.P.; Huseby, D.L.; Crawford, R.W.; Russell, J.M.; Bevins, C.L.; Adams, L.G.; Tsolis, R.M.; et al. Gut Inflammation Provides a Respiratory Electron Acceptor for Salmonella. Nature 2010, 467, 426–429. [Google Scholar] [CrossRef] [PubMed]
- Winter, S.E.; Winter, M.G.; Xavier, M.N.; Thiennimitr, P.; Poon, V.; Keestra, A.M.; Laughlin, R.C.; Gomez, G.; Wu, J.; Lawhon, S.D.; et al. Host-Derived Nitrate Boosts Growth of E. coli in the Inflamed Gut. Science 2013, 339, 708–711. [Google Scholar] [CrossRef]
- Winter, S.E.; Lopez, C.A.; Bäumler, A.J. The Dynamics of Gut-Associated Microbial Communities during Inflammation. EMBO Rep. 2013, 14, 319–327. [Google Scholar] [CrossRef]
- Bradley, P.H.; Pollard, K.S. Proteobacteria Explain Significant Functional Variability in the Human Gut Microbiome. Microbiome 2017, 5, 36. [Google Scholar] [CrossRef]
- Thiennimitr, P.; Winter, S.E.; Winter, M.G.; Xavier, M.N.; Tolstikov, V.; Huseby, D.L.; Sterzenbach, T.; Tsolis, R.M.; Roth, J.R.; Bäumler, A.J. Intestinal Inflammation Allows Salmonella to Use Ethanolamine to Compete with the Microbiota. Proc. Natl. Acad. Sci. USA 2011, 108, 17480–17485. [Google Scholar] [CrossRef] [PubMed]
- Rowley, C.A.; Sauder, A.B.; Kendall, M.M. The Ethanolamine-Sensing Transcription Factor EutR Promotes Virulence and Transmission during Citrobacter Rodentium Intestinal Infection. Infect. Immun. 2020, 88. [Google Scholar] [CrossRef]
- Gillis, C.C.; Hughes, E.R.; Spiga, L.; Winter, M.G.; Zhu, W.; Furtado de Carvalho, T.; Chanin, R.B.; Behrendt, C.L.; Hooper, L.V.; Santos, R.L.; et al. Dysbiosis-Associated Change in Host Metabolism Generates Lactate to Support Salmonella Growth. Cell Host Microbe 2018, 23, 54–64.e6. [Google Scholar] [CrossRef] [PubMed]
- Faber, F.; Tran, L.; Byndloss, M.X.; Lopez, C.A.; Velazquez, E.M.; Kerrinnes, T.; Nuccio, S.P.; Wangdi, T.; Fiehn, O.; Tsolis, R.M.; et al. Host-Mediated Sugar Oxidation Promotes Post-Antibiotic Pathogen Expansion. Nature 2016, 534, 697–699. [Google Scholar] [CrossRef] [PubMed]
- Faber, F.; Thiennimitr, P.; Spiga, L.; Byndloss, M.X.; Litvak, Y.; Lawhon, S.; Andrews-Polymenis, H.L.; Winter, S.E.; Bäumler, A.J. Respiration of Microbiota-Derived 1,2-Propanediol Drives Salmonella Expansion during Colitis. PLoS Pathog. 2017, 13, e1006129. [Google Scholar] [CrossRef]
- Spiga, L.; Winter, M.G.; Furtado de Carvalho, T.; Zhu, W.; Hughes, E.R.; Gillis, C.C.; Behrendt, C.L.; Kim, J.; Chessa, D.; Andrews-Polymenis, H.L.; et al. An Oxidative Central Metabolism Enables Salmonella to Utilize Microbiota-Derived Succinate. Cell Host Microbe 2017, 22, 291–301.e6. [Google Scholar] [CrossRef] [PubMed]
- Kitamoto, S.; Alteri, C.J.; Rodrigues, M.; Nagao-Kitamoto, H.; Sugihara, K.; Himpsl, S.D.; Bazzi, M.; Miyoshi, M.; Nishioka, T.; Hayashi, A.; et al. Dietary L-Serine Confers a Competitive Fitness Advantage to Enterobacteriaceae in the Inflamed Gut. Nat. Microbiol. 2020, 5, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Miyata, N.; Winter, M.G.; Arenales, A.; Hughes, E.R.; Spiga, L.; Kim, J.; Sifuentes-Dominguez, L.; Starokadomskyy, P.; Gopal, P.; et al. Editing of the Gut Microbiota Reduces Carcinogenesis in Mouse Models of Colitis-Associated Colorectal Cancer. J. Exp. Med. 2019, 216, 2378–2393. [Google Scholar] [CrossRef] [PubMed]
- Matamouros, S.; Hayden, H.S.; Hager, K.R.; Brittnacher, M.J.; Lachance, K.; Weiss, E.J.; Pope, C.E.; Imhaus, A.F.; McNally, C.P.; Borenstein, E.; et al. Adaptation of Commensal Proliferating Escherichia coli to the Intestinal Tract of Young Children with Cystic Fibrosis. Proc. Natl. Acad. Sci. USA 2018, 115, 1605–1610. [Google Scholar] [CrossRef]
- Lemons, J.M.S.; Conrad, M.; Tanes, C.; Chen, J.; Friedman, E.S.; Roggiani, M.; Curry, D.; Chau, L.; Hecht, A.L.; Harling, L.; et al. Enterobacteriaceae Growth Promotion by Intestinal Acylcarnitines, a Biomarker of Dysbiosis in Inflammatory Bowel Disease. Cell. Mol. Gastroenterol. Hepatol. 2024, 17, 131–148. [Google Scholar] [CrossRef]
- Wouthuyzen-Bakker, M.; Bodewes, F.A.J.A.; Verkade, H.J. Persistent Fat Malabsorption in Cystic Fibrosis; Lessons from Patients and Mice. J. Cyst. Fibros. 2011, 10, 150–158. [Google Scholar] [CrossRef]
- Caley, L.R.; White, H.; de Goffau, M.C.; Floto, R.A.; Parkhill, J.; Marsland, B.; Peckham, D.G. Cystic Fibrosis-Related Gut Dysbiosis: A Systematic Review. Dig. Dis. Sci. 2023, 68, 1797–1814. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, L.R.; Pope, C.E.; Hayden, H.S.; Heltshe, S.; Levy, R.; McNamara, S.; Jacobs, M.A.; Rohmer, L.; Radey, M.; Ramsey, B.W.; et al. Escherichia coli Dysbiosis Correlates with Gastrointestinal Dysfunction in Children with Cystic Fibrosis. Clin. Infect. Dis. 2014, 58, 396–399. [Google Scholar] [CrossRef] [PubMed]
- Garsin, D.A. Ethanolamine Utilization in Bacterial Pathogens: Roles and Regulation. Nat. Rev. Microbiol. 2010, 8, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.L.; Chassard, C.; Hausmann, M.; Von Itzstein, M.; Hennet, T. Sialic Acid Catabolism Drives Intestinal Inflammation and Microbial Dysbiosis in Mice. Nat. Commun. 2015, 6, 8141. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.M.; Ferreyra, J.A.; Higginbottom, S.K.; Lynch, J.B.; Kashyap, P.C.; Gopinath, S.; Naidu, N.; Choudhury, B.; Weimer, B.C.; Monack, D.M.; et al. Microbiota-Liberated Host Sugars Facilitate Post-Antibiotic Expansion of Enteric Pathogens. Nature 2013, 502, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Tytgat, H.L.P.; Nobrega, F.L.; van der Oost, J.; de Vos, W.M. Bowel Biofilms: Tipping Points between a Healthy and Compromised Gut? Trends Microbiol. 2019, 27, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Ju, T.; Bourrie, B.C.T.; Forgie, A.J.; Pepin, D.M.; Tollenaar, S.; Sergi, C.M.; Willing, B.P. The Gut Commensal Escherichia coli Aggravates High-Fat-Diet-Induced Obesity and Insulin Resistance in Mice. Appl. Environ. Microbiol. 2023, 89, e01628-22. [Google Scholar] [CrossRef]
- Yoo, W.; Zieba, J.K.; Foegeding, N.J.; Torres, T.P.; Shelton, C.D.; Shealy, N.G.; Byndloss, A.J.; Cevallos, S.A.; Gertz, E.; Tiffany, C.R.; et al. High-Fat Diet-Induced Colonocyte Dysfunction Escalates Microbiota-Derived Trimethylamine N-Oxide. Science 2021, 373, 813–818. [Google Scholar] [CrossRef]
- Chervy, M.; Barnich, N.; Denizot, J. Adherent-Invasive E. coli: Update on the Lifestyle of a Troublemaker in Crohn’s Disease. Int. J. Mol. Sci. 2020, 21, 3734. [Google Scholar] [CrossRef]
- Pizer, L.I.; Potochny, M.L. Nutritional and Regulatory Aspects of Serine Metabolism in Escherichia. J. Bacteriol. 1964, 88, 611–619. [Google Scholar] [CrossRef]
- Denizot, J.; Sivignon, A.; Barreau, F.; Darcha, C.; Chan, H.F.C.; Stanners, C.P.; Hofman, P.; Darfeuille-Michaud, A.; Barnich, N. Adherent-Invasive Escherichia coli Induce Claudin-2 Expression and Barrier Defect in CEABAC10 Mice and Crohn’s Disease Patients. Inflamm. Bowel Dis. 2012, 18, 294–304. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Cai, X.; Guo, X.; Xu, Y.; Gong, J.; Li, Y.; Zhu, W. Let-7b Ameliorates Crohn’s Disease-Associated Adherent-Invasive E. coli Induced Intestinal Inflammation via Modulating Toll-Like Receptor 4 Expression in Intestinal Epithelial Cells. Biochem. Pharmacol. 2018, 156, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Glasser, A.L.; Boudeau, J.; Barnich, N.; Perruchot, M.H.; Colombel, J.F.; Darfeuille-Michaud, A. Adherent Invasive Escherichia coli Strains from Patients with Crohn’s Disease Survive and Replicate within Macrophages without Inducing Host Cell Death. Infect. Immun. 2001, 69, 5529–5537. [Google Scholar] [CrossRef] [PubMed]
- Small, C.L.N.; Reid-Yu, S.A.; McPhee, J.B.; Coombes, B.K. Persistent Infection with Crohn’s Disease-Associated Adherent-Invasive Escherichia coli Leads to Chronic Inflammation and Intestinal Fibrosis. Nat. Commun. 2013, 4, 1957. [Google Scholar] [CrossRef]
- Ellermann, M.; Gharaibeh, R.Z.; Fulbright, L.; Dogan, B.; Moore, L.N.; Broberg, C.A.; Lopez, L.R.; Rothemich, A.M.; Herzog, J.W.; Rogala, A.; et al. Yersiniabactin-Producing Adherent/Invasive Escherichia coli Promotes Inflammation-Associated Fibrosis in Gnotobiotic Il10−/− Mice. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef] [PubMed]
- Gilliland, A.; Chan, J.J.; De Wolfe, T.J.; Yang, H.; Vallance, B.A. Pathobionts in Inflammatory Bowel Disease: Origins, Underlying Mechanisms, and Implications for Clinical Care. Gastroenterology 2023, 166, 44–58. [Google Scholar] [CrossRef] [PubMed]
- Hamjane, N.; Mechita, M.B.; Nourouti, N.G.; Barakat, A. Gut Microbiota Dysbiosis–Associated Obesity and Its Involvement in Cardiovascular Diseases and Type 2 Diabetes. A Systematic Review. Microvasc. Res. 2024, 151, 104601. [Google Scholar] [CrossRef]
- Keskitalo, A.; Munukka, E.; Toivonen, R.; Hollmén, M.; Kainulainen, H.; Huovinen, P.; Jalkanen, S.; Pekkala, S. Enterobacter Cloacae Administration Induces Hepatic Damage and Subcutaneous Fat Accumulation in High-Fat Diet Fed Mice. PLoS ONE 2018, 13, e0198262. [Google Scholar] [CrossRef]
- Scheithauer, T.P.M.; Herrema, H.; Yu, H.; Bakker, G.J.; Winkelmeijer, M.; Soukhatcheva, G.; Dai, D.; Ma, C.; Havik, S.R.; Balvers, M.; et al. Gut-Derived Bacterial Flagellin Induces Beta-Cell Inflammation and Dysfunction. Gut Microbes 2022, 14, 2111951. [Google Scholar] [CrossRef]
- Fei, N.; Bruneau, A.; Zhang, X.; Wang, R.; Wang, J.; Rabot, S.; Gérard, P.; Zhao, L. Endotoxin Producers Overgrowing in Human Gut Microbiota as the Causative Agents for Nonalcoholic Fatty Liver Disease. MBio 2020, 11. [Google Scholar] [CrossRef]

| Nutrient | Bacterial Species | References |
|---|---|---|
| Ethanolamine | S. Typhimurium 1 C. rodentium 2 | [51] [52] |
| Lactate | S. Typhimurium | [53] |
| Glucarate/galactarate | S. Typhimurium Commensal E. coli | [54] |
| 1,2-propanediol | S. Typhimurium | [55] |
| Succinate | S. Typhimurium | [56] |
| L-Serine | Adherent-invasive E. coli C. rodentium | [57] [58] |
| Glycerol | Commensal E. coli | [59] |
| Carnitine and Acylcarnitines | E. coli | [60] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreira de Gouveia, M.I.; Bernalier-Donadille, A.; Jubelin, G. Enterobacteriaceae in the Human Gut: Dynamics and Ecological Roles in Health and Disease. Biology 2024, 13, 142. https://doi.org/10.3390/biology13030142
Moreira de Gouveia MI, Bernalier-Donadille A, Jubelin G. Enterobacteriaceae in the Human Gut: Dynamics and Ecological Roles in Health and Disease. Biology. 2024; 13(3):142. https://doi.org/10.3390/biology13030142
Chicago/Turabian StyleMoreira de Gouveia, Maria Ines, Annick Bernalier-Donadille, and Gregory Jubelin. 2024. "Enterobacteriaceae in the Human Gut: Dynamics and Ecological Roles in Health and Disease" Biology 13, no. 3: 142. https://doi.org/10.3390/biology13030142
APA StyleMoreira de Gouveia, M. I., Bernalier-Donadille, A., & Jubelin, G. (2024). Enterobacteriaceae in the Human Gut: Dynamics and Ecological Roles in Health and Disease. Biology, 13(3), 142. https://doi.org/10.3390/biology13030142







