Current Landscape of Cancer Immunotherapy: Harnessing the Immune Arsenal to Overcome Immune Evasion
Abstract
Simple Summary
Abstract
1. Introduction
2. Mechanisms of Immune Suppression
2.1. Immune Checkpoints and Cancer Immunotherapies
2.1.1. PD-1 and PD-L1
2.1.2. Cytotoxic T-Lymphocyte Antigen-4 (CTLA-4)
2.1.3. Lymphocyte-Activation Gene 3 (LAG-3)
2.1.4. T-Cell Immunoglobulin and Mucin Domain-Containing Protein 3 (TIM-3)
2.1.5. T-Cell Immunoreceptor with Immunoglobulin and ITIM Domain (TIGIT)
2.1.6. CD39/CD73/A2AR Pathway
2.1.7. V-Domain Immunoglobulin Suppressor of T-Cell Activation (VISTA)
2.1.8. B7-H3/B7-H4
2.1.9. SIRPA/CD47
2.1.10. NKG2A/HLA-E/CD94
2.1.11. LILRB1/LILRB2: HLA-G
2.1.12. Sialic Acid-Binding Immunoglobulin-Type Lectins (Siglecs)
2.2. Metabolic Reprogramming to Foster an Immunosuppressive Environment
2.3. Recruitment of Immunosuppressive Cells into the Environment
2.4. Antigen Presentation
3. Cancer Immunotherapeutic Strategies
3.1. Antibody-Based Therapy
3.1.1. Immune Checkpoint Blockade (ICB)
3.1.2. Antibody–Drug Conjugates (ADCs)
3.1.3. Bispecific Antibodies (bsAb)
3.1.4. Other Monoclonal Antibody (mAb)-Based Therapies
3.2. Adoptive Cell Transfer (ACT) Therapy
3.2.1. Tumor-Infiltrating Lymphocyte (TIL) Therapy
3.2.2. Chimeric Antigen Receptor (CAR)-Based Therapy
CAR-T
CAR-NK
3.2.3. T-Cell Receptor-Engineered T Cells (TCR-Ts)
3.3. Cancer Vaccines
3.3.1. Cell-Based Cancer Vaccines
3.3.2. Peptide-Based Cancer Vaccines
3.3.3. Viral- and Bacterial-Vector-Driven Cancer Vaccines
3.3.4. Nucleic Acid-Based Cancer Vaccines
3.4. Cytokine-Based Therapies
3.5. Oncolytic Viruses (OVs)
3.6. Combined Approaches to Treat Hot, Altered, and Cold Immune Landscapes
4. Overcoming Cancer Immunotherapeutic Resistance
4.1. Alterations in the Tumor Microenvironment (TME)
4.2. Spatial Immune Cell Heterogeneity
4.3. Alterations in Antigen Presentation
4.4. Altered Signaling Pathways
5. Discussion and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hinshaw, D.C.; Shevde, L.A. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.M.; Simon, M.C. The tumor microenvironment. Curr. Biol. 2020, 30, R921–R925. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Yu, D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol. Ther. 2021, 221, 107753. [Google Scholar] [CrossRef] [PubMed]
- Gajewski, T.F.; Schreiber, H.; Fu, Y.X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013, 14, 1014–1022. [Google Scholar] [CrossRef] [PubMed]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, H.; Hagerling, C.; Werb, Z. Roles of the immune system in cancer: From tumor initiation to metastatic progression. Genes Dev. 2018, 32, 1267–1284. [Google Scholar] [CrossRef] [PubMed]
- Bagchi, S.; Yuan, R.; Engleman, E.G. Immune checkpoint inhibitors for the treatment of cancer: Clinical impact and mechanisms of response and resistance. Annu. Rev. Pathol. 2021, 16, 223–249. [Google Scholar] [CrossRef] [PubMed]
- Hamid, O.; Robert, C.; Daud, A.; Hodi, F.S.; Hwu, W.J.; Kefford, R.; Wolchok, J.D.; Hersey, P.; Joseph, R.W.; Weber, J.S.; et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N. Engl. J. Med. 2013, 369, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef]
- Han, Y.; Liu, D.; Li, L. PD-1/PD-L1 pathway: Current researches in cancer. Am. J. Cancer Res. 2020, 10, 727–742. [Google Scholar]
- Rowshanravan, B.; Halliday, N.; Sansom, D.M. CTLA-4: A moving target in immunotherapy. Blood 2018, 131, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Cristescu, R.; Mogg, R.; Ayers, M.; Albright, A.; Murphy, E.; Yearley, J.; Sher, X.; Liu, X.Q.; Lu, H.; Nebozhyn, M.; et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018, 362, eaar3593. [Google Scholar] [CrossRef]
- Dutta, S.; Ganguly, A.; Chatterjee, K.; Spada, S.; Mukherjee, S. Targets of immune escape mechanisms in cancer: Basis for development and evolution of cancer immune checkpoint inhibitors. Biology 2023, 12, 218. [Google Scholar] [CrossRef]
- He, Y.; Yu, H.; Rozeboom, L.; Rivard, C.J.; Ellison, K.; Dziadziuszko, R.; Suda, K.; Ren, S.; Wu, C.; Hou, L.; et al. LAG-3 Protein Expression in Non-Small Cell Lung Cancer and Its Relationship with PD-1/PD-L1 and Tumor-Infiltrating Lymphocytes. J. Thorac. Oncol. 2017, 12, 814–823. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.Z.; Kim, H.J.; Wu, H.; Jalali, S.; Tang, X.; Krull, J.E.; Ding, W.; Novak, A.J.; Ansell, S.M. TIGIT Expression Is Associated with T-cell Suppression and Exhaustion and Predicts Clinical Outcome and Anti-PD-1 Response in Follicular Lymphoma. Clin. Cancer Res. 2020, 26, 5217–5231. [Google Scholar] [CrossRef]
- Vaddepally, R.K.; Kharel, P.; Pandey, R.; Garje, R.; Chandra, A.B. Review of indications of FDA-approved immune checkpoint inhibitors per NCCN guidelines with the level of evidence. Cancers 2020, 12, 738. [Google Scholar] [CrossRef] [PubMed]
- Guillerey, C.; Harjunpaa, H.; Carrie, N.; Kassem, S.; Teo, T.; Miles, K.; Krumeich, S.; Weulersse, M.; Cuisinier, M.; Stannard, K.; et al. TIGIT immune checkpoint blockade restores CD8+ T-cell immunity against multiple myeloma. Blood 2018, 132, 1689–1694. [Google Scholar] [CrossRef]
- Xu, W.; Hieu, T.; Malarkannan, S.; Wang, L. The structure, expression, and multifaceted role of immune-checkpoint protein VISTA as a critical regulator of anti-tumor immunity, autoimmunity, and inflammation. Cell Mol. Immunol. 2018, 15, 438–446. [Google Scholar] [CrossRef]
- Rafiq, S.; Hackett, C.S.; Brentjens, R.J. Engineering strategies to overcome the current roadblocks in CAR T cell therapy. Nat. Rev. Clin. Oncol. 2020, 17, 147–167. [Google Scholar] [CrossRef]
- Shimasaki, N.; Jain, A.; Campana, D. NK cells for cancer immunotherapy. Nat. Rev. Drug Discov. 2020, 19, 200–218. [Google Scholar] [CrossRef]
- Depil, S.; Duchateau, P.; Grupp, S.A.; Mufti, G.; Poirot, L. ‘Off-the-shelf’ allogeneic CAR T cells: Development and challenges. Nat. Rev. Drug Discov. 2020, 19, 185–199. [Google Scholar] [CrossRef] [PubMed]
- Finck, A.V.; Blanchard, T.; Roselle, C.P.; Golinelli, G.; June, C.H. Engineered cellular immunotherapies in cancer and beyond. Nat. Med. 2022, 28, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Davila, M.L.; Brentjens, R.J. CAR T cell therapy: Looking back and looking forward. Nat. Cancer 2022, 3, 1418–1419. [Google Scholar] [CrossRef] [PubMed]
- Labanieh, L.; Majzner, R.G.; Mackall, C.L. Programming CAR-T cells to kill cancer. Nat. Biomed. Eng. 2018, 2, 377–391. [Google Scholar] [CrossRef] [PubMed]
- Majzner, R.G.; Mackall, C.L. Tumor Antigen Escape from CAR T-cell Therapy. Cancer Discov. 2018, 8, 1219–1226. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Dotti, G.; Savoldo, B. Utilizing cell-based therapeutics to overcome immune evasion in hematologic malignancies. Blood 2016, 127, 3350–3359. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.C.; Balko, J.M. Mechanisms of MHC-I Downregulation and Role in Immunotherapy Response. Front. Immunol. 2022, 13, 844866. [Google Scholar] [CrossRef]
- Kallingal, A.; Olszewski, M.; Maciejewska, N.; Brankiewicz, W.; Baginski, M. Cancer immune escape: The role of antigen presentation machinery. J. Cancer Res. Clin. Oncol. 2023, 149, 8131–8141. [Google Scholar] [CrossRef] [PubMed]
- Piper, M.; Kluger, H.; Ruppin, E.; Hu-Lieskovan, S. Immune Resistance Mechanisms and the Road to Personalized Immunotherapy. Am. Soc. Clin. Oncol. Educ. Book 2023, 43, e390290. [Google Scholar] [CrossRef]
- Kim, H.J.; Cantor, H. CD4 T-cell subsets and tumor immunity: The helpful and the not-so-helpful. Cancer Immunol. Res. 2014, 2, 91–98. [Google Scholar] [CrossRef]
- Maoxi, L.; Haiyi, L. Impact on CD4+ CD25High-CD127low regulatory T (Treg) cells of neoadjuvant therapy for rectal cancer patients. Indian J. Pathol. Microbiol. 2024, 67, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Gubin, M.M.; Zhang, X.; Schuster, H.; Caron, E.; Ward, J.P.; Noguchi, T.; Ivanova, Y.; Hundal, J.; Arthur, C.D.; Krebber, W.J.; et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 2014, 515, 577–581. [Google Scholar] [CrossRef]
- Chen, W.; Yuan, Y.; Jiang, X. Antibody and antibody fragments for cancer immunotherapy. J. Control Release 2020, 328, 395–406. [Google Scholar] [CrossRef]
- Zeller, T.; Lutz, S.; Munnich, I.A.; Windisch, R.; Hilger, P.; Herold, T.; Tahiri, N.; Banck, J.C.; Weigert, O.; Moosmann, A.; et al. Dual checkpoint blockade of CD47 and LILRB1 enhances CD20 antibody-dependent phagocytosis of lymphoma cells by macrophages. Front. Immunol. 2022, 13, 929339. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, T.N.; Schreiber, R.D. Neoantigens in cancer immunotherapy. Science 2015, 348, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.; Shen, G.; Gao, W.; Huang, Z.; Huang, C.; Fu, L. Neoantigens: Promising targets for cancer therapy. Signal Transduct. Target. Ther. 2023, 8, 9. [Google Scholar] [CrossRef]
- Li, J.; Xiao, Z.; Wang, D.; Jia, L.; Nie, S.; Zeng, X.; Hu, W. The screening, identification, design and clinical application of tumor-specific neoantigens for TCR-T cells. Mol. Cancer 2023, 22, 141. [Google Scholar] [CrossRef]
- Zhang, Q.; Jia, Q.; Zhang, J.; Zhu, B. Neoantigens in precision cancer immunotherapy: From identification to clinical applications. Chin. Med. J. Engl. 2022, 135, 1285–1298. [Google Scholar] [CrossRef]
- Okada, M.; Shimizu, K.; Fujii, S.I. Identification of Neoantigens in Cancer Cells as Targets for Immunotherapy. Int. J. Mol. Sci. 2022, 23, 2594. [Google Scholar] [CrossRef]
- Lang, F.; Schrors, B.; Lower, M.; Tureci, O.; Sahin, U. Identification of neoantigens for individualized therapeutic cancer vaccines. Nat. Rev. Drug Discov. 2022, 21, 261–282. [Google Scholar] [CrossRef]
- Badran, Y.R.; Leet, D.E.; Dougan, M. Immune-related adverse events: What we’ve learned and where we’re heading. Expert. Rev. Anticancer Ther. 2020, 20, 727–730. [Google Scholar] [CrossRef] [PubMed]
- Dunn, G.P.; Bruce, A.T.; Ikeda, H.; Old, L.J.; Schreiber, R.D. Cancer immunoediting: From immunosurveillance to tumor escape. Nat. Immunol. 2002, 3, 991–998. [Google Scholar] [CrossRef]
- Ribatti, D. The concept of immune surveillance against tumors. The first theories. Oncotarget 2017, 8, 7175–7180. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Bruni, D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019, 18, 197–218. [Google Scholar] [CrossRef] [PubMed]
- Angell, H.K.; Bruni, D.; Barrett, J.C.; Herbst, R.; Galon, J. The immunoscore: Colon cancer and beyond. Clin. Cancer Res. 2020, 26, 332–339. [Google Scholar] [CrossRef]
- Chew, V.; Chen, J.; Lee, D.; Loh, E.; Lee, J.; Lim, K.H.; Weber, A.; Slankamenac, K.; Poon, R.T.; Yang, H.; et al. Chemokine-driven lymphocyte infiltration: An early intratumoural event determining long-term survival in resectable hepatocellular carcinoma. Gut 2012, 61, 427–438. [Google Scholar] [CrossRef]
- Pai, S.I.; Cesano, A.; Marincola, F.M. The paradox of cancer immune exclusion: Immune oncology next frontier. Cancer Treat. Res. 2020, 180, 173–195. [Google Scholar] [CrossRef]
- Kather, J.N.; Suarez-Carmona, M.; Charoentong, P.; Weis, C.A.; Hirsch, D.; Bankhead, P.; Horning, M.; Ferber, D.; Kel, I.; Herpel, E.; et al. Topography of cancer-associated immune cells in human solid tumors. eLife 2018, 7, e36967. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.T.; Sun, Z.J. Turning cold tumors into hot tumors by improving T-cell infiltration. Theranostics 2021, 11, 5365–5386. [Google Scholar] [CrossRef]
- Mellman, I.; Chen, D.S.; Powles, T.; Turley, S.J. The cancer-immunity cycle: Indication, genotype, and immunotype. Immunity 2023, 56, 2188–2205. [Google Scholar] [CrossRef]
- Giles, J.R.; Globig, A.M.; Kaech, S.M.; Wherry, E.J. CD8(+) T cells in the cancer-immunity cycle. Immunity 2023, 56, 2231–2253. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, A.H.; Pauken, K.E. The diverse functions of the PD1 inhibitory pathway. Nat. Rev. Immunol. 2018, 18, 153–167. [Google Scholar] [CrossRef]
- Patsoukis, N.; Wang, Q.; Strauss, L.; Boussiotis, V.A. Revisiting the PD-1 pathway. Sci. Adv. 2020, 6, eabd2712. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Diaz, A.; Shin, D.S.; Moreno, B.H.; Saco, J.; Escuin-Ordinas, H.; Rodriguez, G.A.; Zaretsky, J.M.; Sun, L.; Hugo, W.; Wang, X.; et al. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 2017, 19, 1189–1201. [Google Scholar] [CrossRef]
- Chen, L.; Han, X. Anti-PD-1/PD-L1 therapy of human cancer: Past, present, and future. J. Clin. Investig. 2015, 125, 3384–3391. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Dai, Z.; Wu, W.; Wang, Z.; Zhang, N.; Zhang, L.; Zeng, W.J.; Liu, Z.; Cheng, Q. Regulatory mechanisms of immune checkpoints PD-L1 and CTLA-4 in cancer. J. Exp. Clin. Cancer Res. 2021, 40, 184. [Google Scholar] [CrossRef] [PubMed]
- Fife, B.T.; Bluestone, J.A. Control of peripheral T-cell tolerance and autoimmunity via the CTLA-4 and PD-1 pathways. Immunol. Rev. 2008, 224, 166–182. [Google Scholar] [CrossRef] [PubMed]
- Seidel, J.A.; Otsuka, A.; Kabashima, K. Anti-PD-1 and Anti-CTLA-4 therapies in cancer: Mechanisms of action, efficacy, and limitations. Front. Oncol. 2018, 8, 86. [Google Scholar] [CrossRef]
- Qureshi, O.S.; Zheng, Y.; Nakamura, K.; Attridge, K.; Manzotti, C.; Schmidt, E.M.; Baker, J.; Jeffery, L.E.; Kaur, S.; Briggs, Z.; et al. Trans-endocytosis of CD80 and CD86: A molecular basis for the cell-extrinsic function of CTLA-4. Science 2011, 332, 600–603. [Google Scholar] [CrossRef]
- Andrews, L.P.; Cillo, A.R.; Karapetyan, L.; Kirkwood, J.M.; Workman, C.J.; Vignali, D.A.A. Molecular pathways and mechanisms of LAG3 in cancer therapy. Clin. Cancer Res. 2022, 28, 5030–5039. [Google Scholar] [CrossRef]
- Maruhashi, T.; Okazaki, I.M.; Sugiura, D.; Takahashi, S.; Maeda, T.K.; Shimizu, K.; Okazaki, T. LAG-3 inhibits the activation of CD4(+) T cells that recognize stable pMHCII through its conformation-dependent recognition of pMHCII. Nat. Immunol. 2018, 19, 1415–1426. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sanmamed, M.F.; Datar, I.; Su, T.T.; Ji, L.; Sun, J.; Chen, L.; Chen, Y.; Zhu, G.; Yin, W.; et al. Fibrinogen-like Protein 1 is a major immune inhibitory ligand of LAG-3. Cell 2019, 176, 334–347. [Google Scholar] [CrossRef] [PubMed]
- Kouo, T.; Huang, L.; Pucsek, A.B.; Cao, M.; Solt, S.; Armstrong, T.; Jaffee, E. Galectin-3 shapes antitumor immune responses by suppressing CD8+ T Cells via LAG-3 and inhibiting expansion of plasmacytoid dendritic cells. Cancer Immunol. Res. 2015, 3, 412–423. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Liu, J.; Liu, D.; Liu, B.; Wang, M.; Hu, Z.; Du, X.; Tang, L.; He, F. LSECtin expressed on melanoma cells promotes tumor progression by inhibiting antitumor T-cell responses. Cancer Res. 2014, 74, 3418–3428. [Google Scholar] [CrossRef] [PubMed]
- Workman, C.J.; Cauley, L.S.; Kim, I.J.; Blackman, M.A.; Woodland, D.L.; Vignali, D.A. Lymphocyte activation gene-3 (CD223) regulates the size of the expanding T cell population following antigen activation in vivo. J. Immunol. 2004, 172, 5450–5455. [Google Scholar] [CrossRef] [PubMed]
- Macon-Lemaitre, L.; Triebel, F. The negative regulatory function of the lymphocyte-activation gene-3 co-receptor (CD223) on human T cells. Immunology 2005, 115, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.Z.; Kim, H.J.; Villasboas, J.C.; Chen, Y.P.; Price-Troska, T.; Jalali, S.; Wilson, M.; Novak, A.J.; Ansell, S.M. Expression of LAG-3 defines exhaustion of intratumoral PD-1+ T cells and correlates with poor outcome in follicular lymphoma. Oncotarget 2017, 8, 61425–61439. [Google Scholar] [CrossRef]
- Takaya, S.; Saito, H.; Ikeguchi, M. Upregulation of Immune Checkpoint Molecules, PD-1 and LAG-3, on CD4+ and CD8+ T Cells after Gastric Cancer Surgery. Yonago Acta Med. 2015, 58, 39–44. [Google Scholar]
- Long, L.; Zhang, X.; Chen, F.; Pan, Q.; Phiphatwatchara, P.; Zeng, Y.; Chen, H. The promising immune checkpoint LAG-3: From tumor microenvironment to cancer immunotherapy. Genes. Cancer 2018, 9, 176–189. [Google Scholar] [CrossRef]
- Xu, B.; Yuan, L.; Gao, Q.; Yuan, P.; Zhao, P.; Yuan, H.; Fan, H.; Li, T.; Qin, P.; Han, L.; et al. Circulating and tumor-infiltrating Tim-3 in patients with colorectal cancer. Oncotarget 2015, 6, 20592–20603. [Google Scholar] [CrossRef]
- Anderson, A.C.; Joller, N.; Kuchroo, V.K. Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity 2016, 44, 989–1004. [Google Scholar] [CrossRef]
- Kandel, S.; Adhikary, P.; Li, G.; Cheng, K. The TIM3/Gal9 signaling pathway: An emerging target for cancer immunotherapy. Cancer Lett. 2021, 510, 67–78. [Google Scholar] [CrossRef]
- Wolf, Y.; Anderson, A.C.; Kuchroo, V.K. TIM3 comes of age as an inhibitory receptor. Nat. Rev. Immunol. 2020, 20, 173–185. [Google Scholar] [CrossRef]
- Fourcade, J.; Sun, Z.; Benallaoua, M.; Guillaume, P.; Luescher, I.F.; Sander, C.; Kirkwood, J.M.; Kuchroo, V.; Zarour, H.M. Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen-specific CD8+ T cell dysfunction in melanoma patients. J. Exp. Med. 2010, 207, 2175–2186. [Google Scholar] [CrossRef] [PubMed]
- Thommen, D.S.; Koelzer, V.H.; Herzig, P.; Roller, A.; Trefny, M.; Dimeloe, S.; Kiialainen, A.; Hanhart, J.; Schill, C.; Hess, C.; et al. A transcriptionally and functionally distinct PD-1+ CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat. Med. 2018, 24, 994–1004. [Google Scholar] [CrossRef]
- Roussel, M.; Le, K.S.; Granier, C.; Llamas Gutierrez, F.; Foucher, E.; Le Gallou, S.; Pangault, C.; Xerri, L.; Launay, V.; Lamy, T.; et al. Functional characterization of PD1+TIM3+ tumor-infiltrating T cells in DLBCL and effects of PD1 or TIM3 blockade. Blood Adv. 2021, 5, 1816–1829. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cai, P.; Liang, T.; Wang, L.; Hu, L. TIM-3 is a potential prognostic marker for patients with solid tumors: A systematic review and meta-analysis. Oncotarget 2017, 8, 31705–31713. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhang, Y.; Zhang, J.P.; Liang, J.; Li, L.; Zheng, L. Tim-3 expression defines regulatory T cells in human tumors. PLoS ONE 2013, 8, e58006. [Google Scholar] [CrossRef]
- da Silva, I.P.; Gallois, A.; Jimenez-Baranda, S.; Khan, S.; Anderson, A.C.; Kuchroo, V.K.; Osman, I.; Bhardwaj, N. Reversal of NK-cell exhaustion in advanced melanoma by Tim-3 blockade. Cancer Immunol. Res. 2014, 2, 410–422. [Google Scholar] [CrossRef]
- Koyama, S.; Akbay, E.A.; Li, Y.Y.; Herter-Sprie, G.S.; Buczkowski, K.A.; Richards, W.G.; Gandhi, L.; Redig, A.J.; Rodig, S.J.; Asahina, H.; et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat. Commun. 2016, 7, 10501. [Google Scholar] [CrossRef]
- Johnston, R.J.; Comps-Agrar, L.; Hackney, J.; Yu, X.; Huseni, M.; Yang, Y.; Park, S.; Javinal, V.; Chiu, H.; Irving, B.; et al. The immunoreceptor TIGIT regulates antitumor and antiviral CD8+ T cell effector function. Cancer Cell 2014, 26, 923–937. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, A.; Campbell, D.; Hannum, C.; Yssel, H.; Franz-Bacon, K.; McClanahan, T.; Kitamura, T.; Nicholl, J.; Sutherland, G.R.; Lanier, L.L.; et al. DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity 1996, 4, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; de Almeida, P.; Manieri, N.; de Almeida Nagata, D.; Wu, T.D.; Harden Bowles, K.; Arumugam, V.; Schartner, J.; Cubas, R.; Mittman, S.; et al. CD226 regulates natural killer cell antitumor responses via phosphorylation-mediated inactivation of transcription factor FOXO1. Proc. Natl. Acad. Sci. USA 2018, 115, E11731–E11740. [Google Scholar] [CrossRef] [PubMed]
- Joller, N.; Hafler, J.P.; Brynedal, B.; Kassam, N.; Spoerl, S.; Levin, S.D.; Sharpe, A.H.; Kuchroo, V.K. Cutting edge: TIGIT has T cell-intrinsic inhibitory functions. J. Immunol. 2011, 186, 1338–1342. [Google Scholar] [CrossRef] [PubMed]
- Worboys, J.D.; Vowell, K.N.; Hare, R.K.; Ambrose, A.R.; Bertuzzi, M.; Conner, M.A.; Patel, F.P.; Zammit, W.H.; Gali-Moya, J.; Hazime, K.S.; et al. TIGIT can inhibit T cell activation via ligation-induced nanoclusters, independent of CD226 co-stimulation. Nat. Commun. 2023, 14, 5016. [Google Scholar] [CrossRef] [PubMed]
- Ostroumov, D.; Duong, S.; Wingerath, J.; Woller, N.; Manns, M.P.; Timrott, K.; Kleine, M.; Ramackers, W.; Roessler, S.; Nahnsen, S.; et al. Transcriptome profiling identifies TIGIT as a marker of T-Cell exhaustion in liver cancer. Hepatology 2021, 73, 1399–1418. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, A.; Liu, X.; Han, S.; Sun, Y.; Zhang, J.; Guo, L.; Zhang, Y. Blocking TIGIT/CD155 signalling reverses CD8+ T cell exhaustion and enhances the antitumor activity in cervical cancer. J. Transl. Med. 2022, 20, 280. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; Tian, W.; Wang, Z.; Zhang, J.; Zhou, R. Co-inhibition of TIGIT and PD-1/PD-L1 in cancer immunotherapy: Mechanisms and clinical trials. Mol. Cancer 2023, 22, 93. [Google Scholar] [CrossRef] [PubMed]
- Vesely, M.D.; Zhang, T.; Chen, L. Resistance mechanisms to Anti-PD cancer immunotherapy. Annu. Rev. Immunol. 2022, 40, 45–74. [Google Scholar] [CrossRef] [PubMed]
- Di Virgilio, F.; Sarti, A.C.; Falzoni, S.; De Marchi, E.; Adinolfi, E. Extracellular ATP and P2 purinergic signalling in the tumour microenvironment. Nat. Rev. Cancer 2018, 18, 601–618. [Google Scholar] [CrossRef]
- Allard, B.; Longhi, M.S.; Robson, S.C.; Stagg, J. The ectonucleotidases CD39 and CD73: Novel checkpoint inhibitor targets. Immunol. Rev. 2017, 276, 121–144. [Google Scholar] [CrossRef] [PubMed]
- Horenstein, A.L.; Quarona, V.; Toscani, D.; Costa, F.; Chillemi, A.; Pistoia, V.; Giuliani, N.; Malavasi, F. Adenosine generated in the bone marrow niche through a CD38-Mediated pathway correlates with progression of human myeloma. Mol. Med. 2016, 22, 694–704. [Google Scholar] [CrossRef]
- Thompson, E.A.; Powell, J.D. Inhibition of the adenosine pathway to potentiate cancer immunotherapy: Potential for combinatorial approaches. Annu. Rev. Med. 2021, 72, 331–348. [Google Scholar] [CrossRef]
- Leone, R.D.; Emens, L.A. Targeting adenosine for cancer immunotherapy. J. Immunother. Cancer 2018, 6, 57. [Google Scholar] [CrossRef]
- Ma, X.L.; Shen, M.N.; Hu, B.; Wang, B.L.; Yang, W.J.; Lv, L.H.; Wang, H.; Zhou, Y.; Jin, A.L.; Sun, Y.F.; et al. CD73 promotes hepatocellular carcinoma progression and metastasis via activating PI3K/AKT signaling by inducing Rap1-mediated membrane localization of P110beta and predicts poor prognosis. J. Hematol. Oncol. 2019, 12, 37. [Google Scholar] [CrossRef]
- Shi, L.; Feng, M.; Du, S.; Wei, X.; Song, H.; Yixin, X.; Song, J.; Wenxian, G. Adenosine generated by regulatory T Cells induces CD8+ T Cell exhaustion in gastric cancer through A2aR pathway. Biomed. Res. Int. 2019, 2019, 4093214. [Google Scholar] [CrossRef]
- Vigano, S.; Alatzoglou, D.; Irving, M.; Menetrier-Caux, C.; Caux, C.; Romero, P.; Coukos, G. Targeting Adenosine in Cancer Immunotherapy to Enhance T-Cell Function. Front. Immunol. 2019, 10, 925. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Tang, N.; Cheng, C.; Hu, T.; Wei, X.; Han, W.; Wang, H. Improving the anti-solid tumor efficacy of CAR-T cells by inhibiting adenosine signaling pathway. Oncoimmunology 2020, 9, 1824643. [Google Scholar] [CrossRef] [PubMed]
- Seifert, M.; Benmebarek, M.R.; Briukhovetska, D.; Markl, F.; Dorr, J.; Cadilha, B.L.; Jobst, J.; Stock, S.; Andreu-Sanz, D.; Lorenzini, T.; et al. Impact of the selective A2(A)R and A2(B)R dual antagonist AB928/etrumadenant on CAR T cell function. Br. J. Cancer 2022, 127, 2175–2185. [Google Scholar] [CrossRef]
- Giuffrida, L.; Sek, K.; Henderson, M.A.; Lai, J.; Chen, A.X.Y.; Meyran, D.; Todd, K.L.; Petley, E.V.; Mardiana, S.; Molck, C.; et al. CRISPR/Cas9 mediated deletion of the adenosine A2A receptor enhances CAR T cell efficacy. Nat. Commun. 2021, 12, 3236. [Google Scholar] [CrossRef]
- Lupia, M.; Angiolini, F.; Bertalot, G.; Freddi, S.; Sachsenmeier, K.F.; Chisci, E.; Kutryb-Zajac, B.; Confalonieri, S.; Smolenski, R.T.; Giovannoni, R.; et al. CD73 regulates stemness and epithelial-mesenchymal transition in ovarian cancer-initiating cells. Stem Cell Rep. 2018, 10, 1412–1425. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.H.; Lin, C.Z.; Cao, W.; Yang, R.; Lu, W.; Liu, Z.Q.; Chen, Y.M.; Yang, X.; Tian, Z.; Wang, L.Z.; et al. CD73 is associated with poor prognosis in HNSCC. Oncotarget 2016, 7, 61690–61702. [Google Scholar] [CrossRef] [PubMed]
- Morello, S.; Capone, M.; Sorrentino, C.; Giannarelli, D.; Madonna, G.; Mallardo, D.; Grimaldi, A.M.; Pinto, A.; Ascierto, P.A. Soluble CD73 as biomarker in patients with metastatic melanoma patients treated with nivolumab. J. Transl. Med. 2017, 15, 244. [Google Scholar] [CrossRef] [PubMed]
- Roh, M.; Wainwright, D.A.; Wu, J.D.; Wan, Y.; Zhang, B. Targeting CD73 to augment cancer immunotherapy. Curr. Opin. Pharmacol. 2020, 53, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Moesta, A.K.; Li, X.Y.; Smyth, M.J. Targeting CD39 in cancer. Nat. Rev. Immunol. 2020, 20, 739–755. [Google Scholar] [CrossRef] [PubMed]
- Mascanfroni, I.D.; Takenaka, M.C.; Yeste, A.; Patel, B.; Wu, Y.; Kenison, J.E.; Siddiqui, S.; Basso, A.S.; Otterbein, L.E.; Pardoll, D.M.; et al. Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-alpha. Nat. Med. 2015, 21, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Ahlmanner, F.; Sundstrom, P.; Akeus, P.; Eklof, J.; Borjesson, L.; Gustavsson, B.; Lindskog, E.B.; Raghavan, S.; Quiding-Jarbrink, M. CD39+ regulatory T cells accumulate in colon adenocarcinomas and display markers of increased suppressive function. Oncotarget 2018, 9, 36993–37007. [Google Scholar] [CrossRef]
- Canale, F.P.; Ramello, M.C.; Nunez, N.; Araujo Furlan, C.L.; Bossio, S.N.; Gorosito Serran, M.; Tosello Boari, J.; Del Castillo, A.; Ledesma, M.; Sedlik, C.; et al. CD39 Expression defines cell exhaustion in tumor-infiltrating CD8+ T Cells. Cancer Res. 2018, 78, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; Thompson, B.; Strange, A.; Amato, C.M.; Vassallo, M.; Dolgalev, I.; Hester-McCullough, J.; Muramatsu, T.; Kimono, D.; Puranik, A.S.; et al. A population of tumor-infiltrating CD4+ T Cells co-expressing CD38 and CD39 is associated with checkpoint inhibitor resistance. Clin. Cancer Res. 2023, 29, 4242–4255. [Google Scholar] [CrossRef]
- Limagne, E.; Euvrard, R.; Thibaudin, M.; Rebe, C.; Derangere, V.; Chevriaux, A.; Boidot, R.; Vegran, F.; Bonnefoy, N.; Vincent, J.; et al. Accumulation of MDSC and Th17 cells in patients with metastatic colorectal cancer predicts the efficacy of a FOLFOX-bevacizumab drug treatment regimen. Cancer Res. 2016, 76, 5241–5252. [Google Scholar] [CrossRef]
- Li, J.; Wang, L.; Chen, X.; Li, L.; Li, Y.; Ping, Y.; Huang, L.; Yue, D.; Zhang, Z.; Wang, F.; et al. CD39/CD73 upregulation on myeloid-derived suppressor cells via TGF-beta-mTOR-HIF-1 signaling in patients with non-small cell lung cancer. Oncoimmunology 2017, 6, e1320011. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, L.; Li, J.; Fan, Z.; Yang, L.; Zhang, Z.; Zhang, C.; Yue, D.; Qin, G.; Zhang, T.; et al. Metformin-induced reduction of CD39 and CD73 blocks myeloid-derived suppressor cell activity in patients with ovarian cancer. Cancer Res. 2018, 78, 1779–1791. [Google Scholar] [CrossRef] [PubMed]
- Lines, J.L.; Pantazi, E.; Mak, J.; Sempere, L.F.; Wang, L.; O’Connell, S.; Ceeraz, S.; Suriawinata, A.A.; Yan, S.; Ernstoff, M.S.; et al. VISTA is an immune checkpoint molecule for human T cells. Cancer Res. 2014, 74, 1924–1932. [Google Scholar] [CrossRef] [PubMed]
- ElTanbouly, M.A.; Croteau, W.; Noelle, R.J.; Lines, J.L. VISTA: A novel immunotherapy target for normalizing innate and adaptive immunity. Semin. Immunol. 2019, 42, 101308. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhang, X.; Li, E.; Zhang, G.; Wang, X.; Tang, T.; Bai, X.; Liang, T. VISTA: An immune regulatory protein checking tumor and immune cells in cancer immunotherapy. J. Hematol. Oncol. 2020, 13, 83. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.S.; Molloy, M.; Ugolkov, A.; von Roemeling, R.W.; Noelle, R.J.; Lewis, L.D.; Johnson, M.; Radvanyi, L.; Martell, R.E. VISTA expression and patient selection for immune-based anticancer therapy. Front. Immunol. 2023, 14, 1086102. [Google Scholar] [CrossRef] [PubMed]
- Johnston, R.J.; Su, L.J.; Pinckney, J.; Critton, D.; Boyer, E.; Krishnakumar, A.; Corbett, M.; Rankin, A.L.; Dibella, R.; Campbell, L.; et al. VISTA is an acidic pH-selective ligand for PSGL-1. Nature 2019, 574, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Tinoco, R.; Carrette, F.; Barraza, M.L.; Otero, D.C.; Magana, J.; Bosenberg, M.W.; Swain, S.L.; Bradley, L.M. PSGL-1 Is an immune checkpoint regulator that promotes T Cell exhaustion. Immunity 2016, 44, 1190–1203. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wu, G.; Manick, B.; Hernandez, V.; Renelt, M.; Erickson, C.; Guan, J.; Singh, R.; Rollins, S.; Solorz, A.; et al. VSIG-3 as a ligand of VISTA inhibits human T-cell function. Immunology 2019, 156, 74–85. [Google Scholar] [CrossRef]
- Chen, W.; Qie, C.; Hu, X.; Wang, L.; Jiang, J.; Liu, W.; Liu, J. A small molecule inhibitor of VSIG-8 prevents its binding to VISTA. Investig. New Drugs 2022, 40, 690–699. [Google Scholar] [CrossRef]
- Hmeljak, J.; Sanchez-Vega, F.; Hoadley, K.A.; Shih, J.; Stewart, C.; Heiman, D.; Tarpey, P.; Danilova, L.; Drill, E.; Gibb, E.A.; et al. Integrative molecular characterization of malignant pleural mesothelioma. Cancer Discov. 2018, 8, 1548–1565. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Ward, J.F.; Pettaway, C.A.; Shi, L.Z.; Subudhi, S.K.; Vence, L.M.; Zhao, H.; Chen, J.; Chen, H.; Efstathiou, E.; et al. VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer. Nat. Med. 2017, 23, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Yuan, Q.; Xia, H.; Zhu, G.; Feng, Y.; Wang, Q.; Zhang, Z.; He, W.; Lu, J.; Dong, C.; et al. Analysis of VISTA expression and function in renal cell carcinoma highlights VISTA as a potential target for immunotherapy. Protein Cell 2019, 10, 840–845. [Google Scholar] [CrossRef] [PubMed]
- Yi, K.H.; Chen, L. Fine tuning the immune response through B7-H3 and B7-H4. Immunol. Rev. 2009, 229, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Kontos, F.; Michelakos, T.; Kurokawa, T.; Sadagopan, A.; Schwab, J.H.; Ferrone, C.R.; Ferrone, S. B7-H3: An attractive target for antibody-based immunotherapy. Clin. Cancer Res. 2021, 27, 1227–1235. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.B.; Stashwick, C.; Powell, D.J. B7-H4 as a potential target for immunotherapy for gynecologic cancers: A closer look. Gynecol. Oncol. 2014, 134, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Guo, X.; Xing, Z.; Tao, Y.; Liang, W.; Shi, Z.; Hu, W.; Zhou, S.; Wang, X. Multi-omics analyses of CD276 in pan-cancer reveals its clinical prognostic value in glioblastoma and other major cancer types. BMC Cancer 2023, 23, 102. [Google Scholar] [CrossRef] [PubMed]
- Si, S.; Wang, L.; Cao, H.; Xu, Y.; Zhan, Q. Co-deficiency of B7-H3 and B7-H4 identifies high CD8+ T cell infiltration and better prognosis in pancreatic cancer. BMC Cancer 2022, 22, 211. [Google Scholar] [CrossRef] [PubMed]
- Carvajal-Hausdorf, D.; Altan, M.; Velcheti, V.; Gettinger, S.N.; Herbst, R.S.; Rimm, D.L.; Schalper, K.A. Expression and clinical significance of PD-L1, B7-H3, B7-H4 and TILs in human small cell lung Cancer (SCLC). J. Immunother. Cancer 2019, 7, 65. [Google Scholar] [CrossRef]
- Brustmann, H.; Igaz, M.; Eder, C.; Brunner, A. Epithelial and tumor-associated endothelial expression of B7-H3 in cervical carcinoma: Relation with CD8+ intraepithelial lymphocytes, FIGO stage, and phosphohistone H3 (PHH3) reactivity. Int. J. Gynecol. Pathol. 2015, 34, 187–195. [Google Scholar] [CrossRef]
- Jin, Y.; Zhang, P.; Li, J.; Zhao, J.; Liu, C.; Yang, F.; Yang, D.; Gao, A.; Lin, W.; Ma, X.; et al. B7-H3 in combination with regulatory T cell is associated with tumor progression in primary human non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 13987–13995. [Google Scholar] [PubMed]
- Fauci, J.M.; Straughn, J.M., Jr.; Ferrone, S.; Buchsbaum, D.J. A review of B7-H3 and B7-H4 immune molecules and their role in ovarian cancer. Gynecol. Oncol. 2012, 127, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Wang, Y.; Shi, X.; Zong, L.; Liu, L.; Zhang, J.; Qian, Q.; Jin, J.; Ma, Y.; Cui, B.; et al. Negative roles of B7-H3 and B7-H4 in the microenvironment of cervical cancer. Exp. Cell Res. 2018, 371, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Chen, L.; Wang, F.; Zhu, D.; Ge, X.; Hua, D.; Sun, J. Cancer cell-expressed B7-H3 regulates the differentiation of tumor-associated macrophages in human colorectal carcinoma. Oncol. Lett. 2017, 14, 6177–6183. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, T.; Murakami, R.; Hamanishi, J.; Tanigaki, K.; Hosoe, Y.; Mise, N.; Takamatsu, S.; Mise, Y.; Ukita, M.; Taki, M.; et al. B7-H3 suppresses antitumor immunity via the CCL2-CCR2-M2 macrophage axis and contributes to ovarian cancer progression. Cancer Immunol. Res. 2022, 10, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.T.; Jin, W.L. B7-H3/CD276: An emerging cancer immunotherapy. Front. Immunol. 2021, 12, 701006. [Google Scholar] [CrossRef] [PubMed]
- Krambeck, A.E.; Thompson, R.H.; Dong, H.; Lohse, C.M.; Park, E.S.; Kuntz, S.M.; Leibovich, B.C.; Blute, M.L.; Cheville, J.C.; Kwon, E.D. B7-H4 expression in renal cell carcinoma and tumor vasculature: Associations with cancer progression and survival. Proc. Natl. Acad. Sci. USA 2006, 103, 10391–10396. [Google Scholar] [CrossRef] [PubMed]
- MacGregor, H.L.; Garcia-Batres, C.; Sayad, A.; Elia, A.; Berman, H.K.; Toker, A.; Katz, S.R.; Shaw, P.A.; Clarke, B.A.; Crome, S.Q.; et al. Tumor cell expression of B7-H4 correlates with higher frequencies of tumor-infiltrating APCs and higher CXCL17 expression in human epithelial ovarian cancer. Oncoimmunology 2019, 8, e1665460. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yang, C.; Liu, X.B.; Wang, L.; Kang, F.B. B7-H4 overexpression contributes to poor prognosis and drug-resistance in triple-negative breast cancer. Cancer Cell Int. 2018, 18, 100. [Google Scholar] [CrossRef]
- Kryczek, I.; Wei, S.; Zhu, G.; Myers, L.; Mottram, P.; Cheng, P.; Chen, L.; Coukos, G.; Zou, W. Relationship between B7-H4, regulatory T cells, and patient outcome in human ovarian carcinoma. Cancer Res. 2007, 67, 8900–8905. [Google Scholar] [CrossRef]
- Dangaj, D.; Lanitis, E.; Zhao, A.; Joshi, S.; Cheng, Y.; Sandaltzopoulos, R.; Ra, H.J.; Danet-Desnoyers, G.; Powell, D.J., Jr.; Scholler, N. Novel recombinant human b7-h4 antibodies overcome tumoral immune escape to potentiate T-cell antitumor responses. Cancer Res. 2013, 73, 4820–4829. [Google Scholar] [CrossRef] [PubMed]
- Sica, G.L.; Choi, I.H.; Zhu, G.; Tamada, K.; Wang, S.D.; Tamura, H.; Chapoval, A.I.; Flies, D.B.; Bajorath, J.; Chen, L. B7-H4, a molecule of the B7 family, negatively regulates T cell immunity. Immunity 2003, 18, 849–861. [Google Scholar] [CrossRef] [PubMed]
- Gray, E.; Ulrich, M.; Epp, A.; Younan, P.; Sahetya, D.; Hensley, K.; Allred, S.; Huang, L.Y.; Hahn, J.; Gahnberg, K.; et al. SGN-B7H4V, an investigational vedotin ADC directed to the immune checkpoint ligand B7-H4, shows promising activity in preclinical models. J. Immunother. Cancer 2023, 11, e007572. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Huang, Q.; Xiao, W.; Zhao, Y.; Pi, J.; Xu, H.; Zhao, H.; Xu, J.; Evans, C.E.; Jin, H. Advances in anti-tumor treatments targeting the CD47/SIRPalpha axis. Front. Immunol. 2020, 11, 18. [Google Scholar] [CrossRef] [PubMed]
- Qu, T.; Li, B.; Wang, Y. Targeting CD47/SIRPalpha as a therapeutic strategy, where we are and where we are headed. Biomark. Res. 2022, 10, 20. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Bang, S.; Jee, S.; Paik, S.S.; Jang, K. Clinicopathological significance of CD47 expression in hepatocellular carcinoma. J. Clin. Pathol. 2021, 74, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Liu, F.; Li, C.; Liang, X.; Li, C.; Liu, Y.; Yi, Z.; Zhang, L.; Fu, S.; Zeng, Y. Role of CD47 in tumor immunity: A potential target for combination therapy. Sci. Rep. 2022, 12, 9803. [Google Scholar] [CrossRef] [PubMed]
- Pai, S.; Bamodu, O.A.; Lin, Y.K.; Lin, C.S.; Chu, P.Y.; Chien, M.H.; Wang, L.S.; Hsiao, M.; Yeh, C.T.; Tsai, J.T. CD47-SIRPalpha signaling induces epithelial-mesenchymal transition and cancer stemness and links to a poor prognosis in patients with oral squamous cell carcinoma. Cells 2019, 8, 1658. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.H.; Pei, X.F.; Zhu, Z.Q.; Lin, Z.; Mao, Y.Y.; Xu, X.L.; Luo, Y.L.; Zhang, L.; Peng, P.J. CD47 overexpression is associated with epstein-barr virus infection and poor prognosis in patients with nasopharyngeal carcinoma. Onco Targets Ther. 2020, 13, 3325–3334. [Google Scholar] [CrossRef]
- Yuan, J.; Shi, X.; Chen, C.; He, H.; Liu, L.; Wu, J.; Yan, H. High expression of CD47 in triple negative breast cancer is associated with epithelial-mesenchymal transition and poor prognosis. Oncol. Lett. 2019, 18, 3249–3255. [Google Scholar] [CrossRef]
- Li, Y.; Lu, S.; Xu, Y.; Qiu, C.; Jin, C.; Wang, Y.; Liu, Z.; Kong, B. Overexpression of CD47 predicts poor prognosis and promotes cancer cell invasion in high-grade serous ovarian carcinoma. Am. J. Transl. Res. 2017, 9, 2901–2910. [Google Scholar] [PubMed]
- Barrera, L.; Montes-Servin, E.; Hernandez-Martinez, J.M.; Garcia-Vicente, M.L.A.; Montes-Servin, E.; Herrera-Martinez, M.; Crispin, J.C.; Borbolla-Escoboza, J.R.; Arrieta, O. CD47 overexpression is associated with decreased neutrophil apoptosis/phagocytosis and poor prognosis in non-small-cell lung cancer patients. Br. J. Cancer 2017, 117, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, L.; Ren, Z.; Yang, K.; Xu, H.; Luan, Y.; Fu, K.; Guo, J.; Peng, H.; Zhu, M.; et al. Dual targeting of innate and adaptive checkpoints on tumor cells limits immune evasion. Cell Rep. 2018, 24, 2101–2111. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.H.; Dominik, P.K.; Stanfield, J.; Ding, S.; Yang, W.; Kurd, N.; Llewellyn, R.; Heyen, J.; Wang, C.; Melton, Z.; et al. Dual checkpoint blockade of CD47 and PD-L1 using an affinity-tuned bispecific antibody maximizes antitumor immunity. J. Immunother. Cancer 2021, 9, e003464. [Google Scholar] [CrossRef] [PubMed]
- Schurch, C.M.; Roelli, M.A.; Forster, S.; Wasmer, M.H.; Bruhl, F.; Maire, R.S.; Di Pancrazio, S.; Ruepp, M.D.; Giger, R.; Perren, A.; et al. Targeting CD47 in anaplastic thyroid carcinoma enhances tumor phagocytosis by macrophages and is a promising therapeutic strategy. Thyroid 2019, 29, 979–992. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Ren, Z.; Tseng, K.F.; Liu, X.; Li, H.; Lu, C.; Cai, Y.; Minna, J.D.; Fu, Y.X. Dual targeting of CTLA-4 and CD47 on T(reg) cells promotes immunity against solid tumors. Sci. Transl. Med. 2021, 13, eabg8693. [Google Scholar] [CrossRef]
- Creelan, B.C.; Antonia, S.J. The NKG2A immune checkpoint—A new direction in cancer immunotherapy. Nat. Rev. Clin. Oncol. 2019, 16, 277–278. [Google Scholar] [CrossRef] [PubMed]
- Borst, L.; van der Burg, S.H.; van Hall, T. The NKG2A-HLA-E Axis as a novel checkpoint in the tumor microenvironment. Clin. Cancer Res. 2020, 26, 5549–5556. [Google Scholar] [CrossRef] [PubMed]
- Bertone, S.; Schiavetti, F.; Bellomo, R.; Vitale, C.; Ponte, M.; Moretta, L.; Mingari, M.C. Transforming growth factor-beta-induced expression of CD94/NKG2A inhibitory receptors in human T lymphocytes. Eur. J. Immunol. 1999, 29, 23–29. [Google Scholar] [CrossRef]
- Andre, P.; Denis, C.; Soulas, C.; Bourbon-Caillet, C.; Lopez, J.; Arnoux, T.; Blery, M.; Bonnafous, C.; Gauthier, L.; Morel, A.; et al. Anti-NKG2A mAb Is a Checkpoint Inhibitor that Promotes Anti-tumor Immunity by Unleashing Both T and NK Cells. Cell 2018, 175, 1731–1743. [Google Scholar] [CrossRef]
- Pereira, B.I.; Devine, O.P.; Vukmanovic-Stejic, M.; Chambers, E.S.; Subramanian, P.; Patel, N.; Virasami, A.; Sebire, N.J.; Kinsler, V.; Valdovinos, A.; et al. Senescent cells evade immune clearance via HLA-E-mediated NK and CD8+ T cell inhibition. Nat. Commun. 2019, 10, 2387. [Google Scholar] [CrossRef] [PubMed]
- Kaulfuss, M.; Mietz, J.; Fabri, A.; Vom Berg, J.; Munz, C.; Chijioke, O. The NK cell checkpoint NKG2A maintains expansion capacity of human NK cells. Sci. Rep. 2023, 13, 10555. [Google Scholar] [CrossRef] [PubMed]
- Bossard, C.; Bezieau, S.; Matysiak-Budnik, T.; Volteau, C.; Laboisse, C.L.; Jotereau, F.; Mosnier, J.F. HLA-E/beta2 microglobulin overexpression in colorectal cancer is associated with recruitment of inhibitory immune cells and tumor progression. Int. J. Cancer 2012, 131, 855–863. [Google Scholar] [CrossRef] [PubMed]
- Sheu, B.C.; Chiou, S.H.; Lin, H.H.; Chow, S.N.; Huang, S.C.; Ho, H.N.; Hsu, S.M. Up-regulation of inhibitory natural killer receptors CD94/NKG2A with suppressed intracellular perforin expression of tumor-infiltrating CD8+ T lymphocytes in human cervical carcinoma. Cancer Res. 2005, 65, 2921–2929. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, T.; Seow, S.V.; Wong, D.; Robinson, M.; Campana, D. Blocking expression of inhibitory receptor NKG2A overcomes tumor resistance to NK cells. J. Clin. Investig. 2019, 129, 2094–2106. [Google Scholar] [CrossRef] [PubMed]
- van Montfoort, N.; Borst, L.; Korrer, M.J.; Sluijter, M.; Marijt, K.A.; Santegoets, S.J.; van Ham, V.J.; Ehsan, I.; Charoentong, P.; Andre, P.; et al. NKG2A blockade potentiates CD8 T cell immunity induced by cancer vaccines. Cell 2018, 175, 1744–1755. [Google Scholar] [CrossRef] [PubMed]
- Levy, E.M.; Bianchini, M.; Von Euw, E.M.; Barrio, M.M.; Bravo, A.I.; Furman, D.; Domenichini, E.; Macagno, C.; Pinsky, V.; Zucchini, C.; et al. Human leukocyte antigen-E protein is overexpressed in primary human colorectal cancer. Int. J. Oncol. 2008, 32, 633–641. [Google Scholar] [CrossRef]
- Eugene, J.; Jouand, N.; Ducoin, K.; Dansette, D.; Oger, R.; Deleine, C.; Leveque, E.; Meurette, G.; Podevin, J.; Matysiak, T.; et al. The inhibitory receptor CD94/NKG2A on CD8+ tumor-infiltrating lymphocytes in colorectal cancer: A promising new druggable immune checkpoint in the context of HLAE/beta2m overexpression. Mod. Pathol. 2020, 33, 468–482. [Google Scholar] [CrossRef]
- Carosella, E.D.; Gregori, S.; Tronik-Le Roux, D. HLA-G/LILRBs: A cancer immunotherapy challenge. Trends Cancer 2021, 7, 389–392. [Google Scholar] [CrossRef]
- Held, W.; Mariuzza, R.A. Cis interactions of immunoreceptors with MHC and non-MHC ligands. Nat. Rev. Immunol. 2008, 8, 269–278. [Google Scholar] [CrossRef]
- Carosella, E.D.; Rouas-Freiss, N.; Tronik-Le Roux, D.; Moreau, P.; LeMaoult, J. HLA-G: An immune checkpoint molecule. Adv. Immunol. 2015, 127, 33–144. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, J.; Wang, S.; Wang, J.; Chen, X.; Zhou, D.; Fang, Y.; Gao, A.; Sun, Y. Overexpressed immunoglobulin-like transcript (ILT) 4 in lung adenocarcinoma is correlated with immunosuppressive T cell subset infiltration and poor patient outcomes. Biomark. Res. 2020, 8, 11. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chen, Y.; Deng, M.; John, S.; Gui, X.; Kansagra, A.; Chen, W.; Kim, J.; Lewis, C.; Wu, G.; et al. Antagonistic anti-LILRB1 monoclonal antibody regulates antitumor functions of natural killer cells. J. Immunother. Cancer 2020, 8, e000515. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.M.; van der Touw, W.; Wang, Y.S.; Kang, K.; Mai, S.; Zhang, J.; Alsina-Beauchamp, D.; Duty, J.A.; Mungamuri, S.K.; Zhang, B.; et al. Blocking immunoinhibitory receptor LILRB2 reprograms tumor-associated myeloid cells and promotes antitumor immunity. J. Clin. Investig. 2018, 128, 5647–5662. [Google Scholar] [CrossRef] [PubMed]
- Barkal, A.A.; Weiskopf, K.; Kao, K.S.; Gordon, S.R.; Rosental, B.; Yiu, Y.Y.; George, B.M.; Markovic, M.; Ring, N.G.; Tsai, J.M.; et al. Engagement of MHC class I by the inhibitory receptor LILRB1 suppresses macrophages and is a target of cancer immunotherapy. Nat. Immunol. 2018, 19, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Crocker, P.R.; Paulson, J.C.; Varki, A. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 2007, 7, 255–266. [Google Scholar] [CrossRef]
- Macauley, M.S.; Crocker, P.R.; Paulson, J.C. Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 2014, 14, 653–666. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Paulson, J.C. Siglecs as immune cell checkpoints in disease. Annu. Rev. Immunol. 2020, 38, 365–395. [Google Scholar] [CrossRef]
- Boelaars, K.; van Kooyk, Y. Targeting myeloid cells for cancer immunotherapy: Siglec-7/9/10/15 and their ligands. Trends Cancer 2024, 10, 230–241. [Google Scholar] [CrossRef]
- Rodriguez, E.; Boelaars, K.; Brown, K.; Eveline Li, R.J.; Kruijssen, L.; Bruijns, S.C.M.; van Ee, T.; Schetters, S.T.T.; Crommentuijn, M.H.W.; van der Horst, J.C.; et al. Sialic acids in pancreatic cancer cells drive tumour-associated macrophage differentiation via the Siglec receptors Siglec-7 and Siglec-9. Nat. Commun. 2021, 12, 1270. [Google Scholar] [CrossRef]
- Wang, J.; Sun, J.; Liu, L.N.; Flies, D.B.; Nie, X.; Toki, M.; Zhang, J.; Song, C.; Zarr, M.; Zhou, X.; et al. Siglec-15 as an immune suppressor and potential target for normalization cancer immunotherapy. Nat. Med. 2019, 25, 656–666. [Google Scholar] [CrossRef] [PubMed]
- Jandus, C.; Boligan, K.F.; Chijioke, O.; Liu, H.; Dahlhaus, M.; Demoulins, T.; Schneider, C.; Wehrli, M.; Hunger, R.E.; Baerlocher, G.M.; et al. Interactions between Siglec-7/9 receptors and ligands influence NK cell-dependent tumor immunosurveillance. J. Clin. Investig. 2014, 124, 1810–1820. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Lu, Q.; Sanmamed, M.F.; Wang, J. Siglec-15 as an emerging target for next-generation cancer immunotherapy. Clin. Cancer Res. 2021, 27, 680–688. [Google Scholar] [CrossRef] [PubMed]
- Barkal, A.A.; Brewer, R.E.; Markovic, M.; Kowarsky, M.; Barkal, S.A.; Zaro, B.W.; Krishnan, V.; Hatakeyama, J.; Dorigo, O.; Barkal, L.J.; et al. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature 2019, 572, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Shi, P.; Dong, Q.; Zhou, X.; Chen, C.; Sui, X.; Tian, W.; Zhu, X.; Wang, X.; Jin, S.; et al. Discovery of a novel dual-targeting D-peptide to block CD24/Siglec-10 and PD-1/PD-L1 interaction and synergize with radiotherapy for cancer immunotherapy. J. Immunother. Cancer 2023, 11, e007068. [Google Scholar] [CrossRef] [PubMed]
- Haas, Q.; Boligan, K.F.; Jandus, C.; Schneider, C.; Simillion, C.; Stanczak, M.A.; Haubitz, M.; Seyed Jafari, S.M.; Zippelius, A.; Baerlocher, G.M.; et al. Siglec-9 regulates an effector memory CD8(+) T-cell subset that congregates in the melanoma tumor microenvironment. Cancer Immunol. Res. 2019, 7, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Stanczak, M.A.; Siddiqui, S.S.; Trefny, M.P.; Thommen, D.S.; Boligan, K.F.; von Gunten, S.; Tzankov, A.; Tietze, L.; Lardinois, D.; Heinzelmann-Schwarz, V.; et al. Self-associated molecular patterns mediate cancer immune evasion by engaging Siglecs on T cells. J. Clin. Investig. 2018, 128, 4912–4923. [Google Scholar] [CrossRef]
- Bull, C.; Boltje, T.J.; Balneger, N.; Weischer, S.M.; Wassink, M.; van Gemst, J.J.; Bloemendal, V.R.; Boon, L.; van der Vlag, J.; Heise, T.; et al. Sialic acid blockade suppresses tumor growth by enhancing T-cell-mediated tumor immunity. Cancer Res. 2018, 78, 3574–3588. [Google Scholar] [CrossRef]
- Yu, L.; Huang, L.; Lin, D.; Lai, X.; Wu, L.; Liao, X.; Liu, J.; Zeng, Y.; Liang, L.; Zhang, G.; et al. GD2-specific chimeric antigen receptor-modified T cells for the treatment of refractory and/or recurrent neuroblastoma in pediatric patients. J. Cancer Res. Clin. Oncol. 2022, 148, 2643–2652. [Google Scholar] [CrossRef]
- Lo, A.S.; Ma, Q.; Liu, D.L.; Junghans, R.P. Anti-GD3 chimeric sFv-CD28/T-cell receptor zeta designer T cells for treatment of metastatic melanoma and other neuroectodermal tumors. Clin. Cancer Res. 2010, 16, 2769–2780. [Google Scholar] [CrossRef]
- Faubert, B.; Solmonson, A.; DeBerardinis, R.J. Metabolic reprogramming and cancer progression. Science 2020, 368, eaaw5473. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Bader, J.E.; Voss, K.; Rathmell, J.C. Targeting metabolism to improve the tumor microenvironment for cancer immunotherapy. Mol. Cell 2020, 78, 1019–1033. [Google Scholar] [CrossRef]
- Kaymak, I.; Williams, K.S.; Cantor, J.R.; Jones, R.G. Immunometabolic interplay in the tumor microenvironment. Cancer Cell 2021, 39, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Arner, E.N.; Rathmell, J.C. Metabolic programming and immune suppression in the tumor microenvironment. Cancer Cell 2023, 41, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Cham, C.M.; Gajewski, T.F. Glucose availability regulates IFN-gamma production and p70S6 kinase activation in CD8+ effector T cells. J. Immunol. 2005, 174, 4670–4677. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.H.; Curtis, J.D.; Maggi, L.B., Jr.; Faubert, B.; Villarino, A.V.; O’Sullivan, D.; Huang, S.C.; van der Windt, G.J.; Blagih, J.; Qiu, J.; et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 2013, 153, 1239–1251. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.H.; Qiu, J.; O’Sullivan, D.; Buck, M.D.; Noguchi, T.; Curtis, J.D.; Chen, Q.; Gindin, M.; Gubin, M.M.; van der Windt, G.J.; et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 2015, 162, 1229–1241. [Google Scholar] [CrossRef] [PubMed]
- Leone, R.D.; Powell, J.D. Metabolism of immune cells in cancer. Nat. Rev. Cancer 2020, 20, 516–531. [Google Scholar] [CrossRef]
- Brand, A.; Singer, K.; Koehl, G.E.; Kolitzus, M.; Schoenhammer, G.; Thiel, A.; Matos, C.; Bruss, C.; Klobuch, S.; Peter, K.; et al. LDHA-Associated lactic acid production blunts tumor immunosurveillance by T and NK Cells. Cell Metab. 2016, 24, 657–671. [Google Scholar] [CrossRef]
- Mendler, A.N.; Hu, B.; Prinz, P.U.; Kreutz, M.; Gottfried, E.; Noessner, E. Tumor lactic acidosis suppresses CTL function by inhibition of p38 and JNK/c-Jun activation. Int. J. Cancer 2012, 131, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Colegio, O.R.; Chu, N.Q.; Szabo, A.L.; Chu, T.; Rhebergen, A.M.; Jairam, V.; Cyrus, N.; Brokowski, C.E.; Eisenbarth, S.C.; Phillips, G.M.; et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 2014, 513, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Gropper, Y.; Feferman, T.; Shalit, T.; Salame, T.M.; Porat, Z.; Shakhar, G. Culturing CTLs under hypoxic conditions enhances their cytolysis and improves their anti-tumor function. Cell Rep. 2017, 20, 2547–2555. [Google Scholar] [CrossRef] [PubMed]
- Palazon, A.; Goldrath, A.W.; Nizet, V.; Johnson, R.S. HIF transcription factors, inflammation, and immunity. Immunity 2014, 41, 518–528. [Google Scholar] [CrossRef] [PubMed]
- Scharping, N.E.; Rivadeneira, D.B.; Menk, A.V.; Vignali, P.D.A.; Ford, B.R.; Rittenhouse, N.L.; Peralta, R.; Wang, Y.; Wang, Y.; DePeaux, K.; et al. Mitochondrial stress induced by continuous stimulation under hypoxia rapidly drives T cell exhaustion. Nat. Immunol. 2021, 22, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Clambey, E.T.; McNamee, E.N.; Westrich, J.A.; Glover, L.E.; Campbell, E.L.; Jedlicka, P.; de Zoeten, E.F.; Cambier, J.C.; Stenmark, K.R.; Colgan, S.P.; et al. Hypoxia-inducible factor-1 alpha-dependent induction of FoxP3 drives regulatory T-cell abundance and function during inflammatory hypoxia of the mucosa. Proc. Natl. Acad. Sci. USA 2012, 109, E2784–E2793. [Google Scholar] [CrossRef]
- Deng, J.; Li, J.; Sarde, A.; Lines, J.L.; Lee, Y.C.; Qian, D.C.; Pechenick, D.A.; Manivanh, R.; Le Mercier, I.; Lowrey, C.H.; et al. Hypoxia-Induced VISTA promotes the suppressive function of myeloid-derived suppressor cells in the tumor microenvironment. Cancer Immunol. Res. 2019, 7, 1079–1090. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.H.; Sun, I.H.; Zhao, L.; Leone, R.D.; Sun, I.M.; Xu, W.; Collins, S.L.; Tam, A.J.; Blosser, R.L.; Patel, C.H.; et al. Targeting glutamine metabolism enhances tumor-specific immunity by modulating suppressive myeloid cells. J. Clin. Investig. 2020, 130, 3865–3884. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, E.M.; Menga, A.; Martin-Perez, R.; Quinto, A.; Riera-Domingo, C.; De Tullio, G.; Hooper, D.C.; Lamers, W.H.; Ghesquiere, B.; McVicar, D.W.; et al. Pharmacologic or genetic targeting of glutamine synthetase skews macrophages toward an M1-like phenotype and inhibits tumor metastasis. Cell Rep. 2017, 20, 1654–1666. [Google Scholar] [CrossRef]
- Leone, R.D.; Zhao, L.; Englert, J.M.; Sun, I.M.; Oh, M.H.; Sun, I.H.; Arwood, M.L.; Bettencourt, I.A.; Patel, C.H.; Wen, J.; et al. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science 2019, 366, 1013–1021. [Google Scholar] [CrossRef]
- Madden, M.Z.; Ye, X.; Chi, C.; Fisher, E.L.; Wolf, M.M.; Needle, G.A.; Bader, J.E.; Patterson, A.R.; Reinfeld, B.I.; Landis, M.D.; et al. Differential effects of glutamine inhibition strategies on antitumor CD8 T Cells. J. Immunol. 2023, 211, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Geiger, R.; Rieckmann, J.C.; Wolf, T.; Basso, C.; Feng, Y.; Fuhrer, T.; Kogadeeva, M.; Picotti, P.; Meissner, F.; Mann, M.; et al. L-Arginine modulates T Cell metabolism and enhances survival and anti-tumor activity. Cell 2016, 167, 829–842.e813. [Google Scholar] [CrossRef] [PubMed]
- Miret, J.J.; Kirschmeier, P.; Koyama, S.; Zhu, M.; Li, Y.Y.; Naito, Y.; Wu, M.; Malladi, V.S.; Huang, W.; Walker, W.; et al. Suppression of myeloid cell arginase activity leads to therapeutic response in a NSCLC mouse model by activating anti-tumor immunity. J. Immunother. Cancer 2019, 7, 32. [Google Scholar] [CrossRef] [PubMed]
- Lamas, B.; Vergnaud-Gauduchon, J.; Goncalves-Mendes, N.; Perche, O.; Rossary, A.; Vasson, M.P.; Farges, M.C. Altered functions of natural killer cells in response to L-Arginine availability. Cell Immunol. 2012, 280, 182–190. [Google Scholar] [CrossRef]
- Frumento, G.; Rotondo, R.; Tonetti, M.; Damonte, G.; Benatti, U.; Ferrara, G.B. Tryptophan-derived catabolites are responsible for inhibition of T and natural killer cell proliferation induced by indoleamine 2,3-dioxygenase. J. Exp. Med. 2002, 196, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Han, Y.; Rodriguez Sillke, Y.; Deng, H.; Siddiqui, S.; Treese, C.; Schmidt, F.; Friedrich, M.; Keye, J.; Wan, J.; et al. Lipid droplet-dependent fatty acid metabolism controls the immune suppressive phenotype of tumor-associated macrophages. EMBO Mol. Med. 2019, 11, e10698. [Google Scholar] [CrossRef] [PubMed]
- Herber, D.L.; Cao, W.; Nefedova, Y.; Novitskiy, S.V.; Nagaraj, S.; Tyurin, V.A.; Corzo, A.; Cho, H.I.; Celis, E.; Lennox, B.; et al. Lipid accumulation and dendritic cell dysfunction in cancer. Nat. Med. 2010, 16, 880–886. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Fang, X.; Wang, H.; Li, D.; Wang, X. Ovarian Cancer-Intrinsic Fatty Acid Synthase Prevents Anti-tumor Immunity by Disrupting Tumor-Infiltrating Dendritic Cells. Front. Immunol. 2018, 9, 2927. [Google Scholar] [CrossRef] [PubMed]
- Field, C.S.; Baixauli, F.; Kyle, R.L.; Puleston, D.J.; Cameron, A.M.; Sanin, D.E.; Hippen, K.L.; Loschi, M.; Thangavelu, G.; Corrado, M.; et al. Mitochondrial Integrity Regulated by Lipid Metabolism Is a Cell-Intrinsic Checkpoint for Treg Suppressive Function. Cell Metab. 2020, 31, 422–437.e425. [Google Scholar] [CrossRef]
- Wang, H.; Franco, F.; Tsui, Y.C.; Xie, X.; Trefny, M.P.; Zappasodi, R.; Mohmood, S.R.; Fernandez-Garcia, J.; Tsai, C.H.; Schulze, I.; et al. CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors. Nat. Immunol. 2020, 21, 298–308. [Google Scholar] [CrossRef]
- Caligiuri, G.; Tuveson, D.A. Activated fibroblasts in cancer: Perspectives and challenges. Cancer Cell 2023, 41, 434–449. [Google Scholar] [CrossRef] [PubMed]
- Feig, C.; Jones, J.O.; Kraman, M.; Wells, R.J.; Deonarine, A.; Chan, D.S.; Connell, C.M.; Roberts, E.W.; Zhao, Q.; Caballero, O.L.; et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 20212–20217. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Wang, Z.; Zhang, Y.; Pradhan, R.N.; Ganguly, D.; Chandra, R.; Murimwa, G.; Wright, S.; Gu, X.; Maddipati, R.; et al. Mesothelial cell-derived antigen-presenting cancer-associated fibroblasts induce expansion of regulatory T cells in pancreatic cancer. Cancer Cell 2022, 40, 656–673.e657. [Google Scholar] [CrossRef] [PubMed]
- Grout, J.A.; Sirven, P.; Leader, A.M.; Maskey, S.; Hector, E.; Puisieux, I.; Steffan, F.; Cheng, E.; Tung, N.; Maurin, M.; et al. Spatial Positioning and matrix programs of cancer-associated fibroblasts promote T-cell exclusion in human lung tumors. Cancer Discov. 2022, 12, 2606–2625. [Google Scholar] [CrossRef] [PubMed]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel, E.E., III; Koeppen, H.; Astarita, J.L.; Cubas, R.; et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Smyth, M.J. Myeloid immunosuppression and immune checkpoints in the tumor microenvironment. Cell Mol. Immunol. 2020, 17, 1–12. [Google Scholar] [CrossRef]
- Hegde, S.; Leader, A.M.; Merad, M. MDSC: Markers, development, states, and unaddressed complexity. Immunity 2021, 54, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Chiu, D.K.; Tse, A.P.; Xu, I.M.; Di Cui, J.; Lai, R.K.; Li, L.L.; Koh, H.Y.; Tsang, F.H.; Wei, L.L.; Wong, C.M.; et al. Hypoxia inducible factor HIF-1 promotes myeloid-derived suppressor cells accumulation through ENTPD2/CD39L1 in hepatocellular carcinoma. Nat. Commun. 2017, 8, 517. [Google Scholar] [CrossRef] [PubMed]
- Obermajer, N.; Muthuswamy, R.; Odunsi, K.; Edwards, R.P.; Kalinski, P. PGE(2)-induced CXCL12 production and CXCR4 expression controls the accumulation of human MDSCs in ovarian cancer environment. Cancer Res. 2011, 71, 7463–7470. [Google Scholar] [CrossRef]
- Chun, E.; Lavoie, S.; Michaud, M.; Gallini, C.A.; Kim, J.; Soucy, G.; Odze, R.; Glickman, J.N.; Garrett, W.S. CCL2 Promotes Colorectal carcinogenesis by enhancing polymorphonuclear myeloid-derived suppressor cell population and function. Cell Rep. 2015, 12, 244–257. [Google Scholar] [CrossRef]
- Beury, D.W.; Parker, K.H.; Nyandjo, M.; Sinha, P.; Carter, K.A.; Ostrand-Rosenberg, S. Cross-talk among myeloid-derived suppressor cells, macrophages, and tumor cells impacts the inflammatory milieu of solid tumors. J. Leukoc. Biol. 2014, 96, 1109–1118. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wei, J.; Guo, G.; Zhou, J. Norepinephrine-induced myeloid-derived suppressor cells block T-cell responses via generation of reactive oxygen species. Immunopharmacol. Immunotoxicol. 2015, 37, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Stiff, A.; Trikha, P.; Mundy-Bosse, B.; McMichael, E.; Mace, T.A.; Benner, B.; Kendra, K.; Campbell, A.; Gautam, S.; Abood, D.; et al. Nitric oxide production by myeloid-derived suppressor cells plays a role in impairing Fc receptor-mediated natural killer cell function. Clin. Cancer Res. 2018, 24, 1891–1904. [Google Scholar] [CrossRef]
- Sade-Feldman, M.; Kanterman, J.; Klieger, Y.; Ish-Shalom, E.; Olga, M.; Saragovi, A.; Shtainberg, H.; Lotem, M.; Baniyash, M. Clinical significance of circulating CD33+CD11b+HLA-DR− myeloid cells in patients with stage IV melanoma treated with ipilimumab. Clin. Cancer Res. 2016, 22, 5661–5672. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Redd, P.S.; Lee, J.R.; Savage, N.; Liu, K. The expression profiles and regulation of PD-L1 in tumor-induced myeloid-derived suppressor cells. Oncoimmunology 2016, 5, e1247135. [Google Scholar] [CrossRef] [PubMed]
- Wculek, S.K.; Cueto, F.J.; Mujal, A.M.; Melero, I.; Krummel, M.F.; Sancho, D. Dendritic cells in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2020, 20, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Spranger, S.; Bao, R.; Gajewski, T.F. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature 2015, 523, 231–235. [Google Scholar] [CrossRef]
- Bottcher, J.P.; Bonavita, E.; Chakravarty, P.; Blees, H.; Cabeza-Cabrerizo, M.; Sammicheli, S.; Rogers, N.C.; Sahai, E.; Zelenay, S.; Reis e Sousa, C. NK Cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control. Cell 2018, 172, 1022–1037.e1014. [Google Scholar] [CrossRef]
- Barry, K.C.; Hsu, J.; Broz, M.L.; Cueto, F.J.; Binnewies, M.; Combes, A.J.; Nelson, A.E.; Loo, K.; Kumar, R.; Rosenblum, M.D.; et al. A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nat. Med. 2018, 24, 1178–1191. [Google Scholar] [CrossRef]
- Chiba, S.; Baghdadi, M.; Akiba, H.; Yoshiyama, H.; Kinoshita, I.; Dosaka-Akita, H.; Fujioka, Y.; Ohba, Y.; Gorman, J.V.; Colgan, J.D.; et al. Tumor-infiltrating DCs suppress nucleic acid-mediated innate immune responses through interactions between the receptor TIM-3 and the alarmin HMGB1. Nat. Immunol. 2012, 13, 832–842. [Google Scholar] [CrossRef]
- Veglia, F.; Tyurin, V.A.; Mohammadyani, D.; Blasi, M.; Duperret, E.K.; Donthireddy, L.; Hashimoto, A.; Kapralov, A.; Amoscato, A.; Angelini, R.; et al. Lipid bodies containing oxidatively truncated lipids block antigen cross-presentation by dendritic cells in cancer. Nat. Commun. 2017, 8, 2122. [Google Scholar] [CrossRef] [PubMed]
- Aspord, C.; Leccia, M.T.; Charles, J.; Plumas, J. Plasmacytoid dendritic cells support melanoma progression by promoting Th2 and regulatory immunity through OX40L and ICOSL. Cancer Immunol. Res. 2013, 1, 402–415. [Google Scholar] [CrossRef] [PubMed]
- Sawant, D.V.; Yano, H.; Chikina, M.; Zhang, Q.; Liao, M.; Liu, C.; Callahan, D.J.; Sun, Z.; Sun, T.; Tabib, T.; et al. Adaptive plasticity of IL-10+ and IL-35+ T(reg) cells cooperatively promotes tumor T cell exhaustion. Nat. Immunol. 2019, 20, 724–735. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Jiang, P.; Wei, S.; Xu, X.; Wang, J. Regulatory T cells in tumor microenvironment: New mechanisms, potential therapeutic strategies and future prospects. Mol. Cancer 2020, 19, 116. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.A.; Vick, S.C.; Iglesia, M.D.; Brickey, W.J.; Midkiff, B.R.; McKinnon, K.P.; Reisdorf, S.; Anders, C.K.; Carey, L.A.; Parker, J.S.; et al. Treg depletion potentiates checkpoint inhibition in claudin-low breast cancer. J. Clin. Investig. 2017, 127, 3472–3483. [Google Scholar] [CrossRef]
- Wen, Y.H.; Lin, H.Q.; Li, H.; Zhao, Y.; Lui, V.W.Y.; Chen, L.; Wu, X.M.; Sun, W.; Wen, W.P. Stromal interleukin-33 promotes regulatory T cell-mediated immunosuppression in head and neck squamous cell carcinoma and correlates with poor prognosis. Cancer Immunol. Immunother. 2019, 68, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Maj, T.; Wang, W.; Crespo, J.; Zhang, H.; Wang, W.; Wei, S.; Zhao, L.; Vatan, L.; Shao, I.; Szeliga, W.; et al. Oxidative stress controls regulatory T cell apoptosis and suppressor activity and PD-L1-blockade resistance in tumor. Nat. Immunol. 2017, 18, 1332–1341. [Google Scholar] [CrossRef]
- Pan, Y.; Yu, Y.; Wang, X.; Zhang, T. Tumor-Associated Macrophages in Tumor Immunity. Front. Immunol. 2020, 11, 583084. [Google Scholar] [CrossRef] [PubMed]
- Cassetta, L.; Fragkogianni, S.; Sims, A.H.; Swierczak, A.; Forrester, L.M.; Zhang, H.; Soong, D.Y.H.; Cotechini, T.; Anur, P.; Lin, E.Y.; et al. Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell 2019, 35, 588–602.e510. [Google Scholar] [CrossRef]
- Loyher, P.L.; Hamon, P.; Laviron, M.; Meghraoui-Kheddar, A.; Goncalves, E.; Deng, Z.; Torstensson, S.; Bercovici, N.; Baudesson de Chanville, C.; Combadiere, B.; et al. Macrophages of distinct origins contribute to tumor development in the lung. J. Exp. Med. 2018, 215, 2536–2553. [Google Scholar] [CrossRef]
- Curiel, T.J.; Coukos, G.; Zou, L.; Alvarez, X.; Cheng, P.; Mottram, P.; Evdemon-Hogan, M.; Conejo-Garcia, J.R.; Zhang, L.; Burow, M.; et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 2004, 10, 942–949. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Halima, A.; Chan, T.A. Antigen presentation in cancer—mechanisms and clinical implications for immunotherapy. Nat. Rev. Clin. Oncol. 2023, 20, 604–623. [Google Scholar] [CrossRef] [PubMed]
- Jhunjhunwala, S.; Hammer, C.; Delamarre, L. Antigen presentation in cancer: Insights into tumour immunogenicity and immune evasion. Nat. Rev. Cancer 2021, 21, 298–312. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Lennerz, V.; Fatho, M.; Gentilini, C.; Frye, R.A.; Lifke, A.; Ferel, D.; Wolfel, C.; Huber, C.; Wolfel, T. The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc. Natl. Acad. Sci. USA 2005, 102, 16013–16018. [Google Scholar] [CrossRef] [PubMed]
- McGranahan, N.; Furness, A.J.; Rosenthal, R.; Ramskov, S.; Lyngaa, R.; Saini, S.K.; Jamal-Hanjani, M.; Wilson, G.A.; Birkbak, N.J.; Hiley, C.T.; et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016, 351, 1463–1469. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; DiNatale, R.G.; Chowell, D.; Krishna, C.; Makarov, V.; Valero, C.; Vuong, L.; Lee, M.; Weiss, K.; Hoen, D.; et al. High response rate and durability driven by HLA genetic diversity in patients with kidney cancer treated with lenvatinib and pembrolizumab. Mol. Cancer Res. 2021, 19, 1510–1521. [Google Scholar] [CrossRef] [PubMed]
- Chowell, D.; Yoo, S.K.; Valero, C.; Pastore, A.; Krishna, C.; Lee, M.; Hoen, D.; Shi, H.; Kelly, D.W.; Patel, N.; et al. Improved prediction of immune checkpoint blockade efficacy across multiple cancer types. Nat. Biotechnol. 2022, 40, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Cuppens, K.; Baas, P.; Geerdens, E.; Cruys, B.; Froyen, G.; Decoster, L.; Thomeer, M.; Maes, B. HLA-I diversity and tumor mutational burden by comprehensive next-generation sequencing as predictive biomarkers for the treatment of non-small cell lung cancer with PD-(L)1 inhibitors. Lung Cancer 2022, 170, 1–10. [Google Scholar] [CrossRef]
- Shukla, S.A.; Rooney, M.S.; Rajasagi, M.; Tiao, G.; Dixon, P.M.; Lawrence, M.S.; Stevens, J.; Lane, W.J.; Dellagatta, J.L.; Steelman, S.; et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat. Biotechnol. 2015, 33, 1152–1158. [Google Scholar] [CrossRef]
- Chowell, D.; Morris, L.G.T.; Grigg, C.M.; Weber, J.K.; Samstein, R.M.; Makarov, V.; Kuo, F.; Kendall, S.M.; Requena, D.; Riaz, N.; et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 2018, 359, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Naranbhai, V.; Viard, M.; Dean, M.; Groha, S.; Braun, D.A.; Labaki, C.; Shukla, S.A.; Yuki, Y.; Shah, P.; Chin, K.; et al. HLA-A*03 and response to immune checkpoint blockade in cancer: An epidemiological biomarker study. Lancet Oncol. 2022, 23, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Pyke, R.M.; Mellacheruvu, D.; Dea, S.; Abbott, C.W.; McDaniel, L.; Bhave, D.P.; Zhang, S.V.; Levy, E.; Bartha, G.; West, J.; et al. A machine learning algorithm with subclonal sensitivity reveals widespread pan-cancer human leukocyte antigen loss of heterozygosity. Nat. Commun. 2022, 13, 1925. [Google Scholar] [CrossRef] [PubMed]
- Lorenzi, S.; Forloni, M.; Cifaldi, L.; Antonucci, C.; Citti, A.; Boldrini, R.; Pezzullo, M.; Castellano, A.; Russo, V.; van der Bruggen, P.; et al. IRF1 and NF-kB restore MHC class I-restricted tumor antigen processing and presentation to cytotoxic T cells in aggressive neuroblastoma. PLoS ONE 2012, 7, e46928. [Google Scholar] [CrossRef] [PubMed]
- Yoshihama, S.; Roszik, J.; Downs, I.; Meissner, T.B.; Vijayan, S.; Chapuy, B.; Sidiq, T.; Shipp, M.A.; Lizee, G.A.; Kobayashi, K.S. NLRC5/MHC class I transactivator is a target for immune evasion in cancer. Proc. Natl. Acad. Sci. USA 2016, 113, 5999–6004. [Google Scholar] [CrossRef] [PubMed]
- Mari, L.; Hoefnagel, S.J.M.; Zito, D.; van de Meent, M.; van Endert, P.; Calpe, S.; Sancho Serra, M.D.C.; Heemskerk, M.H.M.; van Laarhoven, H.W.M.; Hulshof, M.; et al. microRNA 125a regulates MHC-I expression on esophageal adenocarcinoma cells, associated with suppression of antitumor immune response and poor outcomes of patients. Gastroenterology 2018, 155, 784–798. [Google Scholar] [CrossRef] [PubMed]
- Colangelo, T.; Polcaro, G.; Ziccardi, P.; Pucci, B.; Muccillo, L.; Galgani, M.; Fucci, A.; Milone, M.R.; Budillon, A.; Santopaolo, M.; et al. Proteomic screening identifies calreticulin as a miR-27a direct target repressing MHC class I cell surface exposure in colorectal cancer. Cell Death Dis. 2016, 7, e2120. [Google Scholar] [CrossRef] [PubMed]
- Zaretsky, J.M.; Garcia-Diaz, A.; Shin, D.S.; Escuin-Ordinas, H.; Hugo, W.; Hu-Lieskovan, S.; Torrejon, D.Y.; Abril-Rodriguez, G.; Sandoval, S.; Barthly, L.; et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med. 2016, 375, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Venida, A.; Yano, J.; Biancur, D.E.; Kakiuchi, M.; Gupta, S.; Sohn, A.S.W.; Mukhopadhyay, S.; Lin, E.Y.; Parker, S.J.; et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 2020, 581, 100–105. [Google Scholar] [CrossRef]
- Fang, Y.; Wang, L.; Wan, C.; Sun, Y.; Van der Jeught, K.; Zhou, Z.; Dong, T.; So, K.M.; Yu, T.; Li, Y.; et al. MAL2 drives immune evasion in breast cancer by suppressing tumor antigen presentation. J. Clin. Investig. 2021, 131, e140837. [Google Scholar] [CrossRef]
- Tripathi, S.C.; Peters, H.L.; Taguchi, A.; Katayama, H.; Wang, H.; Momin, A.; Jolly, M.K.; Celiktas, M.; Rodriguez-Canales, J.; Liu, H.; et al. Immunoproteasome deficiency is a feature of non-small cell lung cancer with a mesenchymal phenotype and is associated with a poor outcome. Proc. Natl. Acad. Sci. USA 2016, 113, E1555–E1564. [Google Scholar] [CrossRef] [PubMed]
- Kalaora, S.; Lee, J.S.; Barnea, E.; Levy, R.; Greenberg, P.; Alon, M.; Yagel, G.; Bar Eli, G.; Oren, R.; Peri, A.; et al. Immunoproteasome expression is associated with better prognosis and response to checkpoint therapies in melanoma. Nat. Commun. 2020, 11, 896. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Tan, X.; Cen, H. EZH2 is a negative prognostic biomarker associated with immunosuppression in hepatocellular carcinoma. PLoS ONE 2020, 15, e0242191. [Google Scholar] [CrossRef] [PubMed]
- Alspach, E.; Lussier, D.M.; Miceli, A.P.; Kizhvatov, I.; DuPage, M.; Luoma, A.M.; Meng, W.; Lichti, C.F.; Esaulova, E.; Vomund, A.N.; et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 2019, 574, 696–701. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.M.; Huang, J.; Niknafs, N.; Balan, A.; Cherry, C.; White, J.; Velculescu, V.E.; Anagnostou, V.; Karchin, R. HLA class II immunogenic mutation burden predicts response to immune checkpoint blockade. Ann. Oncol. 2022, 33, 728–738. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, L.H.; Najjar, Y.G. Immunotherapy combination approaches: Mechanisms, biomarkers and clinical observations. Nat. Rev. Immunol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Kraehenbuehl, L.; Weng, C.H.; Eghbali, S.; Wolchok, J.D.; Merghoub, T. Enhancing immunotherapy in cancer by targeting emerging immunomodulatory pathways. Nat. Rev. Clin. Oncol. 2022, 19, 37–50. [Google Scholar] [CrossRef]
- Zinn, S.; Vazquez-Lombardi, R.; Zimmermann, C.; Sapra, P.; Jermutus, L.; Christ, D. Advances in antibody-based therapy in oncology. Nat. Cancer 2023, 4, 165–180. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Oldham, R.J.; Teal, E.; Beers, S.A.; Cragg, M.S. Fc-Engineering for modulated effector functions-improving antibodies for cancer treatment. Antibodies 2020, 9, 64. [Google Scholar] [CrossRef]
- Jin, S.; Sun, Y.; Liang, X.; Gu, X.; Ning, J.; Xu, Y.; Chen, S.; Pan, L. Emerging new therapeutic antibody derivatives for cancer treatment. Signal Transduct. Target. Ther. 2022, 7, 39. [Google Scholar] [CrossRef]
- Zhao, B.; Zhao, H.; Zhao, J. Efficacy of PD-1/PD-L1 blockade monotherapy in clinical trials. Ther. Adv. Med. Oncol. 2020, 12, 1758835920937612. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wang, J.; Deng, X.; Xiong, F.; Ge, J.; Xiang, B.; Wu, X.; Ma, J.; Zhou, M.; Li, X.; et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol. Cancer 2019, 18, 10. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.; Zheng, X.; Niu, M.; Zhu, S.; Ge, H.; Wu, K. Combination strategies with PD-1/PD-L1 blockade: Current advances and future directions. Mol. Cancer 2022, 21, 28. [Google Scholar] [CrossRef] [PubMed]
- Wolchok, J.D.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Rutkowski, P.; Lao, C.D.; Cowey, C.L.; Schadendorf, D.; Wagstaff, J.; Dummer, R.; et al. Long-Term Outcomes with Nivolumab Plus Ipilimumab or Nivolumab Alone Versus Ipilimumab in Patients with Advanced Melanoma. J. Clin. Oncol. 2022, 40, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Albiges, L.; Tannir, N.M.; Burotto, M.; McDermott, D.; Plimack, E.R.; Barthelemy, P.; Porta, C.; Powles, T.; Donskov, F.; George, S.; et al. First-line nivolumab plus ipilimumab versus sunitinib in patients without nephrectomy and with an evaluable primary renal tumor in the CheckMate 214 Trial. Eur. Urol. 2022, 81, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, L.; Rodriguez-Abreu, D.; Gadgeel, S.; Esteban, E.; Felip, E.; De Angelis, F.; Domine, M.; Clingan, P.; Hochmair, M.J.; Powell, S.F.; et al. Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 2078–2092. [Google Scholar] [CrossRef] [PubMed]
- Schmid, P.; Rugo, H.S.; Adams, S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Dieras, V.; Henschel, V.; Molinero, L.; Chui, S.Y.; et al. Atezolizumab plus nab-paclitaxel as first-line treatment for unresectable, locally advanced or metastatic triple-negative breast cancer (IMpassion130): Updated efficacy results from a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2020, 21, 44–59. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, A.S.; Kazarnowicz, A.; Karaseva, N.; Sanchez, A.; De Boer, R.; Andric, Z.; Reck, M.; Atagi, S.; Lee, J.S.; Garassino, M.; et al. Safety and patient-reported outcomes of atezolizumab, carboplatin, and etoposide in extensive-stage small-cell lung cancer (IMpower133): A randomized phase I/III trial. Ann. Oncol. 2020, 31, 310–317. [Google Scholar] [CrossRef] [PubMed]
- West, H.; McCleod, M.; Hussein, M.; Morabito, A.; Rittmeyer, A.; Conter, H.J.; Kopp, H.G.; Daniel, D.; McCune, S.; Mekhail, T.; et al. Atezolizumab in combination with carboplatin plus nab-paclitaxel chemotherapy compared with chemotherapy alone as first-line treatment for metastatic non-squamous non-small-cell lung cancer (IMpower130): A multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2019, 20, 924–937. [Google Scholar] [CrossRef]
- Senan, S.; Ozguroglu, M.; Daniel, D.; Villegas, A.; Vicente, D.; Murakami, S.; Hui, R.; Faivre-Finn, C.; Paz-Ares, L.; Wu, Y.L.; et al. Outcomes with durvalumab after chemoradiotherapy in stage IIIA-N2 non-small-cell lung cancer: An exploratory analysis from the PACIFIC trial. ESMO Open 2022, 7, 100410. [Google Scholar] [CrossRef]
- Tawbi, H.A.; Schadendorf, D.; Lipson, E.J.; Ascierto, P.A.; Matamala, L.; Castillo Gutierrez, E.; Rutkowski, P.; Gogas, H.J.; Lao, C.D.; De Menezes, J.J.; et al. Relatlimab and nivolumab versus nivolumab in untreated advanced melanoma. N. Engl. J. Med. 2022, 386, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Curigliano, G.; Gelderblom, H.; Mach, N.; Doi, T.; Tai, D.; Forde, P.M.; Sarantopoulos, J.; Bedard, P.L.; Lin, C.C.; Hodi, F.S.; et al. Phase I/Ib clinical trial of sabatolimab, an Anti-TIM-3 antibody, alone and in combination with spartalizumab, an Anti-PD-1 antibody, in advanced solid tumors. Clin. Cancer Res. 2021, 27, 3620–3629. [Google Scholar] [CrossRef] [PubMed]
- Acharya, N.; Sabatos-Peyton, C.; Anderson, A.C. Tim-3 finds its place in the cancer immunotherapy landscape. J. Immunother. Cancer 2020, 8, e000911. [Google Scholar] [CrossRef] [PubMed]
- Maher, V.E.; Fernandes, L.L.; Weinstock, C.; Tang, S.; Agarwal, S.; Brave, M.; Ning, Y.M.; Singh, H.; Suzman, D.; Xu, J.; et al. Analysis of the association between adverse events and outcome in patients receiving a programmed death Protein 1 or programmed death Ligand 1 antibody. J. Clin. Oncol. 2019, 37, 2730–2737. [Google Scholar] [CrossRef] [PubMed]
- Shankar, B.; Zhang, J.; Naqash, A.R.; Forde, P.M.; Feliciano, J.L.; Marrone, K.A.; Ettinger, D.S.; Hann, C.L.; Brahmer, J.R.; Ricciuti, B.; et al. Multisystem immune-related adverse events associated with immune checkpoint inhibitors for treatment of non-small cell lung cancer. JAMA Oncol. 2020, 6, 1952–1956. [Google Scholar] [CrossRef] [PubMed]
- van der Kooij, M.K.; Suijkerbuijk, K.P.M.; Aarts, M.J.B.; van den Berkmortel, F.; Blank, C.U.; Boers-Sonderen, M.J.; van Breeschoten, J.; van den Eertwegh, A.J.M.; de Groot, J.W.B.; Haanen, J.; et al. Safety and efficacy of checkpoint inhibition in patients with melanoma and preexisting autoimmune disease: A cohort study. Ann. Intern. Med. 2021, 174, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Davar, D.; Dzutsev, A.K.; McCulloch, J.A.; Rodrigues, R.R.; Chauvin, J.M.; Morrison, R.M.; Deblasio, R.N.; Menna, C.; Ding, Q.; Pagliano, O.; et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 2021, 371, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wiesnoski, D.H.; Helmink, B.A.; Gopalakrishnan, V.; Choi, K.; DuPont, H.L.; Jiang, Z.D.; Abu-Sbeih, H.; Sanchez, C.A.; Chang, C.C.; et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat. Med. 2018, 24, 1804–1808. [Google Scholar] [CrossRef] [PubMed]
- Hailemichael, Y.; Johnson, D.H.; Abdel-Wahab, N.; Foo, W.C.; Bentebibel, S.E.; Daher, M.; Haymaker, C.; Wani, K.; Saberian, C.; Ogata, D.; et al. Interleukin-6 blockade abrogates immunotherapy toxicity and promotes tumor immunity. Cancer Cell 2022, 40, 509–523.e506. [Google Scholar] [CrossRef]
- Conroy, M.; Naidoo, J. Immune-related adverse events and the balancing act of immunotherapy. Nat. Commun. 2022, 13, 392. [Google Scholar] [CrossRef]
- De Cecco, M.; Galbraith, D.N.; McDermott, L.L. What makes a good antibody-drug conjugate? Expert. Opin. Biol. Ther. 2021, 21, 841–847. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Li, S.; Han, S.; Shi, C.; Zhang, Y. Antibody drug conjugate: The “biological missile” for targeted cancer therapy. Signal Transduct. Target. Ther. 2022, 7, 93. [Google Scholar] [CrossRef] [PubMed]
- Gogia, P.; Ashraf, H.; Bhasin, S.; Xu, Y. Antibody-drug conjugates: A review of approved drugs and their clinical level of evidence. Cancers 2023, 15, 3886. [Google Scholar] [CrossRef] [PubMed]
- Sheyi, R.; de la Torre, B.G.; Albericio, F. Linkers: An assurance for controlled delivery of antibody-drug conjugate. Pharmaceutics 2022, 14, 396. [Google Scholar] [CrossRef] [PubMed]
- Lewis Phillips, G.D.; Li, G.; Dugger, D.L.; Crocker, L.M.; Parsons, K.L.; Mai, E.; Blattler, W.A.; Lambert, J.M.; Chari, R.V.; Lutz, R.J.; et al. Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody-cytotoxic drug conjugate. Cancer Res. 2008, 68, 9280–9290. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, S.E.; Pearson, C.I.; Gregorio, J.D.; Gonzalez, J.C.; Kenkel, J.A.; Hartmann, F.J.; Luo, A.; Ho, P.Y.; LeBlanc, H.; Blum, L.K.; et al. Immune-stimulating antibody conjugates elicit robust myeloid activation and durable antitumor immunity. Nat. Cancer 2021, 2, 18–33. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Miles, D.; Gianni, L.; Krop, I.E.; Welslau, M.; Baselga, J.; Pegram, M.; Oh, D.Y.; Dieras, V.; Guardino, E.; et al. Trastuzumab emtansine for HER2-positive advanced breast cancer. N. Engl. J. Med. 2012, 367, 1783–1791. [Google Scholar] [CrossRef] [PubMed]
- Powles, T.; Rosenberg, J.E.; Sonpavde, G.P.; Loriot, Y.; Duran, I.; Lee, J.L.; Matsubara, N.; Vulsteke, C.; Castellano, D.; Wu, C.; et al. Enfortumab vedotin in previously treated advanced urothelial carcinoma. N. Engl. J. Med. 2021, 384, 1125–1135. [Google Scholar] [CrossRef]
- Nicolo, E.; Giugliano, F.; Ascione, L.; Tarantino, P.; Corti, C.; Tolaney, S.M.; Cristofanilli, M.; Curigliano, G. Combining antibody-drug conjugates with immunotherapy in solid tumors: Current landscape and future perspectives. Cancer Treat. Rev. 2022, 106, 102395. [Google Scholar] [CrossRef]
- Mullard, A. Claudin-18.2 attracts the cancer crowd. Nat. Rev. Drug Discov. 2023, 22, 683–686. [Google Scholar] [CrossRef]
- Nisonoff, A.; Wissler, F.C.; Lipman, L.N. Properties of the major component of a peptic digest of rabbit antibody. Science 1960, 132, 1770–1771. [Google Scholar] [CrossRef] [PubMed]
- Labrijn, A.F.; Janmaat, M.L.; Reichert, J.M.; Parren, P. Bispecific antibodies: A mechanistic review of the pipeline. Nat. Rev. Drug Discov. 2019, 18, 585–608. [Google Scholar] [CrossRef] [PubMed]
- Holliger, P.; Prospero, T.; Winter, G. “Diabodies”: Small bivalent and bispecific antibody fragments. Proc. Natl. Acad. Sci. USA 1993, 90, 6444–6448. [Google Scholar] [CrossRef] [PubMed]
- Coloma, M.J.; Morrison, S.L. Design and production of novel tetravalent bispecific antibodies. Nat. Biotechnol. 1997, 15, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Dovedi, S.J.; Elder, M.J.; Yang, C.; Sitnikova, S.I.; Irving, L.; Hansen, A.; Hair, J.; Jones, D.C.; Hasani, S.; Wang, B.; et al. Design and efficacy of a monovalent bispecific PD-1/CTLA4 antibody that enhances CTLA4 blockade on PD-1+ activated T Cells. Cancer Discov. 2021, 11, 1100–1117. [Google Scholar] [CrossRef] [PubMed]
- Middleton, M.R.; McAlpine, C.; Woodcock, V.K.; Corrie, P.; Infante, J.R.; Steven, N.M.; Evans, T.R.J.; Anthoney, A.; Shoushtari, A.N.; Hamid, O.; et al. Tebentafusp, A TCR/Anti-CD3 bispecific fusion protein targeting gp100, potently activated antitumor immune responses in patients with metastatic melanoma. Clin. Cancer Res. 2020, 26, 5869–5878. [Google Scholar] [CrossRef] [PubMed]
- Owen, D.H.; Giffin, M.J.; Bailis, J.M.; Smit, M.D.; Carbone, D.P.; He, K. DLL3: An emerging target in small cell lung cancer. J. Hematol. Oncol. 2019, 12, 61. [Google Scholar] [CrossRef] [PubMed]
- Bargou, R.; Leo, E.; Zugmaier, G.; Klinger, M.; Goebeler, M.; Knop, S.; Noppeney, R.; Viardot, A.; Hess, G.; Schuler, M.; et al. Tumor regression in cancer patients by very low doses of a T cell-engaging antibody. Science 2008, 321, 974–977. [Google Scholar] [CrossRef]
- Topp, M.S.; Gokbuget, N.; Stein, A.S.; Zugmaier, G.; O’Brien, S.; Bargou, R.C.; Dombret, H.; Fielding, A.K.; Heffner, L.; Larson, R.A.; et al. Safety and activity of blinatumomab for adult patients with relapsed or refractory B-precursor acute lymphoblastic leukaemia: A multicentre, single-arm, phase 2 study. Lancet Oncol. 2015, 16, 57–66. [Google Scholar] [CrossRef]
- Autio, K.A.; Boni, V.; Humphrey, R.W.; Naing, A. Probody therapeutics: An emerging class of therapies designed to enhance on-target effects with reduced off-tumor toxicity for use in immuno-oncology. Clin. Cancer Res. 2020, 26, 984–989. [Google Scholar] [CrossRef]
- Ahn, M.J.; Cho, B.C.; Felip, E.; Korantzis, I.; Ohashi, K.; Majem, M.; Juan-Vidal, O.; Handzhiev, S.; Izumi, H.; Lee, J.S.; et al. Tarlatamab for patients with previously treated small-cell lung cancer. N. Engl. J. Med. 2023, 389, 2063–2075. [Google Scholar] [CrossRef] [PubMed]
- Fenis, A.; Demaria, O.; Gauthier, L.; Vivier, E.; Narni-Mancinelli, E. New immune cell engagers for cancer immunotherapy. Nat. Rev. Immunol. 2024. [Google Scholar] [CrossRef] [PubMed]
- Leong, S.R.; Sukumaran, S.; Hristopoulos, M.; Totpal, K.; Stainton, S.; Lu, E.; Wong, A.; Tam, L.; Newman, R.; Vuillemenot, B.R.; et al. An anti-CD3/anti-CLL-1 bispecific antibody for the treatment of acute myeloid leukemia. Blood 2017, 129, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Weigelin, B.; Bolanos, E.; Teijeira, A.; Martinez-Forero, I.; Labiano, S.; Azpilikueta, A.; Morales-Kastresana, A.; Quetglas, J.I.; Wagena, E.; Sanchez-Paulete, A.R.; et al. Focusing and sustaining the antitumor CTL effector killer response by agonist anti-CD137 mAb. Proc. Natl. Acad. Sci. USA 2015, 112, 7551–7556. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, L.; Morel, A.; Anceriz, N.; Rossi, B.; Blanchard-Alvarez, A.; Grondin, G.; Trichard, S.; Cesari, C.; Sapet, M.; Bosco, F.; et al. Multifunctional natural killer cell engagers targeting NKp46 trigger protective tumor immunity. Cell 2019, 177, 1701–1713.e1716. [Google Scholar] [CrossRef]
- Mishra, A.K.; Ali, A.; Dutta, S.; Banday, S.; Malonia, S.K. Emerging trends in immunotherapy for cancer. Diseases 2022, 10, 60. [Google Scholar] [CrossRef] [PubMed]
- Plosker, G.L.; Keam, S.J. Trastuzumab: A review of its use in the management of HER2-positive metastatic and early-stage breast cancer. Drugs 2006, 66, 449–475. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Schmitz, K.R.; Jeffrey, P.D.; Wiltzius, J.J.; Kussie, P.; Ferguson, K.M. Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell 2005, 7, 301–311. [Google Scholar] [CrossRef]
- Zahavi, D.; Weiner, L. Monoclonal antibodies in cancer therapy. Antibodies 2020, 9, 34. [Google Scholar] [CrossRef]
- Krejcik, J.; Casneuf, T.; Nijhof, I.S.; Verbist, B.; Bald, J.; Plesner, T.; Syed, K.; Liu, K.; van de Donk, N.W.; Weiss, B.M.; et al. Daratumumab depletes CD38+ immune regulatory cells, promotes T-cell expansion, and skews T-cell repertoire in multiple myeloma. Blood 2016, 128, 384–394. [Google Scholar] [CrossRef]
- Reardon, D.A.; Brandes, A.A.; Omuro, A.; Mulholland, P.; Lim, M.; Wick, A.; Baehring, J.; Ahluwalia, M.S.; Roth, P.; Bahr, O.; et al. Effect of nivolumab vs bevacizumab in patients with recurrent glioblastoma: The CheckMate 143 phase 3 randomized clinical trial. JAMA Oncol. 2020, 6, 1003–1010. [Google Scholar] [CrossRef]
- Oza, A.M.; Dubois, F.; Hegg, R.; Hernandez, C.A.; Finocchiaro, G.; Ghiringhelli, F.; Zamagni, C.; Nick, S.; Irahara, N.; Perretti, T.; et al. A long-term extension study of bevacizumab in patients with solid tumors. Oncologist 2021, 26, e2254–e2264. [Google Scholar] [CrossRef]
- Fousek, K.; Horn, L.A.; Palena, C. Interleukin-8: A chemokine at the intersection of cancer plasticity, angiogenesis, and immune suppression. Pharmacol. Ther. 2021, 219, 107692. [Google Scholar] [CrossRef]
- Feng, L.; Qi, Q.; Wang, P.; Chen, H.; Chen, Z.; Meng, Z.; Liu, L. Serum levels of IL-6, IL-8, and IL-10 are indicators of prognosis in pancreatic cancer. J. Int. Med. Res. 2018, 46, 5228–5236. [Google Scholar] [CrossRef]
- Schalper, K.A.; Carleton, M.; Zhou, M.; Chen, T.; Feng, Y.; Huang, S.P.; Walsh, A.M.; Baxi, V.; Pandya, D.; Baradet, T.; et al. Elevated serum interleukin-8 is associated with enhanced intratumor neutrophils and reduced clinical benefit of immune-checkpoint inhibitors. Nat. Med. 2020, 26, 688–692. [Google Scholar] [CrossRef]
- Kaminski, M.S.; Estes, J.; Zasadny, K.R.; Francis, I.R.; Ross, C.W.; Tuck, M.; Regan, D.; Fisher, S.; Gutierrez, J.; Kroll, S.; et al. Radioimmunotherapy with iodine (131)I tositumomab for relapsed or refractory B-cell non-Hodgkin lymphoma: Updated results and long-term follow-up of the University of Michigan experience. Blood 2000, 96, 1259–1266. [Google Scholar] [CrossRef]
- Rizzieri, D. Zevalin((R)) (ibritumomab tiuxetan): After more than a decade of treatment experience, what have we learned? Crit. Rev. Oncol. Hematol. 2016, 105, 5–17. [Google Scholar] [CrossRef]
- Davis, E.J.; Martin-Liberal, J.; Kristeleit, R.; Cho, D.C.; Blagden, S.P.; Berthold, D.; Cardin, D.B.; Vieito, M.; Miller, R.E.; Hari Dass, P.; et al. First-in-human phase I/II, open-label study of the anti-OX40 agonist INCAGN01949 in patients with advanced solid tumors. J. Immunother. Cancer 2022, 10, e004235. [Google Scholar] [CrossRef]
- Hamid, O.; Chiappori, A.A.; Thompson, J.A.; Doi, T.; Hu-Lieskovan, S.; Eskens, F.; Ros, W.; Diab, A.; Spano, J.P.; Rizvi, N.A.; et al. First-in-human study of an OX40 (ivuxolimab) and 4-1BB (utomilumab) agonistic antibody combination in patients with advanced solid tumors. J. Immunother. Cancer 2022, 10, e005471. [Google Scholar] [CrossRef]
- Barlesi, F.; Isambert, N.; Felip, E.; Cho, B.C.; Lee, D.H.; Peguero, J.; Jerusalem, G.; Penel, N.; Saada-Bouzid, E.; Garrido, P.; et al. Bintrafusp alfa, a bifunctional fusion protein targeting TGF-beta and PD-L1, in patients with non-small cell lung cancer resistant or refractory to immune checkpoint inhibitors. Oncologist 2023, 28, 258–267. [Google Scholar] [CrossRef]
- Kvistborg, P.; Shu, C.J.; Heemskerk, B.; Fankhauser, M.; Thrue, C.A.; Toebes, M.; van Rooij, N.; Linnemann, C.; van Buuren, M.M.; Urbanus, J.H.; et al. TIL therapy broadens the tumor-reactive CD8+ T cell compartment in melanoma patients. Oncoimmunology 2012, 1, 409–418. [Google Scholar] [CrossRef]
- Fernandez-Poma, S.M.; Salas-Benito, D.; Lozano, T.; Casares, N.; Riezu-Boj, J.I.; Mancheno, U.; Elizalde, E.; Alignani, D.; Zubeldia, N.; Otano, I.; et al. Expansion of tumor-infiltrating CD8+ T cells expressing PD-1 improves the efficacy of adoptive T-cell therapy. Cancer Res. 2017, 77, 3672–3684. [Google Scholar] [CrossRef]
- Duhen, T.; Duhen, R.; Montler, R.; Moses, J.; Moudgil, T.; de Miranda, N.F.; Goodall, C.P.; Blair, T.C.; Fox, B.A.; McDermott, J.E.; et al. Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. Nat. Commun. 2018, 9, 2724. [Google Scholar] [CrossRef]
- Ye, Q.; Song, D.G.; Poussin, M.; Yamamoto, T.; Best, A.; Li, C.; Coukos, G.; Powell, D.J., Jr. CD137 accurately identifies and enriches for naturally occurring tumor-reactive T cells in tumor. Clin. Cancer Res. 2014, 20, 44–55. [Google Scholar] [CrossRef]
- Tran, K.Q.; Zhou, J.; Durflinger, K.H.; Langhan, M.M.; Shelton, T.E.; Wunderlich, J.R.; Robbins, P.F.; Rosenberg, S.A.; Dudley, M.E. Minimally cultured tumor-infiltrating lymphocytes display optimal characteristics for adoptive cell therapy. J. Immunother. 2008, 31, 742–751. [Google Scholar] [CrossRef]
- Dudley, M.E.; Gross, C.A.; Langhan, M.M.; Garcia, M.R.; Sherry, R.M.; Yang, J.C.; Phan, G.Q.; Kammula, U.S.; Hughes, M.S.; Citrin, D.E.; et al. CD8+ enriched “young” tumor infiltrating lymphocytes can mediate regression of metastatic melanoma. Clin. Cancer Res. 2010, 16, 6122–6131. [Google Scholar] [CrossRef]
- Hinrichs, C.S.; Spolski, R.; Paulos, C.M.; Gattinoni, L.; Kerstann, K.W.; Palmer, D.C.; Klebanoff, C.A.; Rosenberg, S.A.; Leonard, W.J.; Restifo, N.P. IL-2 and IL-21 confer opposing differentiation programs to CD8+ T cells for adoptive immunotherapy. Blood 2008, 111, 5326–5333. [Google Scholar] [CrossRef]
- Zhao, Y.; Deng, J.; Rao, S.; Guo, S.; Shen, J.; Du, F.; Wu, X.; Chen, Y.; Li, M.; Chen, M.; et al. Tumor Infiltrating Lymphocyte (TIL) therapy for solid tumor treatment: Progressions and challenges. Cancers 2022, 14, 4160. [Google Scholar] [CrossRef]
- Creelan, B.C.; Wang, C.; Teer, J.K.; Toloza, E.M.; Yao, J.; Kim, S.; Landin, A.M.; Mullinax, J.E.; Saller, J.J.; Saltos, A.N.; et al. Tumor-infiltrating lymphocyte treatment for anti-PD-1-resistant metastatic lung cancer: A phase 1 trial. Nat. Med. 2021, 27, 1410–1418. [Google Scholar] [CrossRef]
- Kershaw, M.H.; Westwood, J.A.; Parker, L.L.; Wang, G.; Eshhar, Z.; Mavroukakis, S.A.; White, D.E.; Wunderlich, J.R.; Canevari, S.; Rogers-Freezer, L.; et al. A phase I study on adoptive immunotherapy using gene-modified T cells for ovarian cancer. Clin. Cancer Res. 2006, 12, 6106–6115. [Google Scholar] [CrossRef]
- Melenhorst, J.J.; Chen, G.M.; Wang, M.; Porter, D.L.; Chen, C.; Collins, M.A.; Gao, P.; Bandyopadhyay, S.; Sun, H.; Zhao, Z.; et al. Decade-long leukaemia remissions with persistence of CD4+ CAR T cells. Nature 2022, 602, 503–509. [Google Scholar] [CrossRef]
- van der Stegen, S.J.; Hamieh, M.; Sadelain, M. The pharmacology of second-generation chimeric antigen receptors. Nat. Rev. Drug Discov. 2015, 14, 499–509. [Google Scholar] [CrossRef]
- Ramos, C.A.; Rouce, R.; Robertson, C.S.; Reyna, A.; Narala, N.; Vyas, G.; Mehta, B.; Zhang, H.; Dakhova, O.; Carrum, G.; et al. In Vivo fate and activity of second- versus third-generation CD19-Specific CAR-T Cells in B cell non-hodgkin’s lymphomas. Mol. Ther. 2018, 26, 2727–2737. [Google Scholar] [CrossRef]
- Chmielewski, M.; Abken, H. TRUCKs: The fourth generation of CARs. Expert. Opin. Biol. Ther. 2015, 15, 1145–1154. [Google Scholar] [CrossRef]
- Jan, M.; Scarfo, I.; Larson, R.C.; Walker, A.; Schmidts, A.; Guirguis, A.A.; Gasser, J.A.; Slabicki, M.; Bouffard, A.A.; Castano, A.P.; et al. Reversible ON- and OFF-switch chimeric antigen receptors controlled by lenalidomide. Sci. Transl. Med. 2021, 13, eabb6295. [Google Scholar] [CrossRef]
- Lim, W.A.; June, C.H. The principles of engineering immune cells to treat cancer. Cell 2017, 168, 724–740. [Google Scholar] [CrossRef]
- Kloss, C.C.; Condomines, M.; Cartellieri, M.; Bachmann, M.; Sadelain, M. Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells. Nat. Biotechnol. 2013, 31, 71–75. [Google Scholar] [CrossRef]
- Fedorov, V.D.; Themeli, M.; Sadelain, M. PD-1- and CTLA-4-based inhibitory chimeric antigen receptors (iCARs) divert off-target immunotherapy responses. Sci. Transl. Med. 2013, 5, 215ra172. [Google Scholar] [CrossRef]
- Cappell, K.M.; Kochenderfer, J.N. Long-term outcomes following CAR T cell therapy: What we know so far. Nat. Rev. Clin. Oncol. 2023, 20, 359–371. [Google Scholar] [CrossRef]
- Orlando, E.J.; Han, X.; Tribouley, C.; Wood, P.A.; Leary, R.J.; Riester, M.; Levine, J.E.; Qayed, M.; Grupp, S.A.; Boyer, M.; et al. Genetic mechanisms of target antigen loss in CAR19 therapy of acute lymphoblastic leukemia. Nat. Med. 2018, 24, 1504–1506. [Google Scholar] [CrossRef]
- Choi, B.D.; Yu, X.; Castano, A.P.; Bouffard, A.A.; Schmidts, A.; Larson, R.C.; Bailey, S.R.; Boroughs, A.C.; Frigault, M.J.; Leick, M.B.; et al. CAR-T cells secreting BiTEs circumvent antigen escape without detectable toxicity. Nat. Biotechnol. 2019, 37, 1049–1058. [Google Scholar] [CrossRef]
- Spiegel, J.Y.; Patel, S.; Muffly, L.; Hossain, N.M.; Oak, J.; Baird, J.H.; Frank, M.J.; Shiraz, P.; Sahaf, B.; Craig, J.; et al. CAR T cells with dual targeting of CD19 and CD22 in adult patients with recurrent or refractory B cell malignancies: A phase 1 trial. Nat. Med. 2021, 27, 1419–1431. [Google Scholar] [CrossRef]
- Brudno, J.N.; Lam, N.; Vanasse, D.; Shen, Y.W.; Rose, J.J.; Rossi, J.; Xue, A.; Bot, A.; Scholler, N.; Mikkilineni, L.; et al. Safety and feasibility of anti-CD19 CAR T cells with fully human binding domains in patients with B-cell lymphoma. Nat. Med. 2020, 26, 270–280. [Google Scholar] [CrossRef]
- Martin, T.; Usmani, S.Z.; Berdeja, J.G.; Agha, M.; Cohen, A.D.; Hari, P.; Avigan, D.; Deol, A.; Htut, M.; Lesokhin, A.; et al. Ciltacabtagene autoleucel, an Anti-B-cell maturation antigen chimeric antigen receptor T-Cell therapy, for relapsed/refractory multiple myeloma: CARTITUDE-1 2-year follow-up. J. Clin. Oncol. 2023, 41, 1265–1274. [Google Scholar] [CrossRef]
- Caruana, I.; Savoldo, B.; Hoyos, V.; Weber, G.; Liu, H.; Kim, E.S.; Ittmann, M.M.; Marchetti, D.; Dotti, G. Heparanase promotes tumor infiltration and antitumor activity of CAR-redirected T lymphocytes. Nat. Med. 2015, 21, 524–529. [Google Scholar] [CrossRef]
- Nguyen, R.; Doubrovina, E.; Mousset, C.M.; Jin, B.Y.; Okada, R.; Zhang, X.; Clavel, A.; Reyes-Gonzalez, J.M.; Dyomin, V.; Diaz, L.; et al. Cooperative armoring of CAR and TCR T-cells by T cell-restricted IL-15 and IL-21 universally enhances solid tumor efficacy. Clin. Cancer Res. 2023, 30, 1555–1566. [Google Scholar] [CrossRef]
- Jin, L.; Tao, H.; Karachi, A.; Long, Y.; Hou, A.Y.; Na, M.; Dyson, K.A.; Grippin, A.J.; Deleyrolle, L.P.; Zhang, W.; et al. CXCR1- or CXCR2-modified CAR T cells co-opt IL-8 for maximal antitumor efficacy in solid tumors. Nat. Commun. 2019, 10, 4016. [Google Scholar] [CrossRef]
- Mackensen, A.; Haanen, J.; Koenecke, C.; Alsdorf, W.; Wagner-Drouet, E.; Borchmann, P.; Heudobler, D.; Ferstl, B.; Klobuch, S.; Bokemeyer, C.; et al. CLDN6-specific CAR-T cells plus amplifying RNA vaccine in relapsed or refractory solid tumors: The phase 1 BNT211-01 trial. Nat. Med. 2023, 29, 2844–2853. [Google Scholar] [CrossRef]
- Curio, S.; Jonsson, G.; Marinovic, S. A summary of current NKG2D-based CAR clinical trials. Immunother. Adv. 2021, 1, ltab018. [Google Scholar] [CrossRef]
- Marofi, F.; Motavalli, R.; Safonov, V.A.; Thangavelu, L.; Yumashev, A.V.; Alexander, M.; Shomali, N.; Chartrand, M.S.; Pathak, Y.; Jarahian, M.; et al. CAR T cells in solid tumors: Challenges and opportunities. Stem Cell Res. Ther. 2021, 12, 81. [Google Scholar] [CrossRef]
- Yeku, O.O.; Purdon, T.J.; Koneru, M.; Spriggs, D.; Brentjens, R.J. Armored CAR T cells enhance antitumor efficacy and overcome the tumor microenvironment. Sci. Rep. 2017, 7, 10541. [Google Scholar] [CrossRef]
- Le, R.Q.; Li, L.; Yuan, W.; Shord, S.S.; Nie, L.; Habtemariam, B.A.; Przepiorka, D.; Farrell, A.T.; Pazdur, R. FDA Approval Summary: Tocilizumab for Treatment of Chimeric Antigen Receptor T Cell-Induced Severe or Life-Threatening Cytokine Release Syndrome. Oncologist 2018, 23, 943–947. [Google Scholar] [CrossRef]
- Jones, B.S.; Lamb, L.S.; Goldman, F.; Di Stasi, A. Improving the safety of cell therapy products by suicide gene transfer. Front. Pharmacol. 2014, 5, 254. [Google Scholar] [CrossRef]
- Sommermeyer, D.; Hudecek, M.; Kosasih, P.L.; Gogishvili, T.; Maloney, D.G.; Turtle, C.J.; Riddell, S.R. Chimeric antigen receptor-modified T cells derived from defined CD8+ and CD4+ subsets confer superior antitumor reactivity in vivo. Leukemia 2016, 30, 492–500. [Google Scholar] [CrossRef]
- Lin, H.; Cheng, J.; Mu, W.; Zhou, J.; Zhu, L. Advances in universal CAR-T Cell therapy. Front. Immunol. 2021, 12, 744823. [Google Scholar] [CrossRef]
- Levine, B.L.; Pasquini, M.C.; Connolly, J.E.; Porter, D.L.; Gustafson, M.P.; Boelens, J.J.; Horwitz, E.M.; Grupp, S.A.; Maus, M.V.; Locke, F.L.; et al. Unanswered questions following reports of secondary malignancies after CAR-T cell therapy. Nat. Med. 2024, 30, 338–341. [Google Scholar] [CrossRef]
- Dagher, O.K.; Posey, A.D., Jr. Forks in the road for CAR T and CAR NK cell cancer therapies. Nat. Immunol. 2023, 24, 1994–2007. [Google Scholar] [CrossRef]
- Duan, S.; Guo, W.; Xu, Z.; He, Y.; Liang, C.; Mo, Y.; Wang, Y.; Xiong, F.; Guo, C.; Li, Y.; et al. Natural killer group 2D receptor and its ligands in cancer immune escape. Mol. Cancer 2019, 18, 29. [Google Scholar] [CrossRef]
- Valeri, A.; Garcia-Ortiz, A.; Castellano, E.; Cordoba, L.; Maroto-Martin, E.; Encinas, J.; Leivas, A.; Rio, P.; Martinez-Lopez, J. Overcoming tumor resistance mechanisms in CAR-NK cell therapy. Front. Immunol. 2022, 13, 953849. [Google Scholar] [CrossRef]
- Liu, E.; Tong, Y.; Dotti, G.; Shaim, H.; Savoldo, B.; Mukherjee, M.; Orange, J.; Wan, X.; Lu, X.; Reynolds, A.; et al. Cord blood NK cells engineered to express IL-15 and a CD19-targeted CAR show long-term persistence and potent antitumor activity. Leukemia 2018, 32, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Teng, K.Y.; Mansour, A.G.; Zhu, Z.; Li, Z.; Tian, L.; Ma, S.; Xu, B.; Lu, T.; Chen, H.; Hou, D.; et al. Off-the-Shelf prostate stem cell antigen-directed chimeric antigen receptor natural killer cell therapy to treat pancreatic cancer. Gastroenterology 2022, 162, 1319–1333. [Google Scholar] [CrossRef]
- Rudek, L.S.; Zimmermann, K.; Galla, M.; Meyer, J.; Kuehle, J.; Stamopoulou, A.; Brand, D.; Sandalcioglu, I.E.; Neyazi, B.; Moritz, T.; et al. Generation of an NFkappaB-Driven Alpharetroviral “All-in-One” vector construct as a potent tool for CAR NK cell therapy. Front. Immunol. 2021, 12, 751138. [Google Scholar] [CrossRef]
- Jochems, C.; Hodge, J.W.; Fantini, M.; Fujii, R.; Morillon, Y.M., 2nd; Greiner, J.W.; Padget, M.R.; Tritsch, S.R.; Tsang, K.Y.; Campbell, K.S.; et al. An NK cell line (haNK) expressing high levels of granzyme and engineered to express the high affinity CD16 allele. Oncotarget 2016, 7, 86359–86373. [Google Scholar] [CrossRef]
- Chandran, S.S.; Klebanoff, C.A. T cell receptor-based cancer immunotherapy: Emerging efficacy and pathways of resistance. Immunol. Rev. 2019, 290, 127–147. [Google Scholar] [CrossRef]
- Salter, A.I.; Rajan, A.; Kennedy, J.J.; Ivey, R.G.; Shelby, S.A.; Leung, I.; Templeton, M.L.; Muhunthan, V.; Voillet, V.; Sommermeyer, D.; et al. Comparative analysis of TCR and CAR signaling informs CAR designs with superior antigen sensitivity and in vivo function. Sci. Signal 2021, 14, eabe2606. [Google Scholar] [CrossRef]
- Campillo-Davo, D.; Flumens, D.; Lion, E. The quest for the best: How TCR affinity, avidity, and functional avidity affect TCR-engineered T-Cell antitumor responses. Cells 2020, 9, 1720. [Google Scholar] [CrossRef]
- Legut, M.; Dolton, G.; Mian, A.A.; Ottmann, O.G.; Sewell, A.K. CRISPR-mediated TCR replacement generates superior anticancer transgenic T cells. Blood 2018, 131, 311–322. [Google Scholar] [CrossRef]
- Haga-Friedman, A.; Horovitz-Fried, M.; Cohen, C.J. Incorporation of transmembrane hydrophobic mutations in the TCR enhance its surface expression and T cell functional avidity. J. Immunol. 2012, 188, 5538–5546. [Google Scholar] [CrossRef]
- Tsimberidou, A.M.; Van Morris, K.; Vo, H.H.; Eck, S.; Lin, Y.F.; Rivas, J.M.; Andersson, B.S. T-cell receptor-based therapy: An innovative therapeutic approach for solid tumors. J. Hematol. Oncol. 2021, 14, 102. [Google Scholar] [CrossRef] [PubMed]
- Linette, G.P.; Stadtmauer, E.A.; Maus, M.V.; Rapoport, A.P.; Levine, B.L.; Emery, L.; Litzky, L.; Bagg, A.; Carreno, B.M.; Cimino, P.J.; et al. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood 2013, 122, 863–871. [Google Scholar] [CrossRef] [PubMed]
- Cameron, B.J.; Gerry, A.B.; Dukes, J.; Harper, J.V.; Kannan, V.; Bianchi, F.C.; Grand, F.; Brewer, J.E.; Gupta, M.; Plesa, G.; et al. Identification of a Titin-derived HLA-A1-presented peptide as a cross-reactive target for engineered MAGE A3-directed T cells. Sci. Transl. Med. 2013, 5, 197ra103. [Google Scholar] [CrossRef] [PubMed]
- Baulu, E.; Gardet, C.; Chuvin, N.; Depil, S. TCR-engineered T cell therapy in solid tumors: State of the art and perspectives. Sci. Adv. 2023, 9, eadf3700. [Google Scholar] [CrossRef] [PubMed]
- Idorn, M.; Skadborg, S.K.; Kellermann, L.; Halldorsdottir, H.R.; Holmen Olofsson, G.; Met, O.; Thor Straten, P. Chemokine receptor engineering of T cells with CXCR2 improves homing towards subcutaneous human melanomas in xenograft mouse model. Oncoimmunology 2018, 7, e1450715. [Google Scholar] [CrossRef] [PubMed]
- Hassan, R.; Butler, M.; O’Cearbhaill, R.E.; Oh, D.Y.; Johnson, M.; Zikaras, K.; Smalley, M.; Ross, M.; Tanyi, J.L.; Ghafoor, A.; et al. Mesothelin-targeting T cell receptor fusion construct cell therapy in refractory solid tumors: Phase 1/2 trial interim results. Nat. Med. 2023, 29, 2099–2109. [Google Scholar] [CrossRef] [PubMed]
- Saxena, M.; van der Burg, S.H.; Melief, C.J.M.; Bhardwaj, N. Therapeutic cancer vaccines. Nat. Rev. Cancer 2021, 21, 360–378. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.J.; Svensson-Arvelund, J.; Lubitz, G.S.; Marabelle, A.; Melero, I.; Brown, B.D.; Brody, J.D. Cancer vaccines: The next immunotherapy frontier. Nat. Cancer 2022, 3, 911–926. [Google Scholar] [CrossRef] [PubMed]
- Leko, V.; Rosenberg, S.A. Identifying and targeting human tumor antigens for T Cell-based immunotherapy of solid tumors. Cancer Cell 2020, 38, 454–472. [Google Scholar] [CrossRef] [PubMed]
- Katanasaka, Y.; Kodera, Y.; Kitamura, Y.; Morimoto, T.; Tamura, T.; Koizumi, F. Epidermal growth factor receptor variant type III markedly accelerates angiogenesis and tumor growth via inducing c-myc mediated angiopoietin-like 4 expression in malignant glioma. Mol. Cancer 2013, 12, 31. [Google Scholar] [CrossRef] [PubMed]
- Tomaic, V. Functional roles of E6 and E7 oncoproteins in HPV-induced malignancies at diverse anatomical sites. Cancers 2016, 8, 95. [Google Scholar] [CrossRef]
- Smith, C.C.; Selitsky, S.R.; Chai, S.; Armistead, P.M.; Vincent, B.G.; Serody, J.S. Alternative tumour-specific antigens. Nat. Rev. Cancer 2019, 19, 465–478. [Google Scholar] [CrossRef]
- Roudko, V.; Bozkus, C.C.; Orfanelli, T.; McClain, C.B.; Carr, C.; O’Donnell, T.; Chakraborty, L.; Samstein, R.; Huang, K.L.; Blank, S.V.; et al. Shared immunogenic poly-epitope frameshift mutations in microsatellite unstable tumors. Cell 2020, 183, 1634–1649.e1617. [Google Scholar] [CrossRef]
- Jiang, T.; Shi, T.; Zhang, H.; Hu, J.; Song, Y.; Wei, J.; Ren, S.; Zhou, C. Tumor neoantigens: From basic research to clinical applications. J. Hematol. Oncol. 2019, 12, 93. [Google Scholar] [CrossRef]
- Ahmadzadeh, M.; Pasetto, A.; Jia, L.; Deniger, D.C.; Stevanovic, S.; Robbins, P.F.; Rosenberg, S.A. Tumor-infiltrating human CD4+ regulatory T cells display a distinct TCR repertoire and exhibit tumor and neoantigen reactivity. Sci. Immunol. 2019, 4, eaao4310. [Google Scholar] [CrossRef]
- Liau, L.M.; Ashkan, K.; Brem, S.; Campian, J.L.; Trusheim, J.E.; Iwamoto, F.M.; Tran, D.D.; Ansstas, G.; Cobbs, C.S.; Heth, J.A.; et al. Association of autologous tumor lysate-loaded dendritic cell vaccination with extension of survival among patients with newly diagnosed and recurrent glioblastoma: A Phase 3 prospective externally controlled cohort trial. JAMA Oncol. 2023, 9, 112–121. [Google Scholar] [CrossRef]
- Kantoff, P.W.; Higano, C.S.; Shore, N.D.; Berger, E.R.; Small, E.J.; Penson, D.F.; Redfern, C.H.; Ferrari, A.C.; Dreicer, R.; Sims, R.B.; et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N. Engl. J. Med. 2010, 363, 411–422. [Google Scholar] [CrossRef] [PubMed]
- Nordin, M.L.; Azemi, A.K.; Nordin, A.H.; Nabgan, W.; Ng, P.Y.; Yusoff, K.; Abu, N.; Lim, K.P.; Zakaria, Z.A.; Ismail, N.; et al. Peptide-Based vaccine against breast cancer: Recent advances and prospects. Pharmaceuticals 2023, 16, 923. [Google Scholar] [CrossRef] [PubMed]
- Mittendorf, E.A.; Lu, B.; Melisko, M.; Price Hiller, J.; Bondarenko, I.; Brunt, A.M.; Sergii, G.; Petrakova, K.; Peoples, G.E. Efficacy and safety analysis of Nelipepimut-S vaccine to prevent breast cancer recurrence: A randomized, multicenter, phase III clinical trial. Clin. Cancer Res. 2019, 25, 4248–4254. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Fu, M.; Wang, M.; Wan, D.; Wei, Y.; Wei, X. Cancer vaccines as promising immuno-therapeutics: Platforms and current progress. J. Hematol. Oncol. 2022, 15, 28. [Google Scholar] [CrossRef]
- Lopes, A.; Vandermeulen, G.; Preat, V. Cancer DNA vaccines: Current preclinical and clinical developments and future perspectives. J. Exp. Clin. Cancer Res. 2019, 38, 146. [Google Scholar] [CrossRef]
- Weber, J.S.; Carlino, M.S.; Khattak, A.; Meniawy, T.; Ansstas, G.; Taylor, M.H.; Kim, K.B.; McKean, M.; Long, G.V.; Sullivan, R.J.; et al. Individualised neoantigen therapy mRNA-4157 (V940) plus pembrolizumab versus pembrolizumab monotherapy in resected melanoma (KEYNOTE-942): A randomised, phase 2b study. Lancet 2024, 403, 632–644. [Google Scholar] [CrossRef]
- Rojas, L.A.; Sethna, Z.; Soares, K.C.; Olcese, C.; Pang, N.; Patterson, E.; Lihm, J.; Ceglia, N.; Guasp, P.; Chu, A.; et al. Personalized RNA neoantigen vaccines stimulate T cells in pancreatic cancer. Nature 2023, 618, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [PubMed]
- Crommelin, D.J.A.; Anchordoquy, T.J.; Volkin, D.B.; Jiskoot, W.; Mastrobattista, E. Addressing the cold reality of mRNA vaccine stability. J. Pharm. Sci. 2021, 110, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Laino, A.S.; Woods, D.; Vassallo, M.; Qian, X.; Tang, H.; Wind-Rotolo, M.; Weber, J. Serum interleukin-6 and C-reactive protein are associated with survival in melanoma patients receiving immune checkpoint inhibition. J. Immunother. Cancer 2020, 8, e000842. [Google Scholar] [CrossRef] [PubMed]
- Berraondo, P.; Sanmamed, M.F.; Ochoa, M.C.; Etxeberria, I.; Aznar, M.A.; Perez-Gracia, J.L.; Rodriguez-Ruiz, M.E.; Ponz-Sarvise, M.; Castanon, E.; Melero, I. Cytokines in clinical cancer immunotherapy. Br. J. Cancer 2019, 120, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Lin, J.X.; Leonard, W.J. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity 2013, 38, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ai, X.; Wu, C.; Wang, H.; Zeng, G.; Yang, P.; Liu, G. A novel human IL-2 mutein with minimal systemic toxicity exerts greater antitumor efficacy than wild-type IL-2. Cell Death Dis. 2018, 9, 989. [Google Scholar] [CrossRef] [PubMed]
- Levin, A.M.; Bates, D.L.; Ring, A.M.; Krieg, C.; Lin, J.T.; Su, L.; Moraga, I.; Raeber, M.E.; Bowman, G.R.; Novick, P.; et al. Exploiting a natural conformational switch to engineer an interleukin-2 ‘superkine’. Nature 2012, 484, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Di Trani, C.A.; Cirella, A.; Arrizabalaga, L.; Fernandez-Sendin, M.; Bella, A.; Aranda, F.; Melero, I.; Berraondo, P. Overcoming the limitations of cytokines to improve cancer therapy. Int. Rev. Cell Mol. Biol. 2022, 369, 107–141. [Google Scholar] [CrossRef]
- Propper, D.J.; Balkwill, F.R. Harnessing cytokines and chemokines for cancer therapy. Nat. Rev. Clin. Oncol. 2022, 19, 237–253. [Google Scholar] [CrossRef]
- Oft, M. Immune regulation and cytotoxic T cell activation of IL-10 agonists—Preclinical and clinical experience. Semin. Immunol. 2019, 44, 101325. [Google Scholar] [CrossRef] [PubMed]
- Hecht, J.R.; Lonardi, S.; Bendell, J.; Sim, H.W.; Macarulla, T.; Lopez, C.D.; Van Cutsem, E.; Munoz Martin, A.J.; Park, J.O.; Greil, R.; et al. Randomized phase III study of FOLFOX alone or with pegilodecakin as second-line therapy in patients with metastatic pancreatic cancer that progressed after gemcitabine (SEQUOIA). J. Clin. Oncol. 2021, 39, 1108–1118. [Google Scholar] [CrossRef] [PubMed]
- Tannir, N.M.; Papadopoulos, K.P.; Wong, D.J.; Aljumaily, R.; Hung, A.; Afable, M.; Kim, J.S.; Ferry, D.; Drakaki, A.; Bendell, J.; et al. Pegilodecakin as monotherapy or in combination with anti-PD-1 or tyrosine kinase inhibitor in heavily pretreated patients with advanced renal cell carcinoma: Final results of cohorts A, G, H and I of IVY Phase I study. Int. J. Cancer 2021, 149, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Siefker-Radtke, A.O.; Cho, D.C.; Diab, A.; Sznol, M.; Bilen, M.A.; Balar, A.V.; Grignani, G.; Puente, E.; Tang, L.; Chien, D.; et al. Bempegaldesleukin plus Nivolumab in First-line Metastatic Urothelial Carcinoma: Results from PIVOT-02. Eur. Urol. 2022, 82, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Shalhout, S.Z.; Miller, D.M.; Emerick, K.S.; Kaufman, H.L. Therapy with oncolytic viruses: Progress and challenges. Nat. Rev. Clin. Oncol. 2023, 20, 160–177. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.C.; Coffin, R.S.; Davis, C.J.; Graham, N.J.; Groves, N.; Guest, P.J.; Harrington, K.J.; James, N.D.; Love, C.A.; McNeish, I.; et al. A phase I study of OncoVEXGM-CSF, a second-generation oncolytic herpes simplex virus expressing granulocyte macrophage colony-stimulating factor. Clin. Cancer Res. 2006, 12, 6737–6747. [Google Scholar] [CrossRef] [PubMed]
- Dummer, R.; Gyorki, D.E.; Hyngstrom, J.; Berger, A.C.; Conry, R.; Demidov, L.; Sharma, A.; Treichel, S.A.; Radcliffe, H.; Gorski, K.S.; et al. Neoadjuvant talimogene laherparepvec plus surgery versus surgery alone for resectable stage IIIB-IVM1a melanoma: A randomized, open-label, phase 2 trial. Nat. Med. 2021, 27, 1789–1796. [Google Scholar] [CrossRef] [PubMed]
- Yaguchi, T.; Goto, Y.; Kido, K.; Mochimaru, H.; Sakurai, T.; Tsukamoto, N.; Kudo-Saito, C.; Fujita, T.; Sumimoto, H.; Kawakami, Y. Immune suppression and resistance mediated by constitutive activation of Wnt/beta-catenin signaling in human melanoma cells. J. Immunol. 2012, 189, 2110–2117. [Google Scholar] [CrossRef] [PubMed]
- Hamarsheh, S.; Gross, O.; Brummer, T.; Zeiser, R. Immune modulatory effects of oncogenic KRAS in cancer. Nat. Commun. 2020, 11, 5439. [Google Scholar] [CrossRef]
- Tripathi, P.; Kumari, R.; Pathak, R. Drugging the undruggable: Advances in targeting KRAS signaling in solid tumors. Int. Rev. Cell Mol. Biol. 2024, 385, 1–39. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, D.; Saw, P.E.; Song, E. Turning cold tumors hot: From molecular mechanisms to clinical applications. Trends Immunol. 2022, 43, 523–545. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.R.; Halladay, T.; Yang, L. Immune evasion in cell-based immunotherapy: Unraveling challenges and novel strategies. J. Biomed. Sci. 2024, 31, 5. [Google Scholar] [CrossRef] [PubMed]
- Motzer, R.J.; Tannir, N.M.; McDermott, D.F.; Aren Frontera, O.; Melichar, B.; Choueiri, T.K.; Plimack, E.R.; Barthelemy, P.; Porta, C.; George, S.; et al. Nivolumab plus Ipilimumab versus Sunitinib in Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2018, 378, 1277–1290. [Google Scholar] [CrossRef] [PubMed]
- Eggermont, A.M.M.; Blank, C.U.; Mandala, M.; Long, G.V.; Atkinson, V.; Dalle, S.; Haydon, A.; Lichinitser, M.; Khattak, A.; Carlino, M.S.; et al. Adjuvant pembrolizumab versus placebo in resected stage III melanoma. N. Engl. J. Med. 2018, 378, 1789–1801. [Google Scholar] [CrossRef] [PubMed]
- Sanmamed, M.F.; Pastor, F.; Rodriguez, A.; Perez-Gracia, J.L.; Rodriguez-Ruiz, M.E.; Jure-Kunkel, M.; Melero, I. Agonists of Co-stimulation in cancer immunotherapy directed against CD137, OX40, GITR, CD27, CD28, and ICOS. Semin. Oncol. 2015, 42, 640–655. [Google Scholar] [CrossRef]
- Esensten, J.H.; Helou, Y.A.; Chopra, G.; Weiss, A.; Bluestone, J.A. CD28 Costimulation: From mechanism to therapy. Immunity 2016, 44, 973–988. [Google Scholar] [CrossRef] [PubMed]
- Hunig, T. The storm has cleared: Lessons from the CD28 superagonist TGN1412 trial. Nat. Rev. Immunol. 2012, 12, 317–318. [Google Scholar] [CrossRef] [PubMed]
- Nagarsheth, N.; Wicha, M.S.; Zou, W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 2017, 17, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Briere, D.M.; Li, S.; Calinisan, A.; Sudhakar, N.; Aranda, R.; Hargis, L.; Peng, D.H.; Deng, J.; Engstrom, L.D.; Hallin, J.; et al. The KRAS(G12C) inhibitor MRTX849 reconditions the tumor immune microenvironment and sensitizes tumors to checkpoint inhibitor therapy. Mol. Cancer Ther. 2021, 20, 975–985. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, G.; Chen, J.; Zhuang, L.; Du, Y.; Yu, Q.; Zhuang, W.; Zhao, Y.; Zhou, M.; Zhang, W.; et al. AK112, a novel PD-1/VEGF bispecific antibody, in combination with chemotherapy in patients with advanced non-small cell lung cancer (NSCLC): An open-label, multicenter, phase II trial. EClinicalMedicine 2023, 62, 102106. [Google Scholar] [CrossRef]
- Sabat, R.; Grutz, G.; Warszawska, K.; Kirsch, S.; Witte, E.; Wolk, K.; Geginat, J. Biology of interleukin-10. Cytokine Growth Factor. Rev. 2010, 21, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Yi, M.; Jiao, Y.; Chu, Q.; Wu, K. Blocking TGF-beta Signaling To Enhance The Efficacy Of Immune Checkpoint Inhibitor. Onco Targets Ther. 2019, 12, 9527–9538. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, M.H.; O’Reilly, E.M.; Varadhachary, G.; Wolff, R.A.; Wainberg, Z.A.; Ko, A.H.; Fisher, G.; Rahma, O.; Lyman, J.P.; Cabanski, C.R.; et al. CD40 agonistic monoclonal antibody APX005M (sotigalimab) and chemotherapy, with or without nivolumab, for the treatment of metastatic pancreatic adenocarcinoma: An open-label, multicentre, phase 1b study. Lancet Oncol. 2021, 22, 118–131. [Google Scholar] [CrossRef] [PubMed]
- Mangsbo, S.M.; Broos, S.; Fletcher, E.; Veitonmaki, N.; Furebring, C.; Dahlen, E.; Norlen, P.; Lindstedt, M.; Totterman, T.H.; Ellmark, P. The human agonistic CD40 antibody ADC-1013 eradicates bladder tumors and generates T-cell-dependent tumor immunity. Clin. Cancer Res. 2015, 21, 1115–1126. [Google Scholar] [CrossRef] [PubMed]
- Duan, Q.; Zhang, H.; Zheng, J.; Zhang, L. Turning Cold into Hot: Firing up the Tumor Microenvironment. Trends Cancer 2020, 6, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Schadt, L.; Sparano, C.; Schweiger, N.A.; Silina, K.; Cecconi, V.; Lucchiari, G.; Yagita, H.; Guggisberg, E.; Saba, S.; Nascakova, Z.; et al. Cancer-Cell-Intrinsic cGAS expression mediates tumor immunogenicity. Cell Rep. 2019, 29, 1236–1248.e1237. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Altman, M.D.; Lesburg, C.A.; Perera, S.A.; Piesvaux, J.A.; Schroeder, G.K.; Wyss, D.F.; Cemerski, S.; Chen, Y.; DiNunzio, E.; et al. Discovery of MK-1454: A Potent Cyclic dinucleotide stimulator of interferon genes agonist for the treatment of cancer. J. Med. Chem. 2022, 65, 5675–5689. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Jang, M.; Tarhan, Y.E.; Katagiri, T.; Sasa, M.; Miyoshi, Y.; Kalari, K.R.; Suman, V.J.; Weinshilboum, R.; Wang, L.; et al. Clonal expansion of antitumor T cells in breast cancer correlates with response to neoadjuvant chemotherapy. Int. J. Oncol. 2016, 49, 471–478. [Google Scholar] [CrossRef]
- Curigliano, G.; Mueller, V.; Borges, V.; Hamilton, E.; Hurvitz, S.; Loi, S.; Murthy, R.; Okines, A.; Paplomata, E.; Cameron, D.; et al. Tucatinib versus placebo added to trastuzumab and capecitabine for patients with pretreated HER2+ metastatic breast cancer with and without brain metastases (HER2CLIMB): Final overall survival analysis. Ann. Oncol. 2022, 33, 321–329. [Google Scholar] [CrossRef]
- Spranger, S.; Dai, D.; Horton, B.; Gajewski, T.F. Tumor-Residing Batf3 dendritic cells are required for effector T Cell trafficking and adoptive T Cell therapy. Cancer Cell 2017, 31, 711–723.e714. [Google Scholar] [CrossRef]
- Kaufman, H.L.; Kohlhapp, F.J.; Zloza, A. Oncolytic viruses: A new class of immunotherapy drugs. Nat. Rev. Drug Discov. 2015, 14, 642–662. [Google Scholar] [CrossRef] [PubMed]
- Chesney, J.; Puzanov, I.; Collichio, F.; Singh, P.; Milhem, M.M.; Glaspy, J.; Hamid, O.; Ross, M.; Friedlander, P.; Garbe, C.; et al. Randomized, open-label phase ii study evaluating the efficacy and safety of talimogene laherparepvec in combination with ipilimumab versus ipilimumab alone in patients with advanced, unresectable melanoma. J. Clin. Oncol. 2018, 36, 1658–1667. [Google Scholar] [CrossRef] [PubMed]
- Ribas, A.; Dummer, R.; Puzanov, I.; VanderWalde, A.; Andtbacka, R.H.I.; Michielin, O.; Olszanski, A.J.; Malvehy, J.; Cebon, J.; Fernandez, E.; et al. Oncolytic virotherapy promotes intratumoral T Cell infiltration and improves Anti-PD-1 immunotherapy. Cell 2017, 170, 1109–1119.e1110. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, A.I.; Chong, C.; Huber, F.; Pak, H.; Stevenson, B.J.; Muller, M.; Michaux, J.; Altimiras, E.R.; Rusakiewicz, S.; Simo-Riudalbas, L.; et al. The immunopeptidome landscape associated with T cell infiltration, inflammation and immune editing in lung cancer. Nat. Cancer 2023, 4, 608–628. [Google Scholar] [CrossRef] [PubMed]
- Saez-Ibanez, A.R.; Upadhaya, S.; Campbell, J. Immuno-oncology clinical trials take a turn beyond PD1/PDL1 inhibitors. Nat. Rev. Drug Discov. 2023, 22, 442–443. [Google Scholar] [CrossRef] [PubMed]
- Saez-Ibanez, A.R.; Sommers, E.; Upadhaya, S.; Campbell, J. PD1/PDL1 clinical trials adapt to a growing landscape of patients resistant to PDx. Nat. Rev. Drug Discov. 2023, 22, 944–945. [Google Scholar] [CrossRef] [PubMed]
- van der Burg, S.H. Correlates of immune and clinical activity of novel cancer vaccines. Semin. Immunol. 2018, 39, 119–136. [Google Scholar] [CrossRef] [PubMed]
- Said, S.S.; Ibrahim, W.N. Cancer resistance to immunotherapy: Comprehensive insights with future perspectives. Pharmaceutics 2023, 15, 1143. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef]
- Wu, P.; Gao, W.; Su, M.; Nice, E.C.; Zhang, W.; Lin, J.; Xie, N. Adaptive mechanisms of tumor therapy resistance driven by tumor microenvironment. Front. Cell Dev. Biol. 2021, 9, 641469. [Google Scholar] [CrossRef]
- Bazzichetto, C.; Conciatori, F.; Luchini, C.; Simionato, F.; Santoro, R.; Vaccaro, V.; Corbo, V.; Falcone, I.; Ferretti, G.; Cognetti, F.; et al. From Genetic alterations to tumor microenvironment: The Ariadne’s string in pancreatic cancer. Cells 2020, 9, 309. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Li, F.; Li, F.; Gong, A. Targeting tumor vascularization: Promising strategies for vascular normalization. J. Cancer Res. Clin. Oncol. 2021, 147, 2489–2505. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, T.; Sheta, M.; Fujii, M.; Calderwood, S.K. Cancer extracellular vesicles, tumoroid models, and tumor microenvironment. Semin. Cancer Biol. 2022, 86, 112–126. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.N.; Higashi, R.M.; Fan, T.W. Metabolic reprogramming in tumors: Contributions of the tumor microenvironment. Genes Dis. 2020, 7, 185–198. [Google Scholar] [CrossRef] [PubMed]
- van Herk, E.H.; Te Velde, A.A. Treg subsets in inflammatory bowel disease and colorectal carcinoma: Characteristics, role, and therapeutic targets. J. Gastroenterol. Hepatol. 2016, 31, 1393–1404. [Google Scholar] [CrossRef] [PubMed]
- Olguin, J.E.; Medina-Andrade, I.; Rodriguez, T.; Rodriguez-Sosa, M.; Terrazas, L.I. Relevance of Regulatory T Cells during colorectal cancer development. Cancers 2020, 12, 1888. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y. Spatial heterogeneity in the tumor microenvironment. Cold Spring Harb. Perspect. Med. 2016, 6, a026583. [Google Scholar] [CrossRef]
- Ramon, Y.C.S.; Sese, M.; Capdevila, C.; Aasen, T.; De Mattos-Arruda, L.; Diaz-Cano, S.J.; Hernandez-Losa, J.; Castellvi, J. Clinical implications of intratumor heterogeneity: Challenges and opportunities. J. Mol. Med. 2020, 98, 161–177. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Tang, L.H.; Klimstra, D.S. Effect of tumor heterogeneity on the assessment of Ki67 labeling index in well-differentiated neuroendocrine tumors metastatic to the liver: Implications for prognostic stratification. Am. J. Surg. Pathol. 2011, 35, 853–860. [Google Scholar] [CrossRef]
- Hockel, M.; Schlenger, K.; Aral, B.; Mitze, M.; Schaffer, U.; Vaupel, P. Association between tumor hypoxia and malignant progression in advanced cancer of the uterine cervix. Cancer Res. 1996, 56, 4509–4515. [Google Scholar]
- Tang, S.; Ning, Q.; Yang, L.; Mo, Z.; Tang, S. Mechanisms of immune escape in the cancer immune cycle. Int. Immunopharmacol. 2020, 86, 106700. [Google Scholar] [CrossRef]
- Zhang, A.; Miao, K.; Sun, H.; Deng, C.X. Tumor heterogeneity reshapes the tumor microenvironment to influence drug resistance. Int. J. Biol. Sci. 2022, 18, 3019–3033. [Google Scholar] [CrossRef]
- Marusyk, A.; Janiszewska, M.; Polyak, K. Intratumor heterogeneity: The rosetta stone of therapy resistance. Cancer Cell 2020, 37, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Sobti, A.; Sakellariou, C.; Nilsson, J.S.; Askmyr, D.; Greiff, L.; Lindstedt, M. Exploring spatial heterogeneity of immune cells in nasopharyngeal cancer. Cancers 2023, 15, 2165. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Ni, Y.; Wang, Y.; Feng, J.; Zhuang, N.; Li, J.; Liu, L.; Shen, W.; Zheng, J.; Zheng, W.; et al. Spatial heterogeneity of tumor microenvironment influences the prognosis of clear cell renal cell carcinoma. J. Transl. Med. 2023, 21, 489. [Google Scholar] [CrossRef]
- Ganguly, A.; Mukherjee, S.; Spada, S. Editorial: Spatial immune cell heterogeneity in the tumor microenvironment. Front. Immunol. 2024, 15, 1377532. [Google Scholar] [CrossRef]
- Dhatchinamoorthy, K.; Colbert, J.D.; Rock, K.L. Cancer immune evasion through loss of MHC Class I antigen presentation. Front. Immunol. 2021, 12, 636568. [Google Scholar] [CrossRef] [PubMed]
- Rock, K.L.; Gramm, C.; Rothstein, L.; Clark, K.; Stein, R.; Dick, L.; Hwang, D.; Goldberg, A.L. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 1994, 78, 761–771. [Google Scholar] [CrossRef]
- Wei, J.; Zanker, D.; Di Carluccio, A.R.; Smelkinson, M.G.; Takeda, K.; Seedhom, M.O.; Dersh, D.; Gibbs, J.S.; Yang, N.; Jadhav, A.; et al. Varied role of ubiquitylation in generating MHC Class I peptide ligands. J. Immunol. 2017, 198, 3835–3845. [Google Scholar] [CrossRef]
- Shen, L.; Rock, K.L. Cellular protein is the source of cross-priming antigen in vivo. Proc. Natl. Acad. Sci. USA 2004, 101, 3035–3040. [Google Scholar] [CrossRef]
- Bai, J.; Gao, Z.; Li, X.; Dong, L.; Han, W.; Nie, J. Regulation of PD-1/PD-L1 pathway and resistance to PD-1/PD-L1 blockade. Oncotarget 2017, 8, 110693–110707. [Google Scholar] [CrossRef] [PubMed]
- Sade-Feldman, M.; Jiao, Y.J.; Chen, J.H.; Rooney, M.S.; Barzily-Rokni, M.; Eliane, J.P.; Bjorgaard, S.L.; Hammond, M.R.; Vitzthum, H.; Blackmon, S.M.; et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat. Commun. 2017, 8, 1136. [Google Scholar] [CrossRef] [PubMed]
- Reis, B.; Attig, J.; Dziadek, S.; Graefe, N.; Heller, A.; Rieder, N.; Gomes, B. Tumor beta2-microglobulin and HLA-A expression is increased by immunotherapy and can predict response to CIT in association with other biomarkers. Front. Immunol. 2024, 15, 1285049. [Google Scholar] [CrossRef] [PubMed]
- Roehle, K.; Qiang, L.; Ventre, K.S.; Heid, D.; Ali, L.R.; Lenehan, P.; Heckler, M.; Crowley, S.J.; Stump, C.T.; Ro, G.; et al. cIAP1/2 antagonism eliminates MHC class I-negative tumors through T cell-dependent reprogramming of mononuclear phagocytes. Sci. Transl. Med. 2021, 13, eabf5058. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.H.; Keam, B.; Ahn, Y.O.; Park, H.R.; Kim, M.; Kim, T.M.; Kim, D.W.; Heo, D.S. Inhibition of MEK with trametinib enhances the efficacy of anti-PD-L1 inhibitor by regulating anti-tumor immunity in head and neck squamous cell carcinoma. Oncoimmunology 2019, 8, e1515057. [Google Scholar] [CrossRef]
- Liu, C.; Liu, X.; Xiang, X.; Pang, X.; Chen, S.; Zhang, Y.; Ren, E.; Zhang, L.; Liu, X.; Lv, P.; et al. A nanovaccine for antigen self-presentation and immunosuppression reversal as a personalized cancer immunotherapy strategy. Nat. Nanotechnol. 2022, 17, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Bao, P.; Gu, H.Y.; Ye, J.J.; He, J.L.; Zhong, Z.; Yu, A.X.; Zhang, X.Z. Chimeric Exosomes Functionalized with STING Activation for Personalized Glioblastoma Immunotherapy. Adv. Sci. 2024, 11, e2306336. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Shi, L.Z.; Zhao, H.; Chen, J.; Xiong, L.; He, Q.; Chen, T.; Roszik, J.; Bernatchez, C.; Woodman, S.E.; et al. Loss of IFN-gamma Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy. Cell 2016, 167, 397–404.e399. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Allison, J.P. The future of immune checkpoint therapy. Science 2015, 348, 56–61. [Google Scholar] [CrossRef]
- Jiang, Y.; Zheng, Y.; Zhang, Y.W.; Kong, S.; Dong, J.; Wang, F.; Ziman, B.; Gery, S.; Hao, J.J.; Zhou, D.; et al. Reciprocal inhibition between TP63 and STAT1 regulates anti-tumor immune response through interferon-gamma signaling in squamous cancer. Nat. Commun. 2024, 15, 2484. [Google Scholar] [CrossRef]
- Taube, J.M.; Anders, R.A.; Young, G.D.; Xu, H.; Sharma, R.; McMiller, T.L.; Chen, S.; Klein, A.P.; Pardoll, D.M.; Topalian, S.L.; et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci. Transl. Med. 2012, 4, 127ra137. [Google Scholar] [CrossRef] [PubMed]
- Platanias, L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.S.; Zaretsky, J.M.; Escuin-Ordinas, H.; Garcia-Diaz, A.; Hu-Lieskovan, S.; Kalbasi, A.; Grasso, C.S.; Hugo, W.; Sandoval, S.; Torrejon, D.Y.; et al. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov. 2017, 7, 188–201. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H.; Old, L.J.; Schreiber, R.D. The roles of IFN gamma in protection against tumor development and cancer immunoediting. Cytokine Growth Factor. Rev. 2002, 13, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Hotchkiss, K.M.; Patel, K.K.; Wilkinson, D.S.; Mohan, A.A.; Cook, S.L.; Sampson, J.H. Enhancing T Cell Chemotaxis and Infiltration in Glioblastoma. Cancers 2021, 13, 5367. [Google Scholar] [CrossRef] [PubMed]
- Prendergast, G.C.; Smith, C.; Thomas, S.; Mandik-Nayak, L.; Laury-Kleintop, L.; Metz, R.; Muller, A.J. Indoleamine 2,3-dioxygenase pathways of pathogenic inflammation and immune escape in cancer. Cancer Immunol. Immunother. 2014, 63, 721–735. [Google Scholar] [CrossRef] [PubMed]
- Toulmonde, M.; Penel, N.; Adam, J.; Chevreau, C.; Blay, J.Y.; Le Cesne, A.; Bompas, E.; Piperno-Neumann, S.; Cousin, S.; Grellety, T.; et al. Use of PD-1 targeting, macrophage infiltration, and IDO pathway activation in sarcomas: A Phase 2 clinical trial. JAMA Oncol. 2018, 4, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Fares, C.M.; Van Allen, E.M.; Drake, C.G.; Allison, J.P.; Hu-Lieskovan, S. Mechanisms of resistance to immune checkpoint blockade: Why does checkpoint inhibitor immunotherapy not work for all patients? Am. Soc. Clin. Oncol. Educ. Book. 2019, 39, 147–164. [Google Scholar] [CrossRef] [PubMed]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef]
- Jia, J.; Zhou, X.; Chu, Q. Mechanisms and therapeutic prospect of the JAK-STAT signaling pathway in liver cancer. Mol. Cell Biochem. 2024. [Google Scholar] [CrossRef]
- Sucker, A.; Zhao, F.; Pieper, N.; Heeke, C.; Maltaner, R.; Stadtler, N.; Real, B.; Bielefeld, N.; Howe, S.; Weide, B.; et al. Acquired IFNgamma resistance impairs anti-tumor immunity and gives rise to T-cell-resistant melanoma lesions. Nat. Commun. 2017, 8, 15440. [Google Scholar] [CrossRef] [PubMed]
- Kalbasi, A.; Tariveranmoshabad, M.; Hakimi, K.; Kremer, S.; Campbell, K.M.; Funes, J.M.; Vega-Crespo, A.; Parisi, G.; Champekar, A.; Nguyen, C.; et al. Uncoupling interferon signaling and antigen presentation to overcome immunotherapy resistance due to JAK1 loss in melanoma. Sci. Transl. Med. 2020, 12, eabb0152. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Kumar, M.; Pathak, R.; Bala, K.; Kumar, A. Exploring TLR signaling pathways as promising targets in cervical cancer: The road less traveled. Int. Rev. Cell Mol. Biol. 2024, 385, 227–261. [Google Scholar] [CrossRef]
- Topalian, S.L.; Drake, C.G.; Pardoll, D.M. Targeting the PD-1/B7-H1(PD-L1) pathway to activate anti-tumor immunity. Curr. Opin. Immunol. 2012, 24, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Volovat, S.R.; Scripcariu, D.V.; Vasilache, I.A.; Stolniceanu, C.R.; Volovat, C.; Augustin, I.G.; Volovat, C.C.; Ostafe, M.R.; Andreea-Voichita, S.G.; Bejusca-Vieriu, T.; et al. Oncolytic virotherapy: A new paradigm in cancer immunotherapy. Int. J. Mol. Sci. 2024, 25, 1180. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Che, X.; Wang, X.; Ma, C.; Wu, G. Tumor vaccines: Unleashing the power of the immune system to fight cancer. Pharmaceuticals 2023, 16, 1384. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yan, Q.; Zeng, Z.; Fan, C.; Xiong, W. Advances and prospects of mRNA vaccines in cancer immunotherapy. Biochim. Biophys. Acta Rev. Cancer 2024, 1879, 189068. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yang, M.; Zhang, D.; Chen, M.; Zhu, D. Clinical cancer immunotherapy: Current progress and prospects. Front. Immunol. 2022, 13, 961805. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Singleton, S.S.; Bhuiyan, U.; Krammer, L.; Mazumder, R. Multi-omics approaches to studying gastrointestinal microbiome in the context of precision medicine and machine learning. Front. Mol. Biosci. 2023, 10, 1337373. [Google Scholar] [CrossRef]
- Voith von Voithenberg, L.; Fomitcheva Khartchenko, A.; Huber, D.; Schraml, P.; Kaigala, G.V. Spatially multiplexed RNA in situ hybridization to reveal tumor heterogeneity. Nucleic Acids Res. 2020, 48, e17. [Google Scholar] [CrossRef]
- Kumar, G.; Pandurengan, R.K.; Parra, E.R.; Kannan, K.; Haymaker, C. Spatial modelling of the tumor microenvironment from multiplex immunofluorescence images: Methods and applications. Front. Immunol. 2023, 14, 1288802. [Google Scholar] [CrossRef] [PubMed]
- Kong, B.; Kim, Y.; Kim, E.H.; Suk, J.S.; Yang, Y. mRNA: A promising platform for cancer immunotherapy. Adv. Drug Deliv. Rev. 2023, 199, 114993. [Google Scholar] [CrossRef] [PubMed]
- Blanco, E.; Shen, H.; Ferrari, M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 2015, 33, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Jin, Y.; Jin, C. A new approach to overcoming resistance to immunotherapy: Nanotechnology. Front. Oncol. 2023, 13, 1210245. [Google Scholar] [CrossRef]
- Thakur, N.; Thakur, S.; Chatterjee, S.; Das, J.; Sil, P.C. Nanoparticles as smart carriers for enhanced cancer immunotherapy. Front. Chem. 2020, 8, 597806. [Google Scholar] [CrossRef]
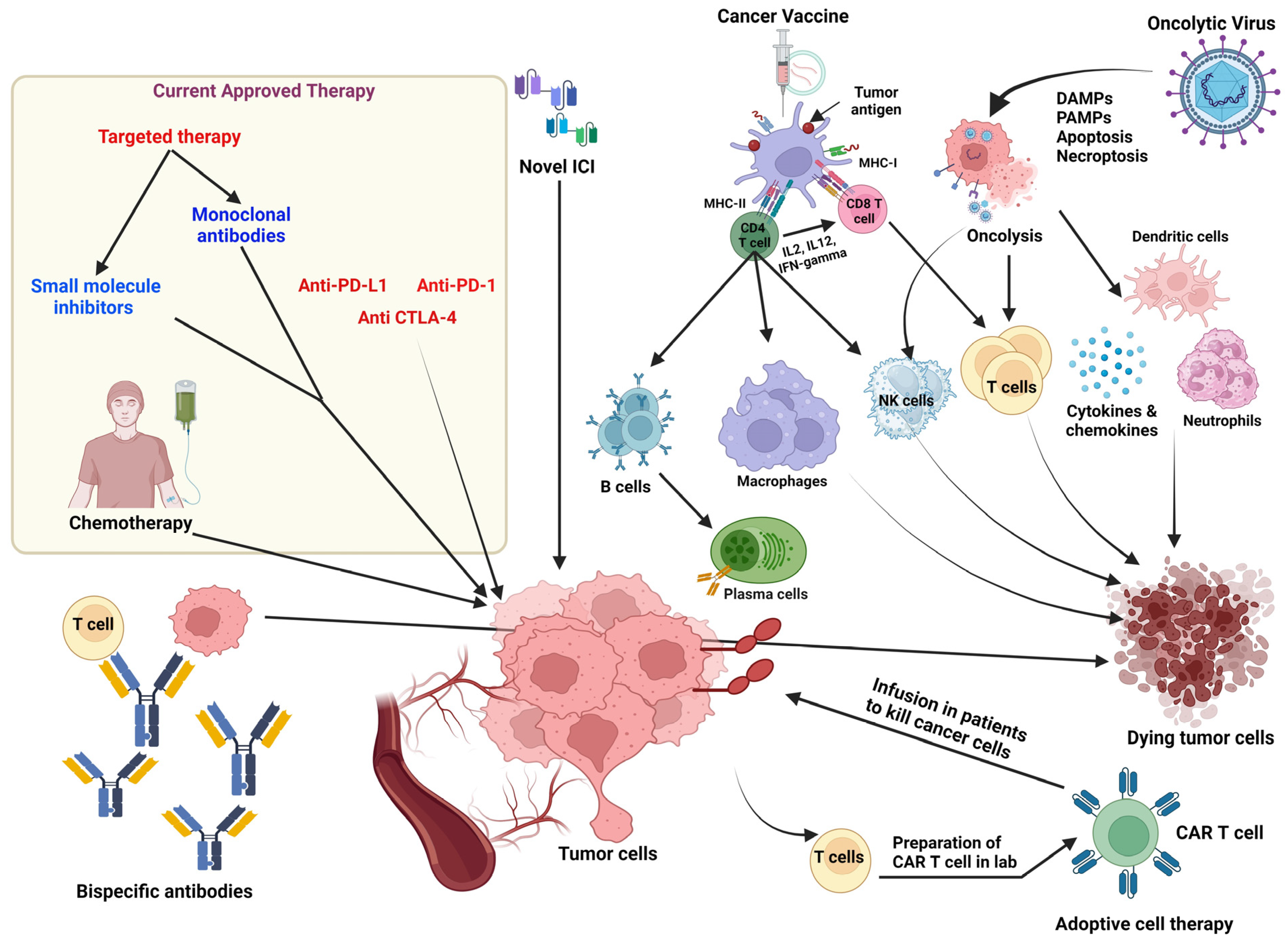
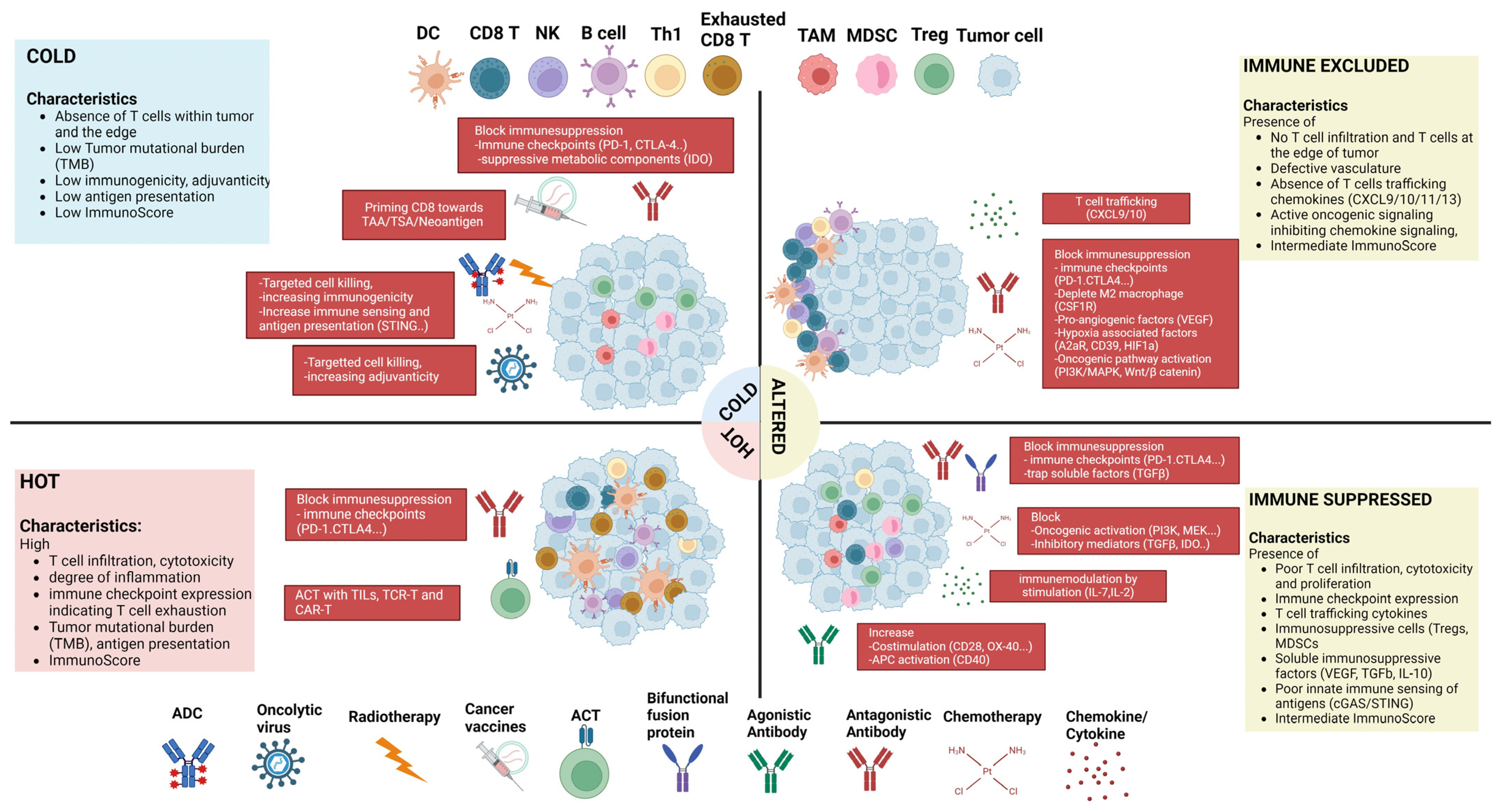
| Condition | Intervention | Format | Gov. Identifier | Phase | Status | Enrollment |
|---|---|---|---|---|---|---|
| Immune checkpoint inhibitors | ||||||
| Metastatic hepatocellular carcinoma (HCC) | Cobolimab (anti Tim3), Dostarlimab (anti PD-1) | Humanized IgG4 | NCT03680508 | II | Recruiting | 42 |
| Advanced solid tumor | BND-22 (anti-LILRB1) monotherapy or in combination with Pembrolizumab (anti-PD-1) or Cetuximab (anti EGFR) | Humanized IgG4 | NCT04717375 | I/II | Recruiting | 456 |
| Stage IV NSCLC | Nivolumab Ipilimumab plus chemotherapy (Carboplatin Paclitaxel Pemetrexed Cisplatin) | -- | NCT03215706 | III | Active, not recruiting | 719 |
| Metastatic squamous HNSCC | Durvalumab in combination with small-molecule antagonists of CXCR2 and STAT3 | IgG1 | NCT02499328 | I/II | Active, not recruiting | 340 |
| Hormone-sensitive prostate cancer | Nivolumab monotherapy or in combination with BMS-986253 (anti-IL8) | -- | NCT03689699 | I/II | Active, not recruiting | 60 |
| Advanced solid tumor | Uliledlimab (anti- CD73) Toripalimab (anti-PD-1) | Humanized mAb | NCT04322006 | I/II | Recruiting | 376 |
| Antibody–drug conjugates | ||||||
| HER2-positive and PD-L1-positive locally advanced or metastatic breast cancer | T-DM1 (Trastuzumab Emtansine) Atelizumab (anti PD-L 1) | - | NCT04740918 | III | Active, not recruiting | 96 |
| Pre-treated HER2 breast cancer | T-Dxd (Trastuzumab Deruxtecan) Capecitabine Lapatinib Trastuzumab | - | NCT03523585 | III | Active, not recruiting | 608 |
| Advanced solid tumor | SGN-B7H4V (B7-H4 targeted ADC) | - | NCT05194072 | I | Recruiting | 430 |
| Relapsed or refractory DLBCL | Loncastuximab Tesirine in combination with Rituximab | NCT04384484 | III | Recruiting | 350 | |
| Bispecific Antibodies (bsAb) | ||||||
| Advanced solid tumors | IOS-1002 (anti- LILRB1, LILRB2, KIR3DL1) monotherapy or in combination with Pembrolizumab | HLA-B57-Fc fusion protein | NCT05763004 | Ia/Ib | Recruiting | 140 |
| CDX585 (anti-LILRB2, PD-1) | IgG1k | NCT05788484 | I | Recruiting | 130 | |
| Advanced hepatobiliary cancer | Volrustomig (anti-PD-1, CTLA-4) Or Rilvegostomig (anti-PD-1, TIGIT) as monotherapy or in combination With Carboplatin Gemcitabine Cisplatin |
IgG1 Humanized IgG1 | NCT05775159 | II | Recruiting | 260 |
| High risk locally advanced cervical cancer | Volrustomig (anti- PD-1, CTLA-4) | IgG1 | NCT06079671 | III | Recruiting | 1000 |
| EGFR-mutant locally advanced or metastatic non-squamous NSCLC | Ivonescimab (anti PD-1/VEGF) | Humanized IgG1 | NCT05184712 | III | Recruiting | 470 |
| Refractory small cell lung cancer (SCLC) | Tarlatamab (engages DLL3, CD3) | bsTCE | NCT05060016 | II | Active, not recruiting | 222 |
| Metastatic castration-resistant prostate cancer (mCRPC) | Gammabody® (engages Vγ2Vδ9, PSMA) | Bs γδTCE | NCT05369000 | I/II | Recruiting | 66 |
| Other monoclonal antibodies (mAbs) | ||||||
| Metastatic HCC | Humax-IL8 (anti IL-8) or Cabiralizumab (anti-CSF1-R) in combination with Nivolumab |
Humanized IgG1K -- | NCT04050462 | II | Active, not recruiting | 23 |
| Treatment naïve advanced melanoma or RCC | Sotigalimab (CD40 agonist mAb)\ Nivolumab Ipilimumab | -- | NCT04495257 | I | Recruiting | 36 |
| Condition | Intervention | Gov. Identifier | Phase | Status | Enrollment |
|---|---|---|---|---|---|
| CAR-T | |||||
| GD2-expressing brain tumor | GD2 CAR-T | NCT03373097 | I | Recruiting | 34 |
| CLDN6-positive relapsed or refractory advanced solid tumors | CLDN6 CAR-T CLDN6 uRNA-LPX/CLDN6 modRNA-LPX (mRNA vaccine) | NCT04503278 | I/IIa | Recruiting | 145 |
| Children with recurrent/refractory malignant brain tumors | IL13Ralpha2 targeting Hinge-optimized 41BB-co-stimulatory CD19-CAR-T Fludarabine Cyclophosphamide | NCT04510051 | I | Recruiting | 18 |
| Multiple myeloma | SLAMF7 CAR-T BCMA CAR-T Bortezomib Dexamethasone Lenalidomide Cyclophosphamide Fludarabine | NCT04499339 NCT04923893 | I/IIa III | Recruiting | 38 650 |
| CD30+ refractory/relapsed Hodgkin lymphoma (HL) and non-Hodgkin lymphoma (NHL) | CD30 CAR-T | NCT02690545 | Ib/II | Recruiting | 40 |
| Relapsed or refractory B-cell lymphoma | CD19-CD22 CAR-T | NCT04715217 | I/II | Recruiting | 24 |
| Malignant pleural disease | Mesothelin CAR-T Cyclophosphamide Pembrolizumab | NCT02414269 | I/II | Active, not recruiting | 113 |
| CD70-expressing cancers | CD70 CAR-T | NCT02830724 | I/II | Recruiting | 124 |
| CAR-NK | |||||
| Stage IV ovarian cancer, refractory testis cancer, recurrent endometrial cancer CAR NK | CLDN6 targeting CAR-NK cells | NCT05410717 | I/IIa | Recruiting | 200 |
| Metastatic locally advanced gastric/GEJ cancer or HNSCC | PD-L1 t-haNK N-803 Pembrolizumab (anti-PD-1) | NCT04847466 | II | Recruiting | 55 |
| Relapsed/refractory NHL, CLL) or B-cell acute lymphoblastic leukemia (B-ALL) | Allogenic CD19-CAR-NK | NCT05020678 | I | Recruiting | 150 |
| Refractory metastatic colorectal cancer | NKG2D CAR-NK | NCT05213195 | I | Recruiting | 38 |
| CAR-Gamma Delta T cells | |||||
| B-cell malignancy | CD20 Allogenic Gamma Delta CAR-T Fludarabine Cyclophosphamide | NCT04735471 | I | Recruiting | 78 |
| TCR-T | |||||
| Induction therapy prior to definitive treatment (chemoradiation or surgery) of locoregionally advanced HPV-associated cancers | Conditioning, E7 TCR-T cells Aldesleukin (IL-2) | NCT05639972 | I/II | Recruiting | 15 |
| TCR against neoantigens in subjects with relapsed/refractory solid tumors (gynecologic colorectal, pancreatic, NSCLC, ovarian cancer) | Neoantigen-specific TCR-T cell Aldesleukin | NCT05194735 | I/II | Active, not recruiting | 180 |
| Previously untreated advanced melanoma | PRAM (PReferentially expressed antigen in Melanoma) TCR-T Nivolumab (anti PD-1) Nivolumab + Relatlimab (anti LAG3) | NCT05122221 | III | Recruiting | 12 |
| Refractory mesothelin-expressing mesothelioma (MPM), ovarian cancer (OvC), cholangiocarcinoma (CHO), or NSCLC | Gavocabtagene autoleucel (Mesothelin targeted TCR-T) Fludarabine Cyclophosphamide Nivolumab Ipilimumab | NCT03907852 | II | Recruiting | 175 |
| KRAS G12V-expressing solid tumors | KRAS G12V targeted TCR-T | NCT06105021 | I/II | Recruiting | 100 |
| Tumor-Infiltrating Lymphocyte (TIL) therapy | |||||
| Metastatic CRC, ovarian, pancreatic, and breast cancer | Young TIL Pembrolizumab Aldesleukin Chemotherapy | NCT01174121 | II | Recruiting | 332 |
| Recurrent, metastatic, or persistent cervical carcinoma | Autologous TIL (IL-145) Pembrolizumab IL-2 | NCT03108495 | II | Recruiting | 189 |
| Condition | Intervention | Adjuvant | Covt. Identifier | Phase | Status | Enrollment |
|---|---|---|---|---|---|---|
| mRNA vaccine | ||||||
| Advanced melanoma | Neoantigen mRNA Pembrolizumab | - | NCT05933577 | III | Recruiting | 1089 |
| TriMix DC (MAGE-A3, MAGE-C2, tyrosinase and gp100) Ipilimumab | CD70, CD40 ligand, TLR4 | NCT01302496 | II | Recruiting | 39 | |
| Neoantigen mRNA Pembrolizumab | -- | NCT03815058 | II | Recruiting | 131 | |
| Resected Stage II (High Risk) and Stage III CRC | Neoantigen mRNA Pembrolizumab | - | NCT04486378 | II | Recruiting | 201 |
| Unresectable recurrent or metastatic HPV16+ HNSCC | HPV 16 E6 and E7 mRNA Pembrolizumab | - | NCT04534205 | II | Recruiting | 285 |
| NSCLC | NY-ESO-1, MAGEC1, MAGEC2, 5 T4, survivin, and MUC1 mRNA Durvalumab | - | NCT03164772 | I/II | Recruiting | 61 |
| Advanced malignant solid tumors. | Neoantigen mRNA | - | NCT05198752 | I | Recruiting | 30 |
| DNA vaccine | ||||||
| Advanced hepatocellular carcinoma | Neoantigen DNA | Plasmid Encoded IL-12 | NCT04251117 | I/IIa | Recruiting | 36 |
| CRC | OncoMimics™ peptides, UCP2 Nivolumab | Montanide | NCT05350501 | II | Recruiting | 34 |
| Resectable HPV Type 16- and/or 18-positive head and neck cancer | HPV16/18 E6/E7 DNA | Flt3L | NCT05286060 | II | Recruiting | 25 |
| Early stage TNBC | MDM2, YB1, SOX2, CDC25B, CD105 plasmid | GM-CSF | NCT05455658 | II | Recruiting | 33 |
| Peptide vaccine | ||||||
| IIIC-IV melanoma or hormone receptor-positive Her2-negative metastatic refractory breast cancer | Neoantigen peptide Nivolumab | Poly ICLC | NCT05098210 | I | Recruiting | 20 |
| Advanced solid tumor | Neoantigen peptide Pembrolizumab | -- | NCT05269381 | I | Recruiting | 36 |
| Condition | Intervention | Govt. Identifier | Phase | Status | Enrollment |
|---|---|---|---|---|---|
| Oncolytic viruses | |||||
| Pancreatic adenocarcinoma, ovarian, biliary, and colorectal cancer | LOAd703 (oAD/CD40L-4-1BBL) | NCT03225989 | I/II | Active not, recruiting | 46 |
| Metastatic pancreatic cancer | VCN-01(oAD/HA) Nab-Paclitaxel Gemcitabine | NCT05673811 | II | Active not, recruiting | 96 |
| Localized prostate cancer | ProstAtak (aglatimagene besadenovec + Valacyclovir | NCT01436968 | III | Active, not recruiting | 711 |
| Engineered cytokines | |||||
| Metastatic castration sensitive and castration-resistant prostate cancer | NHS-IL12 (IL-12 molecules fused to anti-NHS76) | NCT04633252 | I/II | Recruiting | 86 |
| Advanced Kaposi sarcoma | NHS-IL12 monotherapy or in combination with M7824 (anti PD-L1/TGFβ TRAP) | NCT04303117 | I/II | Recruiting | 64 |
| Locally advanced or metastatic solid tumors | IL-15–IL-15Rα (sushi) heterodimer (IL15 superagonist) | NCT04250155 | I | Recruiting | 250 |
| Advanced solid tumor | Pegilodecakin (PEG-rIL-10) | NCT02009449 | I | Active, not recruiting | 350 |
| Combination Agent | Conditions |
|---|---|
| Nivolumab + Ipilimumab | Metastatic melanoma, advanced RCC, MSI-H or dMMR metastatic CRC, advanced HCC, metastatic NSCLC (PD-L1 tumor expression ≥1%), unresectable malignant pleural mesothelioma, metastatic ESCC |
| Nivolumab + Chemotherapy | Metastatic ESCC, metastatic gastric cancer Neo |
| Nivolumab + Ipilimumab + 2 Cycles of Platinum-Doublet chemotherapy | First-line metastatic or recurrent NSCLC, esophageal or GEJ carcinoma |
| Nivolumab + Relatlimab-rmbw | Unresectable/metastatic melanoma |
| Pembrolizumab + Chemotherapy | HNSCC, metastatic SCLC, high-risk early-stage and locally recurrent unresectable/metastatic TNBC, advanced unresectable/metastatic HER2-negative GEJ adenocarcinoma, first-line metastatic non-squamous NSCLC, locally advanced unresectable/metastatic BTC |
| Pembrolizumab + Chemoradiotherapy | FIGO 2014 Stage III-IVA cervical cancer |
| Pembrolizumab + Axitinib | First-line advanced RCC |
| Pembrolizumab + Lenvatinib | Non-MSI-H or dMMR advanced endometrial carcinoma, first-line advanced RCC |
| Pembrolizumab + Chemotherapy + Bevacizumab | Metastatic cervical cancer |
| Pembrolizumab + Trastuzumab + Chemotherapy | First-line locally advanced unresectable/metastatic PD-L1-positive HER2-positive GEJ adenocarcinoma |
| toripalimab-tpzi + Chemotherapy | Metastatic or recurrent, locally advanced NPC |
| Durvalumab + Chemotherapy | Locally advanced or metastatic BTC |
| Durvalumab + Chemoradiotherapy | Unresectable Stage III NSCLC |
| Atezolizumab + Bevacizumab + Chemotherapy | First-line metastatic non-squamous NSCLC |
| Atezolizumab + Bevacizumab | First-line unresectable HCC |
| Atezolizumab + chemotherapy | First-line Extensive stage SCLC, metastatic NSCLC without EGFR/ALK aberrations Metastatic TNBC |
| Atezolizumab + Cobimetinib + Vemurafenib | BRAFV600 mutation-positive advanced melanoma |
| Avelumab + Axitinib | Advanced RCC |
| Avelumab + Chemotherapy | Locally advanced or metastatic UC |
| Tremelimumab + Durvalumab | Unresectable HCC |
| Tremelimumab + Durvalumab + Chemotherapy | Metastatic NSCLC |
| Daratumumab + bortezomib + dexamethasone + thalidomide | MM |
| Elotuzumab + Lenalidomide + Dexamethasone | Newly diagnosed MM |
| Efortumab vedotin-ejfv (ADC) + Pembrolizumab | Locally advanced or metastatic UC |
| Polatuzumab vedotin + Rituximab + Chemotherapy | previously untreated DLBCL |
| Anti-PD-1 (Nivolumab, Pembrolizumab, Toripalimab-tpzi), anti-CTLA-4 (Ipilimumab, Tremelimumab), anti-PD-L1 (Atezolizumab, Avelumab, Durvalumab), anti-LAG3 (Relatlimab), anti-VEGF (Bevacizumab), anti-HER2 (Trastuzumab), anti-CD38 (Daratumumab), anti-SLAMF7 (Elutuzumab), CD-79b directed ADC (Polatuzumab vedotin) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mitra, A.; Kumar, A.; Amdare, N.P.; Pathak, R. Current Landscape of Cancer Immunotherapy: Harnessing the Immune Arsenal to Overcome Immune Evasion. Biology 2024, 13, 307. https://doi.org/10.3390/biology13050307
Mitra A, Kumar A, Amdare NP, Pathak R. Current Landscape of Cancer Immunotherapy: Harnessing the Immune Arsenal to Overcome Immune Evasion. Biology. 2024; 13(5):307. https://doi.org/10.3390/biology13050307
Chicago/Turabian StyleMitra, Ankita, Anoop Kumar, Nitin P. Amdare, and Rajiv Pathak. 2024. "Current Landscape of Cancer Immunotherapy: Harnessing the Immune Arsenal to Overcome Immune Evasion" Biology 13, no. 5: 307. https://doi.org/10.3390/biology13050307
APA StyleMitra, A., Kumar, A., Amdare, N. P., & Pathak, R. (2024). Current Landscape of Cancer Immunotherapy: Harnessing the Immune Arsenal to Overcome Immune Evasion. Biology, 13(5), 307. https://doi.org/10.3390/biology13050307







