Effects of Rice–Frog Co-Cropping on the Soil Microbial Community Structure in Reclaimed Paddy Fields
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Overview of the Study Area
2.2. Experimental Design
2.3. Sample Collection
2.4. Methods of Measurement
2.4.1. Determination of Soil Chemical Properties
2.4.2. Extraction of the Total DNA from the Soil
2.4.3. Construction of the Library
2.5. Sequencing Analysis and Microbial Taxonomic Identification
2.6. Statistical Analysis
3. Results
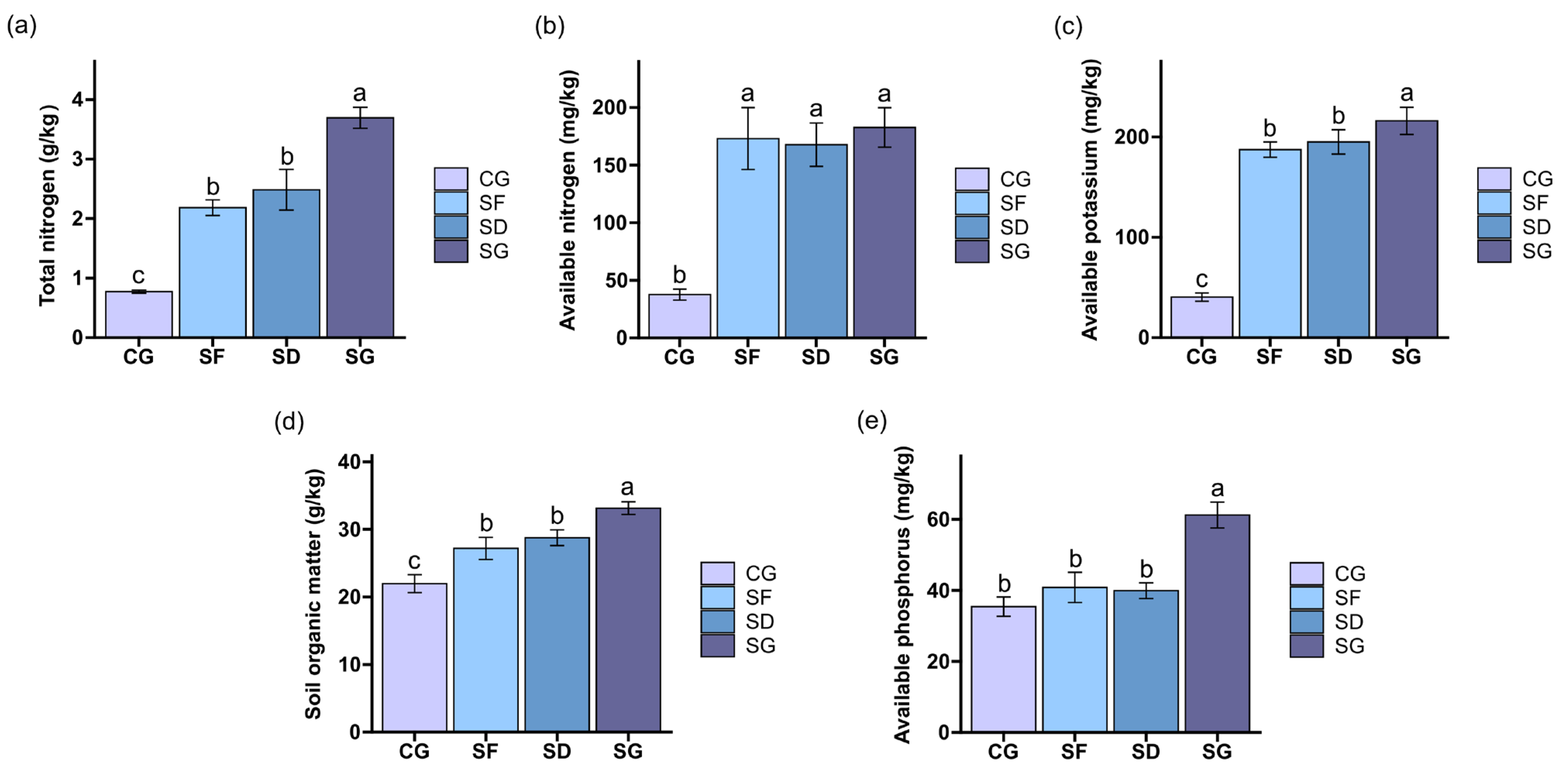
3.1. Properties of the Soils under Three Farming Modes
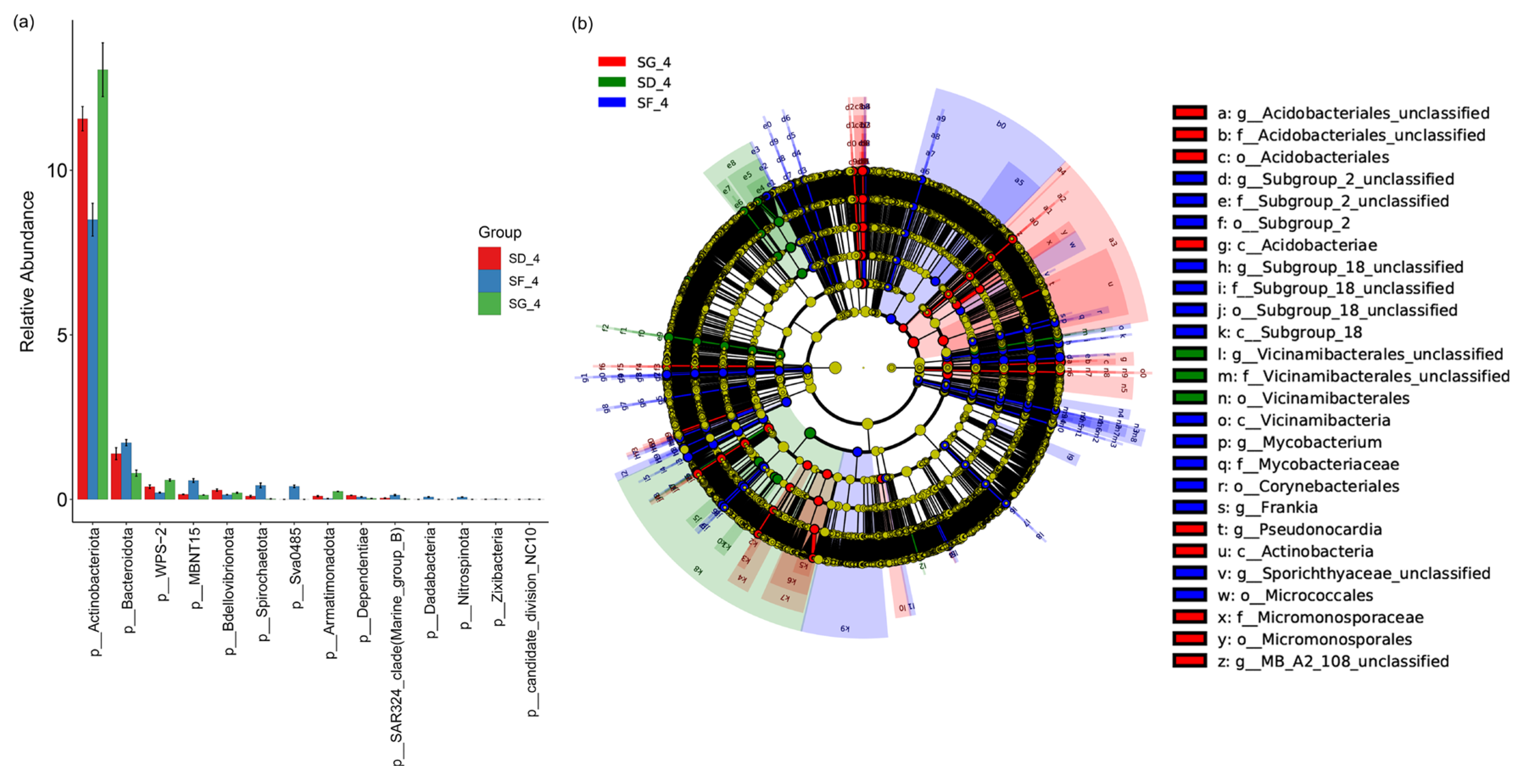
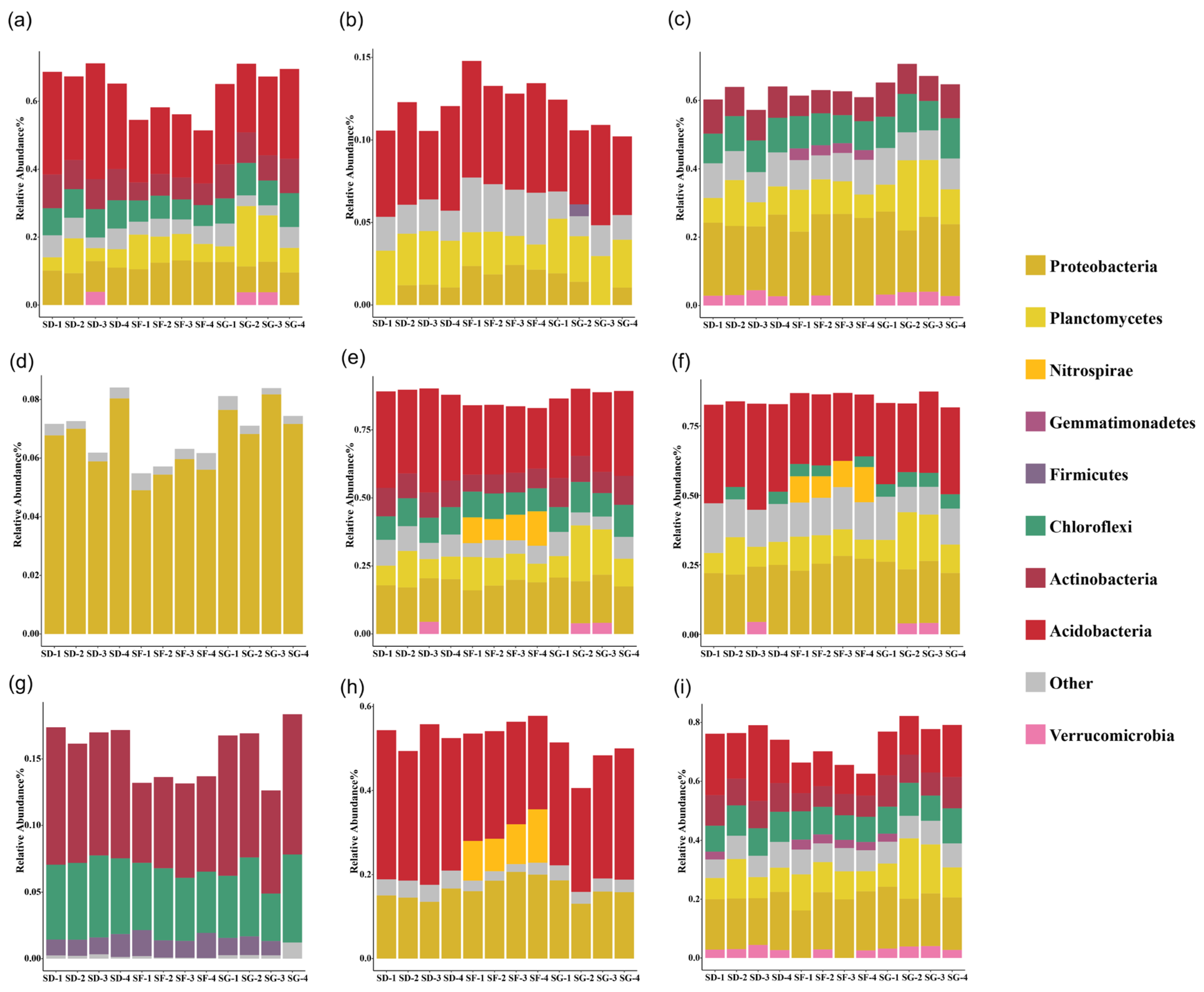
3.2. Bacterial Community Composition of the Soils
3.3. Alpha-Diversity Analysis of the Soil Bacterial Communities under Three Different Farming Modes
3.4. Beta-Diversity Analysis of the Soil Bacterial Community Structure under Three Different Farming Modes
3.5. Analysis of Significant Differences among the Groups of Soil Samples
3.6. Predictive Analysis of the Microbial Functions of the Soil Samples from the Rice–Frog Fields at Different Densities
3.7. Correlations of the Soil Chemical Properties with the Microbial Communities
4. Discussion
4.1. Effects of Different Frog Densities on the Soil Chemical Properties of Reclaimed Paddy Fields
4.2. Effects of Different Frog Densities on the Soil Microorganisms in Reclaimed Paddy Fields
4.3. Soil Chemical Properties and Microbial Correlation in Reclaimed Paddy Fields
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fuss, S.; Havlík, P.; Szolgayová, J.; Schmid, E.; Reuter, W.H.; Khabarov, N.; Obersteiner, M.; Ermoliev, Y.; Ermolieva, T.; Kraxner, F. Global food security & adaptation under crop yield volatility. Technol. Forecast. Soc. Change 2015, 98, 223–233. [Google Scholar] [CrossRef]
- Hasegawa, T.; Sakurai, G.; Fujimori, S.; Takahashi, K.; Hijioka, Y.; Masui, T. Extreme climate events increase risk of global food insecurity and adaptation needs. Nat. Food 2021, 2, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Albaladejo, J.; Díaz-Pereira, E.; Vente, D.J. Eco-holistic soil conservation to support land degradation neutrality and the sustainable development goals. Catena 2021, 196, 104823. [Google Scholar] [CrossRef]
- Kołodziej, B.; Bryk, M.; Antonkiewicz, J. Temporal and spatial variability of physical and chemical properties in reclaimed soil after sulphur borehole mining. Soil Tillage Res. 2024, 237, 105980. [Google Scholar] [CrossRef]
- Liu, M.; Ma, Q.; He, B.; Ni, M.; Zhou, D.; Zhou, S.; Yuan, J. Assessing nutrient budgets and N2O emission of newly constructed rice-giant freshwater prawn co-culture on reclaimed land. Agr. Ecosyst. Environ. 2023, 357, 108686. [Google Scholar] [CrossRef]
- Sanchez, S.P.; Schmidt, O.; Frouz, J.; Čápová, K.; Courtney, R. Effects of earthworms on microbial community structure, functionality and soil properties in soil cover treatments for mine tailings rehabilitation. Eur. J. Soil Biol. 2024, 120, 103603. [Google Scholar] [CrossRef]
- Wang, J.; Zhai, B.; Shi, D.; Chen, A.; Liu, C. How does bio-organic fertilizer combined with biochar affect chinese small cabbage’s growth and quality on newly reclaimed land? Plants 2024, 13, 598. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Sun, Y.; Li, Y.; Wang, L.; Guo, G.; Song, L.; Wang, C.; Wu, G.; Zang, X.; Cai, X.; et al. Ecosystem sustainability of rice and aquatic animal co-culture systems and a synthesis of its underlying mechanisms. Sci. Total Environ. 2023, 880, 163314. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Caspersen, S.; Yong, J.W.H. Growing together gives more rice and aquatic food. Elife 2022, 11, e77202. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Zhang, J.; Ren, W.; Guo, L.; Cheng, Y.; Li, J.; Li, K.; Zhu, Z.; Zhang, J.; Luo, S.; et al. Can the co-cultivation of rice and fish help sustain rice production? Sci. Rep. 2016, 6, 28728. [Google Scholar] [CrossRef]
- Guo, L.; Zhao, L.; Ye, J.; Ji, Z.; Tang, J.J.; Bai, K.; Zheng, S.; Hu, L.; Chen, X. Using aquatic animals as partners to increase yield and maintain soil nitrogen in the paddy ecosystems. eLife 2022, 11, e73869. [Google Scholar] [CrossRef] [PubMed]
- Khatiwada, J.R.; Ghimire, S.; Khatiwada, S.P.; Paudel, B.; Bischof, R.; Jiang, J.; Haugaasen, T. Frogs as potential biological control agents in the rice fields of Chitwan, Nepal. Agric. Ecosyst. Environ. 2016, 230, 307–314. [Google Scholar] [CrossRef]
- Li, P.; Wu, G.; Li, Y.; Hu, C.; Ge, L.; Zheng, X.; Zhang, J.; Chen, J.; Zhang, H.; Bai, N.; et al. Long-term rice-crayfish-turtle co-culture maintains high crop yields by improving soil health and increasing soil microbial community stability. Geoderma 2022, 413, 115745. [Google Scholar] [CrossRef]
- Fang, K.; Dai, W.; Chen, H.; Wang, J.; Gao, H.; Sha, Z.; Cao, L. The effect of integrated rice-frog ecosystem on rice morphological traits and methane emission from paddy fields. Sci. Total Environ. 2021, 783, 147123. [Google Scholar] [CrossRef] [PubMed]
- Fang, K.; Chen, H.; Dai, W.; Wang, J.; Cao, L.; Sha, Z. Microbe-mediated reduction of methane emission in rice-frog crop ecosystem. Appl. Soil Ecol. 2022, 174, 104415. [Google Scholar] [CrossRef]
- Sha, Z.; Chu, Q.; Zhao, Z.; Yue, Y.; Lu, L.; Yuan, J.; Cao, L. Variations in nutrient and trace element composition of rice in an organic rice-frog coculture system. Sci. Rep. 2017, 7, 15706. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Jiang, M.; Zhang, P.; Shu, H.; Li, Y.; Guo, Z.; Li, Y. Amelioration of Cd-induced bioaccumulation, oxidative stress and intestinal microbiota by Bacillus cereus in Carassius auratus gibelio. Chemosphere 2020, 245, 125613. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Yuan, J.; Zhu, Y.; Yi, X.; Zhao, Q.; Fang, K.; Cao, L. Comparison of the abundance and community structure of N-Cycling bacteria in paddy Rhizosphere soil under different rice cultivation patterns. Int. J. Mol. Sci. 2018, 19, 3772. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.; Six, J. Soil structure and microbiome functions in agroecosystems. Nat. Rev. Earth Environ. 2022, 4, 4–18. [Google Scholar] [CrossRef]
- Xiong, C.; Lu, Y. Microbiomes in agroecosystem: Diversity, function and assembly mechanisms. Environ. Microbiol. Rep. 2022, 14, 833–849. [Google Scholar] [CrossRef] [PubMed]
- Bodelier, P.L. Towards understanding, managing and protecting microbial ecosystems. Front. Microbiol. 2011, 2, 80. [Google Scholar] [CrossRef] [PubMed]
- Crowther, T.W.; van den Hoogen, J.; Wan, J.; Mayes, M.A.; Keiser, A.D.; Mo, L.; Averill, C.; Maynard, D.S. The global soil community and its influence on biogeochemistry. Science 2019, 365, eaav0550. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, R.; Peukert, M.; Succurro, A.; Koprivova, A.; Kopriva, S. The role of soil microorganisms in plant mineral Nutrition-current knowledge and future directions. Front. Plant Sci. 2017, 8, 1617. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.C.; Li, H.Y.; Lin, Z.A.; Zhao, B.Q.; Sun, Z.B.; Yuan, L.; Xu, J.K.; Li, Y.Q. Long-term fertilization alters soil properties and fungal community composition in fluvo-aquic soil of the North China Plain. Sci. Rep. 2020, 10, 7198. [Google Scholar] [CrossRef] [PubMed]
- Falkowski, P.G.; Fenchel, T.; Delong, E.F. The microbial engines that drive earth’s biogeochemical cycles. Science 2008, 320, 1034–1039. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Xiao, Y.; Lv, F.; Hu, L.; Wei, L.; Yuan, Z.; Lin, H. Bacterial community structure and functional potential of rhizosphere soils as influenced by nitrogen addition and bacterial wilt disease under continuous sesame cropping. Appl. Soil Ecol. 2018, 125, 117–127. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Anslan, S.; Riit, T.; Kõljalg, U.; Nilsson, R.H.; Hildebrand, F.; Abarenkov, K. Response to comment on “global diversity and geography of soil fungi”. Science 2015, 349, 936. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.Y.; Yao, X.B.; Zhang, X.C.; Fan, M.Y.; Xue, J.F.; Cao, J.L.; Virk, A.L.; Pan, S.G.; Tang, X.R. Effects of tillage practices on soil organic carbon, microbial community and necromass in a double rice cropping system. Appl. Soil Ecol. 2024, 194, 105190. [Google Scholar] [CrossRef]
- Zhang, M.; Li, S.; Wu, X.; Zheng, F.; Song, X.; Lu, J.; Liu, X.; Wang, B.; Abdelrhman, A.A.; Degré, A. Nitrogen addition mediates the effect of soil microbial diversity on microbial carbon use efficiency under long-term tillage practices. Land Degrad. Dev. 2022, 33, 2258–2275. [Google Scholar] [CrossRef]
- Gellie, N.J.C.; Mills, J.G.; Breed, M.F.; Lowe, A.J. Revegetation rewilds the soil bacterial microbiome of an old field. Mol. Ecol. 2017, 26, 2895–2904. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Chu, C.; Zhou, D.; Wang, Q.; Wu, S.; Zheng, X.; Song, K.; Lv, W. Soil bacterial community composition in rice-fish integrated farming systems with different planting years. Sci. Rep. 2021, 11, 10855. [Google Scholar] [CrossRef] [PubMed]
- Gautam, A.; Sekaran, U.; Guzman, J.; Kovács, P.; Hernandez, J.L.G.; Kumar, S. Responses of soil microbial community structure and enzymatic activities to long-term application of mineral fertilizer and beef manure. Environ. Sustain. Ind. 2020, 8, 100073. [Google Scholar] [CrossRef]
- Si, G.; Peng, C.; Yuan, J.; Xu, X.; Zhao, S.; Xu, D.; Wu, J. Changes in soil microbial community composition and organic carbon fractions in an integrated rice-crayfish farming system in subtropical China. Sci. Rep. 2017, 7, 2856. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Yi, K.; Fang, K.; Gao, H.; Dai, W.; Cao, L. Microbial community structures and important associations between soil nutrients and the responses of specific taxa to rice-frog cultivation. Front. Microbiol. 2019, 10, 1752. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xu, J.; Wan, W.; Xu, Z.; Hu, R.; Zhang, Y.; Zheng, J.; Gu, Z. The microbiome structure of a rice-crayfish integrated breeding model and its association with crayfish growth and water quality. Microbiol. Spectr. 2022, 10, e0220421. [Google Scholar] [CrossRef] [PubMed]
- Arunrat, N.; Sansupa, C.; Kongsurakan, P.; Sereenonchai, S.; Hatano, R. Soil microbial diversity and community composition in rice-fish co-culture and rice monoculture farming system. Biology 2022, 11, 1242. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Ma, W.; Wu, D.; Zhang, Y.; Ma, X.; Lv, G.; Ding, J.; Fu, Z.; Chen, C.; Huang, H. Soil bacterial community composition in rice-turtle coculture systems with different planting years. Sci. Rep. 2023, 13, 22708. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Guo, S.; Chen, F.; Chen, L.; Sun, F.; Ying, L. Efficient ecological cultivation technology of rice-frog symbiosis. China Fish. 2016, 4, 73–75. [Google Scholar]
- Wei, Q.; Wang, D.; Wei, K.; Xu, B.; Xu, J. The Mechanism of Elizabethkingia miricola infection of the black spotted frog as revealed by multi-omics analysis. Fishes 2024, 9, 91. [Google Scholar] [CrossRef]
- Li, Y.; Fang, F.; Wei, J.; Wu, X.; Cui, R.; Li, G.; Zheng, F.; Tan, D. Humic acid fertilizer improved soil properties and soil microbial diversity of continuous cropping peanut: A three-year experiment. Sci. Rep. 2019, 9, 12014. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Tan, G.; Wang, H.; Gai, X. Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 2016, 74, 1–8. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; Green, E.D.; et al. Topographical and temporal diversity of the human skin microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef] [PubMed]
- Nagumo, T.; Tajima, S.; Chikushi, S.; Yamashita, A. Phosphorus balance and soil phosphorus status in paddy rice fields with various fertilizer practices. Plant Prod. Sci. 2015, 16, 69–76. [Google Scholar] [CrossRef]
- Teng, Q.; Hu, X.; Luo, F.; Cheng, C.; Ge, X.; Yang, M.; Liu, L. Influences of introducing frogs in the paddy fields on soil properties and rice growth. J. Soils Sediments 2016, 16, 51–61. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Y.; Ouyang, D.; Lei, J.; Tan, Q.; Xie, L.; Li, Z.; Liu, T.; Xiao, Y.; Farooq, T.H.; et al. Systematical review of interactions between microplastics and microorganisms in the soil environment. J. Hazard. Mater. 2021, 418, 126288. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Sun, J.; Wang, C.; Li, S.; Li, Z.; Luo, W.; Wei, G.; Chen, W. Residue quality drives SOC sequestration by altering microbial taxonomic composition and ecophysiological function in desert ecosystem. Environ. Res. 2024, 251, 118518. [Google Scholar] [CrossRef] [PubMed]
- Suolang, Y.; Luo, W.; Ma, J.; Zan, Y.; Yu, Y.; Wan, M.; Yuan, J.; Liu, J.; Tao, J. Extreme precipitation alters soil nitrogen cycling related microbial community in karst abandoned farmland. Appl. Soil Ecol. 2024, 197, 105345. [Google Scholar] [CrossRef]
- Ma, L.; Zhou, G.; Zhang, J.; Jia, Z.; Zou, H.; Chen, L.; Zhang, C.; Ma, D.; Han, C.; Duan, Y. Long-term conservation tillage enhances microbial carbon use efficiency by altering multitrophic interactions in soil. Sci. Total Environ. 2024, 915, 170018. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, L.; Chen, Q.; Wen, X.; Liao, Y. Conservation tillage increases soil bacterial diversity in the dryland of northern China. Agron. Sustain. Dev. 2016, 36, 28. [Google Scholar] [CrossRef]
- Hu, B.; Flemetakis, E.; Liu, Z.; Hänsch, R.; Rennenberg, H. Significance of nitrogen-fixing actinorhizal symbioses for restoration of depleted, degraded, and contaminated soil. Trends Plant Sci. 2023, 28, 752–764. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xiang, W.; Wu, H.; Ouyang, S.; Zhou, B.; Zeng, Y.; Chen, Y.; Kuzyakov, Y. Tree species identity surpasses richness in affecting soil microbial richness and community composition in subtropical forests. Soil Biol. Biochem. 2019, 130, 113–121. [Google Scholar] [CrossRef]
- Shi, Y.; Delgado-Baquerizo, M.; Li, Y.; Yang, Y.; Zhu, Y.-G.; Peñuelas, J.; Chu, H. Abundance of kinless hubs within soil microbial networks are associated with high functional potential in agricultural ecosystems. Environ. Int. 2020, 142, 105869. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, K.; Yan, F.; Xiang, Q.; Zhao, X.; Ji, L.; Xin, Y.; Sun, J.; Liu, C.; Xu, X.; et al. Soil nitrogen content and key functional microorganisms influence the response of wetland anaerobic oxidation of methane to trivalent iron input. Chemosphere 2023, 322, 138183. [Google Scholar] [CrossRef]
- Williams, T.J.; Allen, M.A.; Ray, A.E.; Benaud, N.; Chelliah, D.S.; Albanese, D.; Donati, C.; Selbmann, L.; Coleine, C.; Ferrari, B.C. Novel endolithic bacteria of phylum Chloroflexota reveal a myriad of potential survival strategies in the Antarctic desert. Appl. Environ. Microbiol. 2024, 90, e0226423. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, J.M.; Shaw, N.A.; Nagashima, S.; Venkiteswaran, J.J.; Schiff, S.L.; Watanabe, T.; Fukui, M.; Hanada, S.; Tank, M.; Neufeld, J.D. Anoxygenic phototroph of the Chloroflexota uses a type I reaction centre. Nature 2024, 627, 915–922. [Google Scholar] [CrossRef] [PubMed]
- Xue, P.P.; Carrillo, Y.; Pino, V.; Minasny, B.; McBratney, A.B. Soil properties drive microbial community structure in a large scale transect in south eastern Australia. Sci. Rep. 2018, 8, 11725. [Google Scholar] [CrossRef] [PubMed]
- Sui, X.; Zhang, R.; Frey, B.; Yang, L.; Liu, Y.; Ni, H.; Li, M.H. Soil physicochemical properties drive the variation in soil microbial communities along a forest successional series in a degraded wetland in northeastern China. Ecol. Evol. 2021, 11, 2194–2208. [Google Scholar] [CrossRef] [PubMed]
- Viruel, E.; Fontana, C.A.; Puglisi, E.; Nasca, J.A.; Banegas, N.R.; Cocconcelli, P.S. Land-use change affects the diversity and functionality of soil bacterial communities in semi-arid Chaco region, Argentina. Appl. Soil Ecol. 2022, 172, 104362. [Google Scholar] [CrossRef]

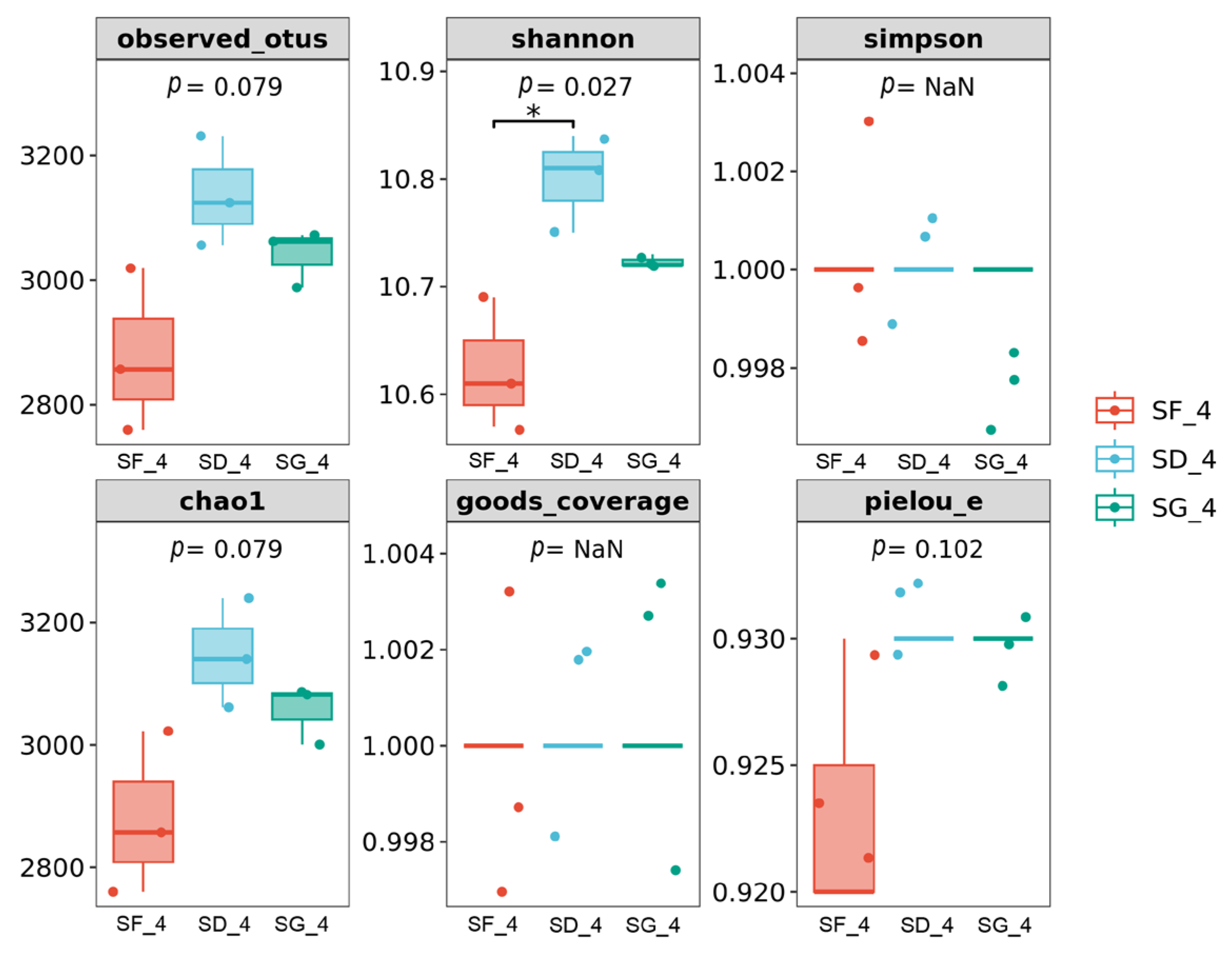


| Sample ID | Observed Otus | Shannon | Simpson | Chao 1 | Coverage | Pielou_e |
|---|---|---|---|---|---|---|
| SF_1 | 3013 ± 115 ab | 10.67 ± 0.06 ab | 1 | 3013 ± 115 ab | 1 | 0.92 ± 0.00 a |
| SF_2 | 3076 ± 80 ab | 10.71 ± 0.04 ab | 1 | 3077 ± 88 ab | 1 | 0.92 ± 0.01 ab |
| SF_3 | 2971 ± 152 ab | 10.64 ± 0.10 ab | 1 | 2975 ± 154 ab | 1 | 0.92 ± 0.01 ab |
| SF_4 | 2877 ± 129 ab | 10.62 ± 0.06 ab | 1 | 2879 ± 132 ab | 1 | 0.92 ± 0.01 ab |
| SD_1 | 2998 ± 218 ab | 10.64 ± 0.10 ab | 1 | 3006 ± 224 ab | 1 | 0.92 ± 0.00 a |
| SD_2 | 3437 ± 463 b | 10.91 ± 0.20 b | 1 | 3451 ± 482 b | 1 | 0.93 ± 0.00 c |
| SD_3 | 2636 ± 318 a | 10.44 ± 0.19 a | 1 | 2648 ± 321 a | 1 | 0.92 ± 0.00 a |
| SD_4 | 3135 ± 90 ab | 10.80 ± 0.04 b | 1 | 3147 ± 92 ab | 1 | 0.93 ± 0.00 c |
| SG_1 | 3055 ± 402 ab | 10.77 ± 0.96 ab | 1 | 3066 ± 416 ab | 1 | 0.93 ± 0.00 c |
| SG_2 | 3064 ± 912 ab | 10.69 ± 0.50 ab | 1 | 3087 ± 925 ab | 1 | 0.93 ± 0.01 bc |
| SG_3 | 3141 ± 182 ab | 10.79 ± 0.05 ab | 1 | 3160 ± 186 ab | 1 | 0.93 ± 0.00 c |
| SG_4 | 3043 ± 44 ab | 10.72 ± 0.01 ab | 1 | 3061 ± 40 ab | 1 | 0.93 ± 0.00 c |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Yu, A.; Zhang, L.; Zheng, R. Effects of Rice–Frog Co-Cropping on the Soil Microbial Community Structure in Reclaimed Paddy Fields. Biology 2024, 13, 396. https://doi.org/10.3390/biology13060396
Ma Y, Yu A, Zhang L, Zheng R. Effects of Rice–Frog Co-Cropping on the Soil Microbial Community Structure in Reclaimed Paddy Fields. Biology. 2024; 13(6):396. https://doi.org/10.3390/biology13060396
Chicago/Turabian StyleMa, Yunshuang, Anran Yu, Liangliang Zhang, and Rongquan Zheng. 2024. "Effects of Rice–Frog Co-Cropping on the Soil Microbial Community Structure in Reclaimed Paddy Fields" Biology 13, no. 6: 396. https://doi.org/10.3390/biology13060396
APA StyleMa, Y., Yu, A., Zhang, L., & Zheng, R. (2024). Effects of Rice–Frog Co-Cropping on the Soil Microbial Community Structure in Reclaimed Paddy Fields. Biology, 13(6), 396. https://doi.org/10.3390/biology13060396






