The Impact of Nitrogen-Fixing Bacteria-Based Biostimulant Alone or in Combination with Commercial Inoculum on Tomato Native Rhizosphere Microbiota and Production: An Open-Field Trial
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
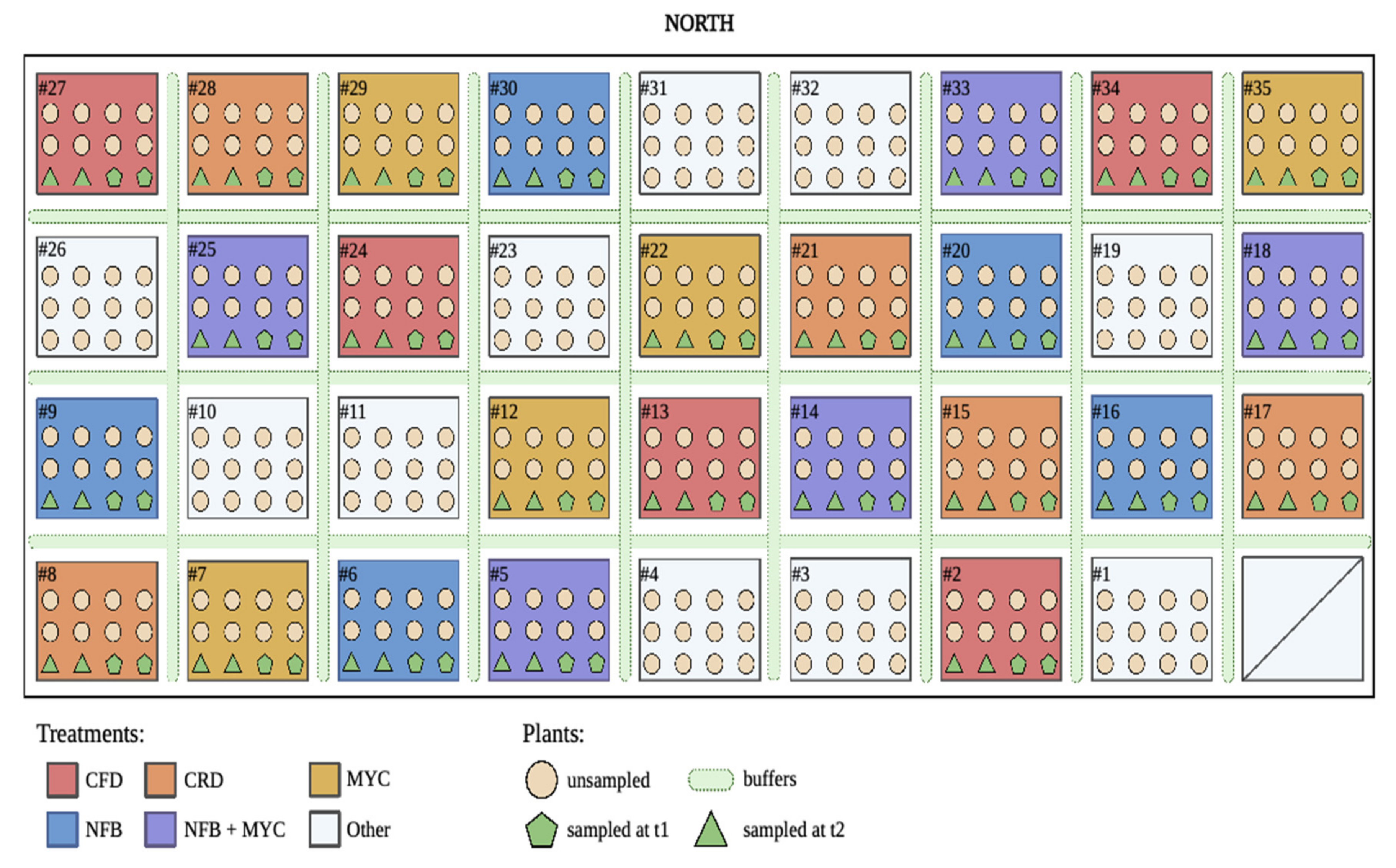
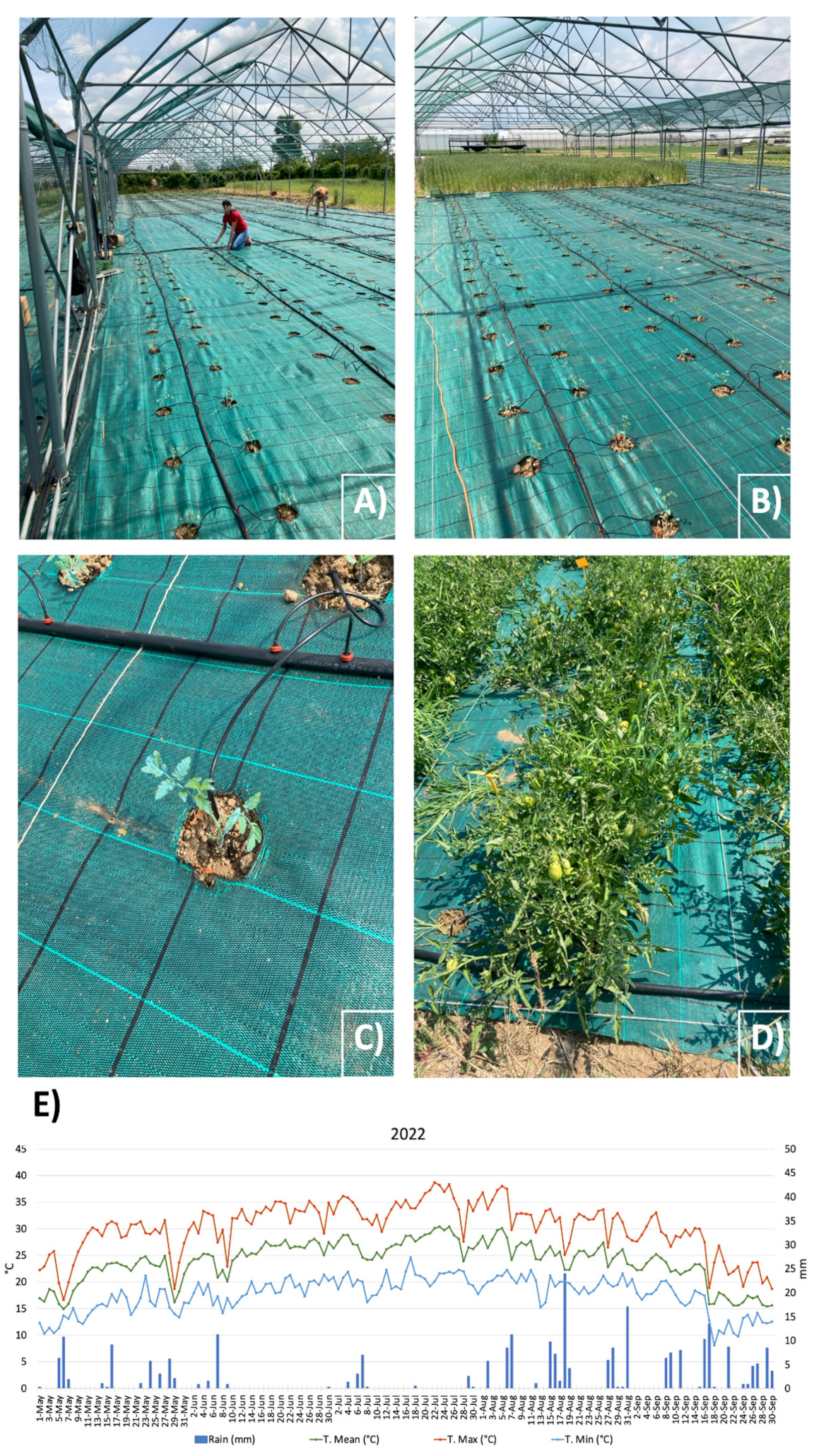
2.1. Experimental Design and Field Conditions
2.2. Soil Collection and Analyses
2.3. Root AM Colonization Assay
2.4. Microbial Community Characterization and Bioinformatic and Statistical Analyses
2.5. Tomato Fruit Sampling and Analyses
3. Results
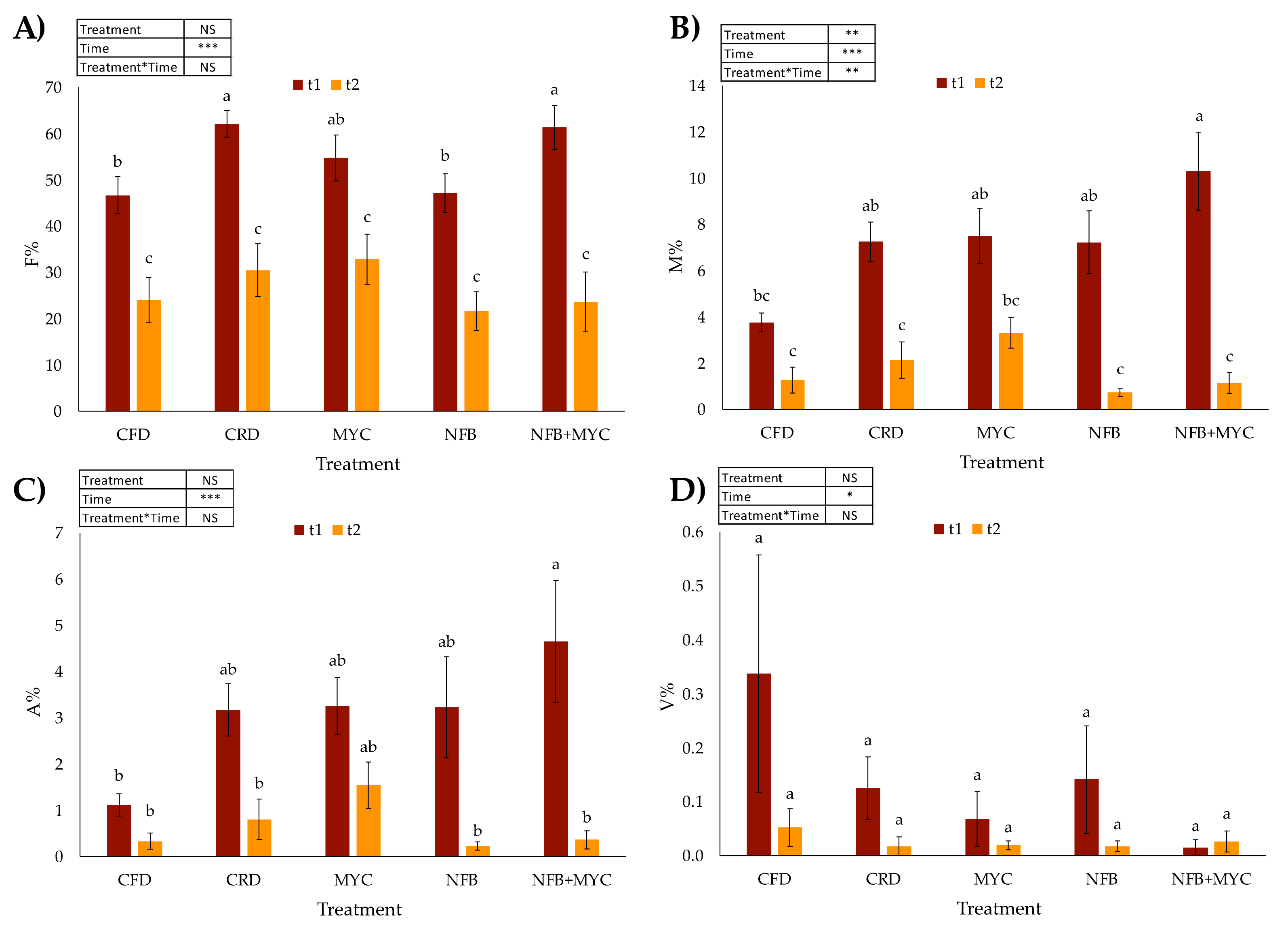
3.1. Mycorrhizal Degree
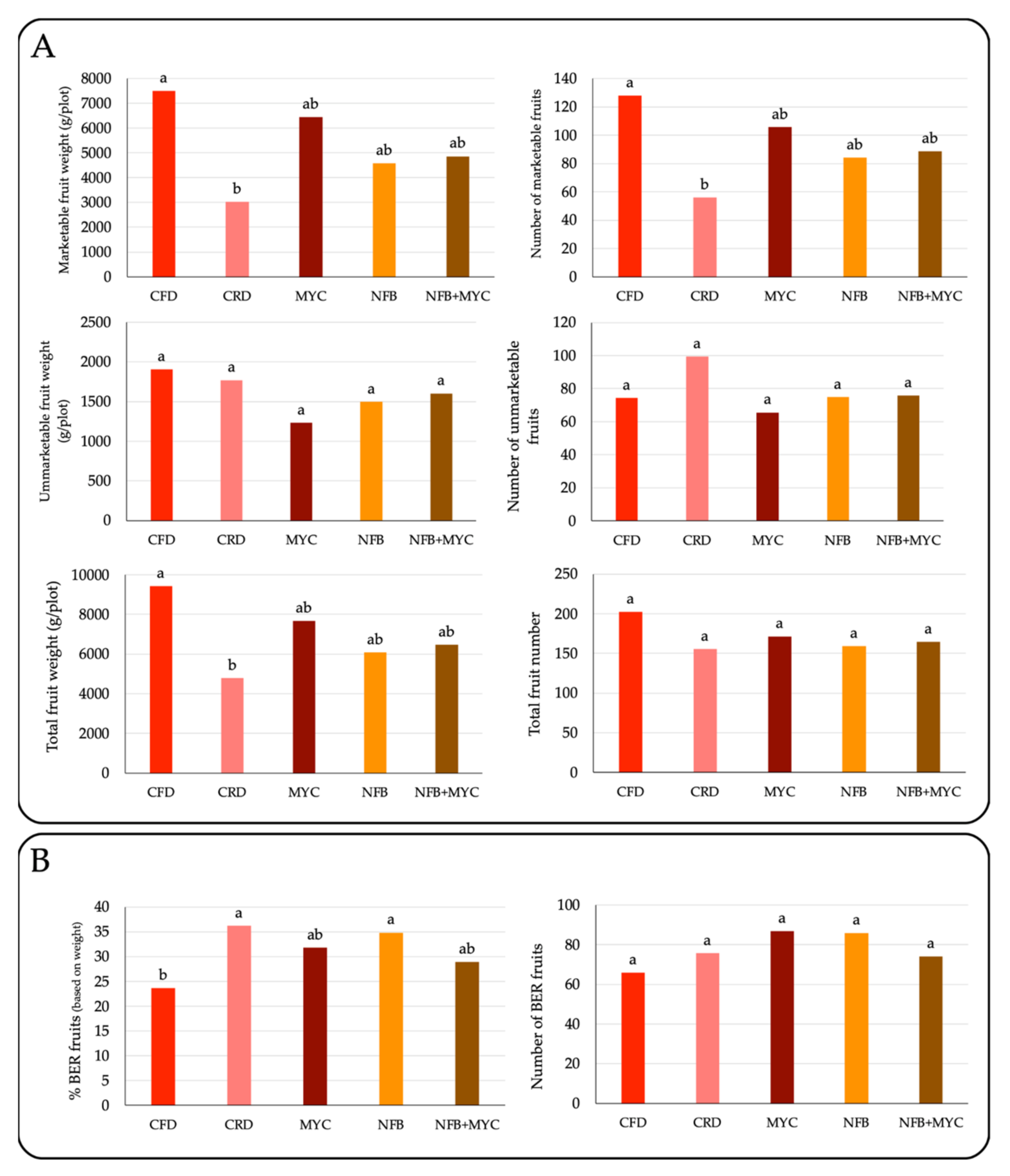
3.2. Tomato Production
3.3. Soil Microbiota Profiling
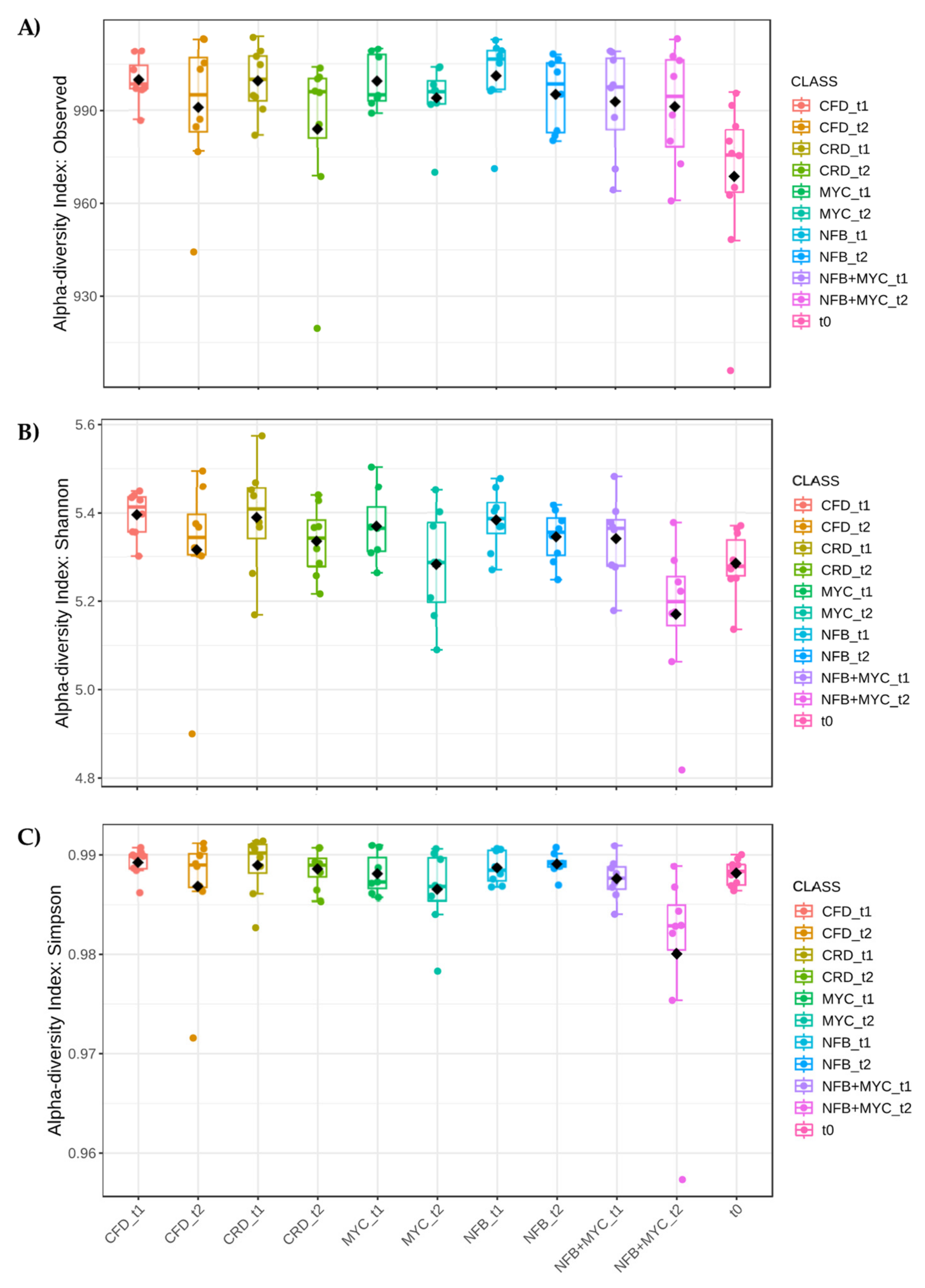
3.3.1. Plant Effect
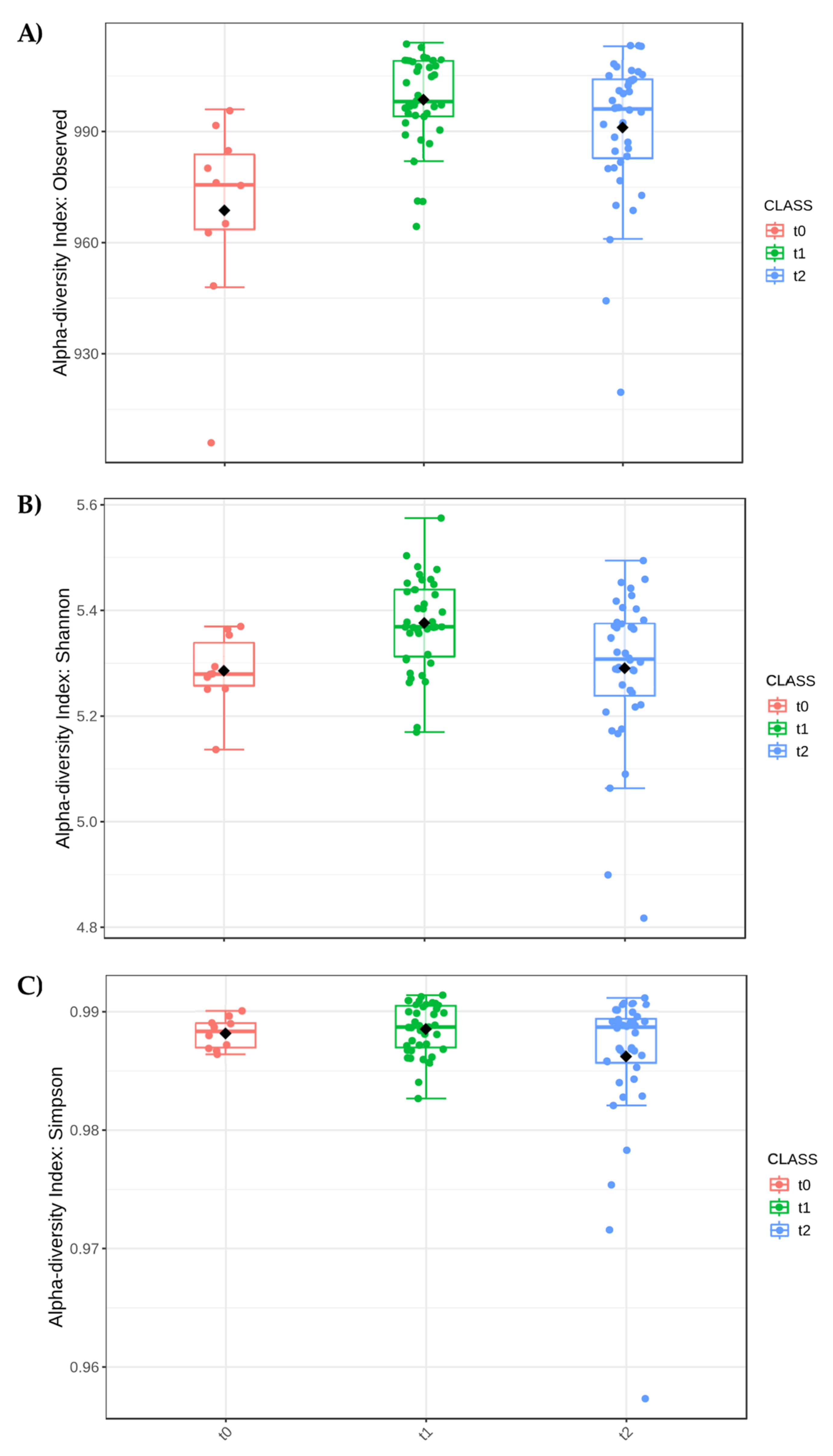
3.3.2. Effect of Phenological Stage of the Plant
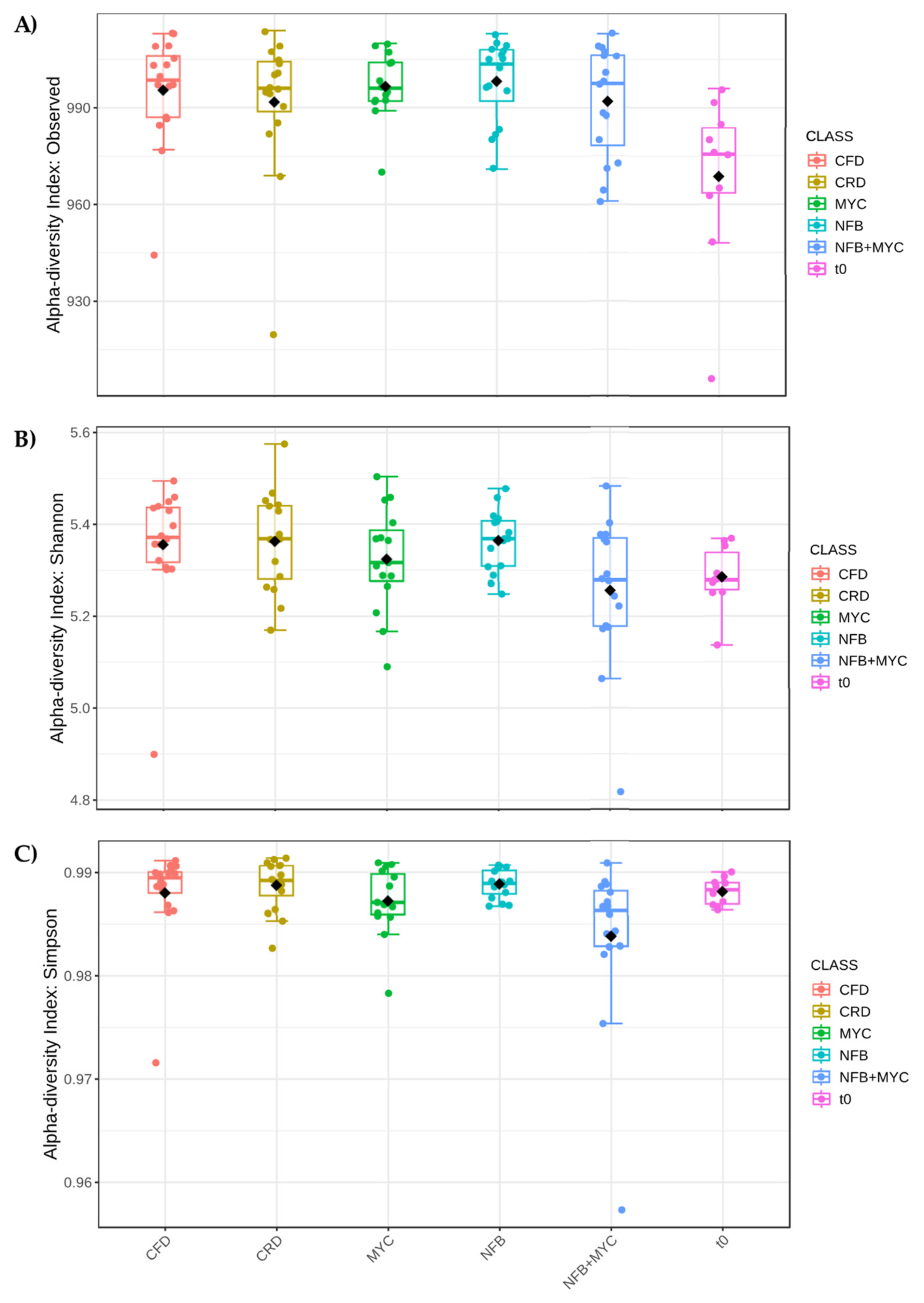
3.3.3. Bioinocula Effect
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. Climate Change and Food Security: Risks and Responses; FAO: Roma, Italy, 2015. [Google Scholar]
- Del Carmen Orozco-Mosqueda, M.; Glick, B.R.; Santoyo, G. ACC Deaminase in Plant Growth-Promoting Bacteria (PGPB): An Efficient Mechanism to Counter Salt Stress in Crops. Microbiol. Res. 2020, 235, 126439. [Google Scholar] [CrossRef] [PubMed]
- Fadiji, A.E.; Babalola, O.O.; Santoyo, G.; Perazzolli, M. The Potential Role of Microbial Biostimulants in the Amelioration of Climate Change-Associated Abiotic Stresses on Crops. Front. Microbiol. 2022, 12, 829099. [Google Scholar] [CrossRef] [PubMed]
- Antar, M.; Lyu, D.; Nazari, M.; Shah, A.; Zhou, X.; Smith, D.L. Biomass for a Sustainable Bioeconomy: An Overview of World Biomass Production and Utilization. Renew. Sustain. Energy Rev. 2021, 139, 110691. [Google Scholar] [CrossRef]
- Santoyo, G.; Gamalero, E.; Glick, B.R. Mycorrhizal-Bacterial Amelioration of Plant Abiotic and Biotic Stress. Front. Sustain. Food Syst. 2021, 5, 18. [Google Scholar] [CrossRef]
- Adesemoye, A.O.; Torbert, H.A.; Kloepper, J.W. Plant Growth-Promoting Rhizobacteria Allow Reduced Application Rates of Chemical Fertilizers. Microb. Ecol. 2009, 58, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Van der Heijden, M.G.A.; Klironomos, J.N.; Ursic, M.; Moutoglis, P.; Streitwolf-Engel, R.; Boller, T.; Wiemken, A.; Sanders, I.R. Mycorrhizal Fungal Diversity Determines Plant Biodiversity, Ecosystem Variability and Productivity. Nature 1998, 396, 69–72. [Google Scholar] [CrossRef]
- Van Loon, L.C.; Glick, B.R. Increased Plant Fitness by Rhizobacteria. In Molecular ecotoxicology of plants; Springer: Berlin/Heidelberg, Germany, 2004; pp. 177–205. [Google Scholar]
- Yadegari, M.; Rahmani, H.A.; Noormohammadi, G.; Ayneband, A. Evaluation of Bean (Phaseolus vulgaris) Seeds Inoculation with Rhizobium Phaseoli and Plant Growth Promoting Rhizobacteria on Yield and Yield Components. Pak. J. Biol. Sci. 2008, 11, 1935–1939. [Google Scholar] [CrossRef] [PubMed]
- Emmanuel, O.C.; Babalola, O.O. Productivity and Quality of Horticultural Crops through Co-Inoculation of Arbuscular Mycorrhizal Fungi and Plant Growth Promoting Bacteria. Microbiol. Res. 2020, 239, 126569. [Google Scholar] [CrossRef] [PubMed]
- Saia, S.; Aissa, E.; Luziatelli, F.; Ruzzi, M.; Colla, G.; Ficca, A.G.; Cardarelli, M.; Rouphael, Y. Growth-Promoting Bacteria and Arbuscular Mycorrhizal Fungi Differentially Benefit Tomato and Corn Depending upon the Supplied form of Phosphorus. Mycorrhiza 2020, 30, 133–147. [Google Scholar] [CrossRef]
- Forni, C.; Duca, D.; Glick, B.R. Mechanisms of Plant Response to Salt and Drought Stress and Their Alteration by Rhizobacteria. Plant Soil 2017, 410, 335–356. [Google Scholar] [CrossRef]
- Gamalero, E.; Glick, B.R. Plant Growth-Promoting Bacteria in Agriculture and Stress Environments. In Modern Soil Microbiology, 3rd ed.; Van Elsas, J.D., Trevors, J.T., Rosado, A.S., Nannipieri, P., Eds.; CRC Press: Boca Raton, FL, USA, 2019; pp. 361–380. [Google Scholar]
- Chandra, P.; Wunnava, A.; Verma, P.; Chandra, A.; Sharma, R.K. Strategies to Mitigate the Adverse Effect of Drought Stress on Crop Plants—Influences of Soil Bacteria: A Review. Pedosphere 2021, 31, 496–509. [Google Scholar] [CrossRef]
- Ali, S.; Charles, T.C.; Glick, B.R. Amelioration of High Salinity Stress Damage by Plant Growth-Promoting Bacterial Endophytes That Contain ACC Deaminase. Plant Physiol. Biochem. 2014, 80, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Brilli, F.; Pollastri, S.; Raio, A.; Baraldi, R.; Neri, L.; Bartolini, P.; Podda, A.; Loreto, F.; Maserti, B.E.; Balestrini, R. Root Colonization by Pseudomonas chlororaphis Primes Tomato (Lycopersicum esculentum) Plants for Enhanced Tolerance to Water Stress. J. Plant Physiol. 2019, 232, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Prasad, H.; Sajwan, P.; Kumari, M.; Solanki, S. Effect of Organic Manures and Biofertilizer on Plant Growth, Yield and Quality of Horticultural Crop: A Review. Int. J. Chem. Stud. 2017, 5, 217–221. [Google Scholar]
- Qiao, C.; Penton, C.R.; Xiong, W.; Liu, C.; Wang, R.; Liu, Z.; Xu, X.; Li, R.; Shen, Q. Reshaping the Rhizosphere Microbiome by Bio-Organic Amendment to Enhance Crop Yield in a Maize-Cabbage Rotation System. Appl. Soil Ecol. 2019, 142, 136–146. [Google Scholar] [CrossRef]
- Tsegaye, Z.; Alemu, T.; Desta, F.A.; Assefa, F. Plant Growth-Promoting Rhizobacterial Inoculation to Improve Growth, Yield, and Grain Nutrient Uptake of Teff Varieties. Front. Microbiol. 2022, 13, 896770. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Bastida, F.; Liu, Y.; Zhou, Y.; He, J.; Song, P.; Kuang, N.; Li, Y. Nanobubble Oxygenated Increases Crop Production via Soil Structure Improvement: The Perspective of Microbially Mediated Effects. Agric. Water Manag. 2023, 282, 108263. [Google Scholar] [CrossRef]
- Bona, E.; Todeschini, V.; Cantamessa, S.; Cesaro, P.; Copetta, A.; Lingua, G.; Gamalero, E.; Berta, G.; Massa, N. Combined Bacterial and Mycorrhizal Inocula Improve Tomato Quality at Reduced Fertilization. Sci. Hortic. 2018, 234, 160–165. [Google Scholar] [CrossRef]
- Bona, E.; Cantamessa, S.; Massa, N.; Manassero, P.; Marsano, F.; Copetta, A.; Lingua, G.; D’Agostino, G.; Gamalero, E.; Berta, G. Arbuscular Mycorrhizal Fungi and Plant Growth-Promoting Pseudomonads Improve Yield, Quality and Nutritional Value of Tomato: A Field Study. Mycorrhiza 2017, 27, 1–11. [Google Scholar] [CrossRef]
- Bona, E.; Scarafoni, A.; Marsano, F.; Boatti, L.; Copetta, A.; Massa, N.; Gamalero, E.; D’Agostino, G.; Cesaro, P.; Cavaletto, M.; et al. Arbuscular Mycorrhizal Symbiosis Affects the Grain Proteome of Zea mays: A Field Study. Sci. Rep. 2016, 6, 26439. [Google Scholar] [CrossRef]
- Vishwakarma, K.; Kumar, N.; Shandilya, C.; Mohapatra, S.; Bhayana, S.; Varma, A. Revisiting Plant-Microbe Interactions and Microbial Consortia Application for Enhancing Sustainable Agriculture: A Review. Front. Microbiol. 2020, 11, 560406. [Google Scholar] [CrossRef] [PubMed]
- Sandrini, M.; Moffa, L.; Velasco, R.; Balestrini, R.; Chitarra, W.; Nerva, L. Microbe-Assisted Crop Improvement: A Sustainable Weapon to Restore Holobiont Functionality and Resilience. Hortic. Res. 2022, 9, uhac160. [Google Scholar] [CrossRef] [PubMed]
- Großkopf, T.; Soyer, O.S. Synthetic Microbial Communities. Curr. Opin. Microbiol. 2014, 18, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Ahkami, A.H.; Allen White, R.; Handakumbura, P.P.; Jansson, C. Rhizosphere Engineering: Enhancing Sustainable Plant Ecosystem Productivity. Rhizosphere 2017, 3, 233–243. [Google Scholar] [CrossRef]
- Nasuelli, M.; Novello, G.; Gamalero, E.; Massa, N.; Gorrasi, S.; Sudiro, C.; Hochart, M.; Altissimo, A.; Vuolo, F.; Bona, E. PGPB and/or AM Fungi Consortia Affect Tomato Native Rhizosphere Microbiota. Microorganisms 2023, 11, 1891. [Google Scholar] [CrossRef] [PubMed]
- Bona, E.; Lingua, G.; Todeschini, V. Effect of Bioinoculants on the Quality of Crops. In Bioformulations: For Sustainable Agriculture; Arora, N.K., Mehnaz, S., Balestrini, R., Eds.; Springer: New Delhi, India, 2016; pp. 93–124. ISBN 978-81-322-2777-9. [Google Scholar]
- Vuolo, F.; Novello, G.; Bona, E.; Gorrasi, S.; Gamalero, E. Impact of Plant-Beneficial Bacterial Inocula on the Resident Bacteriome: Current Knowledge and Future Perspectives. Microorganisms 2022, 10, 2462. [Google Scholar] [CrossRef] [PubMed]
- Trouvelot, A.; Kough, J.; Gianinazzi-Pearson, V. Mesure Du Taux de Mycorrhization VA d’un Système Radiculaire. Recherche de Méthodes d’estimation Ayant Une Signification Functionnelle. In Physiological and Genetical Aspects of Mycorrhizae; INRA: Paris, France, 1986; pp. 217–221. [Google Scholar]
- FAO. Agricultural Production Statistics 2000–2021; FAOSTAT Analytical Brief Series No. 41; FAO: Rome, Italy, 2022. [Google Scholar]
- Gianinazzi, S.; Gollotte, A.; Binet, M.-N.; Van Tuinen, D.; Redecker, D.; Wipf, D. Agroecology: The Key Role of Arbuscular Mycorrhizas in Ecosystem Services. Mycorrhiza 2010, 20, 519–530. [Google Scholar] [CrossRef] [PubMed]
- European Commission. Directorate General for Research and Innovation. In Directive of the European Parliament and of the Council on Soil Monitoring and Resilience (Soil Monitoring Law); Publications Office: Luxembourg, 2023. [Google Scholar]
- Johnson, C.R.; Graham, J.H.; Leonard, R.T.; Menge, J.A. Effect of Flower Bud Development in Chrysanthemum on Vesicular-Arbuscular Mycorrhiza Formation. New Phytol. 1982, 90, 671–675. [Google Scholar] [CrossRef]
- Tahir, M.; Mirza, M.S.; Hameed, S.; Dimitrov, M.R.; Smidt, H. Cultivation-Based and Molecular Assessment of Bacterial Diversity in the Rhizosheath of Wheat under Different Crop Rotations. PLoS ONE 2015, 10, e0130030. [Google Scholar] [CrossRef]
- Zhao, M.; Yuan, J.; Shen, Z.; Dong, M.; Liu, H.; Wen, T.; Li, R.; Shen, Q. Predominance of Soil vs Root Effect in Rhizosphere Microbiota Reassembly. FEMS Microbiol. Ecol. 2019, 95, fiz139. [Google Scholar] [CrossRef]
- Dams, R.I.; Paton, G.I.; Killham, K. Rhizoremediation of Pentachlorophenol by Sphingobium chlorophenolicum ATCC 39723. Chemosphere 2007, 68, 864–870. [Google Scholar] [CrossRef] [PubMed]
- Lal, R.; Dadhwal, M.; Kumari, K.; Sharma, P.; Singh, A.; Kumari, H.; Jit, S.; Gupta, S.K.; Nigam, A.; Lal, D.; et al. Pseudomonas sp. to Sphingobium indicum: A Journey of Microbial Degradation and Bioremediation of Hexachlorocyclohexane. Indian J. Microbiol. 2008, 48, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chiarucci, A.; Fang, H.; Chen, M. An Interspecific Variation in Rhizosphere Effects on Soil Anti-Erodibility. Sci. Rep. 2020, 10, 2411. [Google Scholar] [CrossRef] [PubMed]
- López-Aladid, R.; Fernández-Barat, L.; Alcaraz-Serrano, V.; Bueno-Freire, L.; Vázquez, N.; Pastor-Ibáñez, R.; Palomeque, A.; Oscanoa, P.; Torres, A. Determining the Most Accurate 16S rRNA Hypervariable Region for Taxonomic Identification from Respiratory Samples. Sci. Rep. 2023, 13, 3974. [Google Scholar] [CrossRef] [PubMed]
- Dias, V.H.G.; Gomes, P.D.S.F.C.; Azevedo-Martins, A.C.; Cabral, B.C.A.; Woerner, A.E.; Budowle, B.; Moura-Neto, R.S.; Silva, R. Evaluation of 16S rRNA Hypervariable Regions for Bioweapon Species Detection by Massively Parallel Sequencing. Int. J. Microbiol. 2020, 2020, 8865520. [Google Scholar] [CrossRef] [PubMed]
- Bona, E.; Massa, N.; Toumatia, O.; Novello, G.; Cesaro, P.; Todeschini, V.; Boatti, L.; Mignone, F.; Titouah, H.; Zitouni, A.; et al. Climatic Zone and Soil Properties Determine the Biodiversity of the Soil Bacterial Communities Associated to Native Plants from Desert Areas of North-Central Algeria. Microorganisms 2021, 9, 1359. [Google Scholar] [CrossRef] [PubMed]
- Myer, P.R.; McDaneld, T.G.; Kuehn, L.A.; Dedonder, K.D.; Apley, M.D.; Capik, S.F.; Lubbers, B.V.; Harhay, G.P.; Harhay, D.M.; Keele, J.W.; et al. Classification of 16S rRNA Reads Is Improved Using a Niche-Specific Database Constructed by near-Full Length Sequencing. PLoS ONE 2020, 15, e0235498. [Google Scholar] [CrossRef]
- Chakravorty, S.; Helb, D.; Burday, M.; Connell, N.; Alland, D. A Detailed Analysis of 16S Ribosomal RNA Gene Segments for the Diagnosis of Pathogenic Bacteria. J. Microbiol. Methods 2007, 69, 330–339. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Novello, G.; Bona, E.; Nasuelli, M.; Massa, N.; Sudiro, C.; Campana, D.C.; Gorrasi, S.; Hochart, M.L.; Altissimo, A.; Vuolo, F.; et al. The Impact of Nitrogen-Fixing Bacteria-Based Biostimulant Alone or in Combination with Commercial Inoculum on Tomato Native Rhizosphere Microbiota and Production: An Open-Field Trial. Biology 2024, 13, 400. https://doi.org/10.3390/biology13060400
Novello G, Bona E, Nasuelli M, Massa N, Sudiro C, Campana DC, Gorrasi S, Hochart ML, Altissimo A, Vuolo F, et al. The Impact of Nitrogen-Fixing Bacteria-Based Biostimulant Alone or in Combination with Commercial Inoculum on Tomato Native Rhizosphere Microbiota and Production: An Open-Field Trial. Biology. 2024; 13(6):400. https://doi.org/10.3390/biology13060400
Chicago/Turabian StyleNovello, Giorgia, Elisa Bona, Martina Nasuelli, Nadia Massa, Cristina Sudiro, Daniela Cristina Campana, Susanna Gorrasi, Marie Louise Hochart, Adriano Altissimo, Francesco Vuolo, and et al. 2024. "The Impact of Nitrogen-Fixing Bacteria-Based Biostimulant Alone or in Combination with Commercial Inoculum on Tomato Native Rhizosphere Microbiota and Production: An Open-Field Trial" Biology 13, no. 6: 400. https://doi.org/10.3390/biology13060400
APA StyleNovello, G., Bona, E., Nasuelli, M., Massa, N., Sudiro, C., Campana, D. C., Gorrasi, S., Hochart, M. L., Altissimo, A., Vuolo, F., & Gamalero, E. (2024). The Impact of Nitrogen-Fixing Bacteria-Based Biostimulant Alone or in Combination with Commercial Inoculum on Tomato Native Rhizosphere Microbiota and Production: An Open-Field Trial. Biology, 13(6), 400. https://doi.org/10.3390/biology13060400











