Network Pharmacology Reveals Curcuma aeruginosa Roxb. Regulates MAPK and HIF-1 Pathways to Treat Androgenetic Alopecia
Abstract
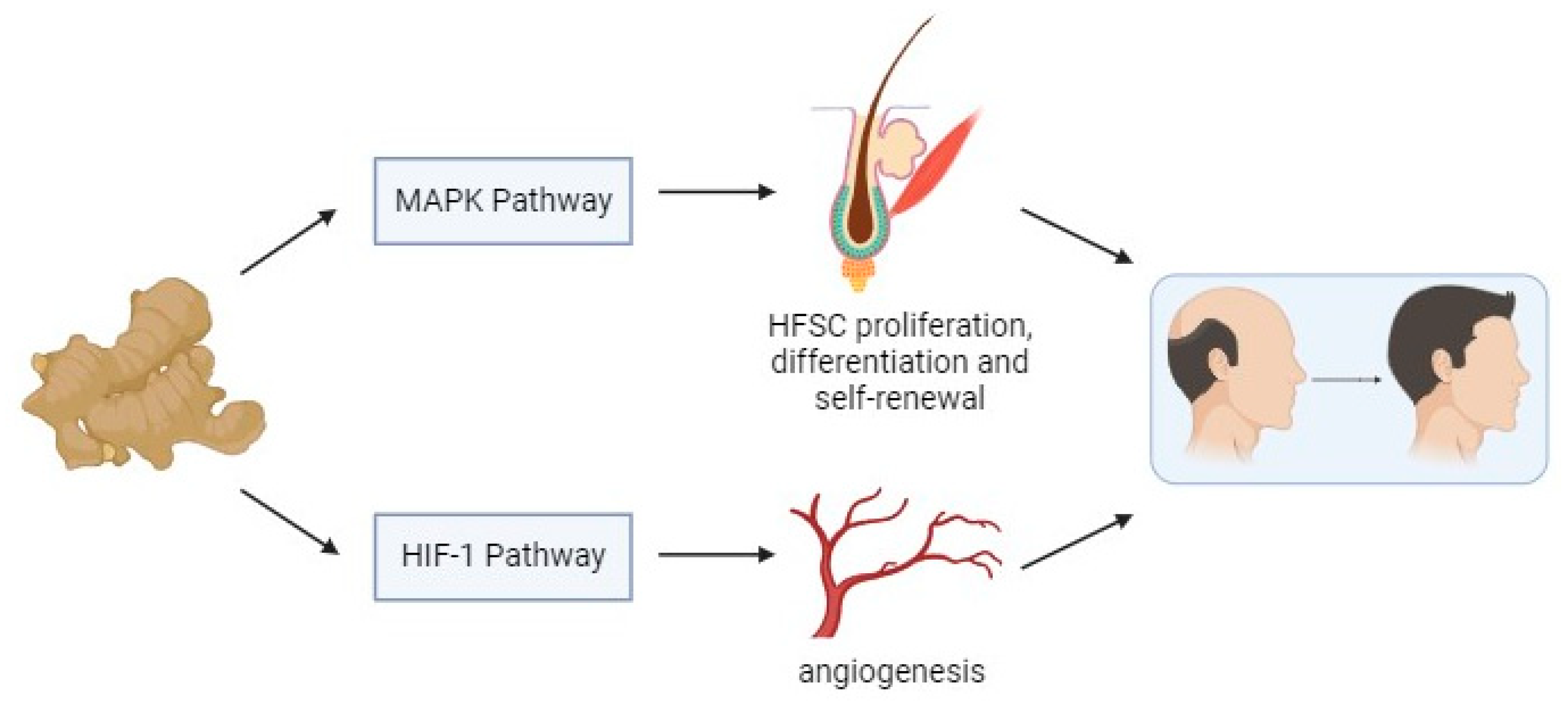
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Screening of Bioactive Compounds in C. aeruginosa
2.2. Target Gene Prediction in C. aeruginosa
2.3. Target Gene Prediction in Androgenetic Alopecia
2.4. Protein–Protein Interaction Network
2.5. Gene Ontology and Pathway Enrichment Analysis
2.6. Components–Targets–Pathways Network Construction
2.7. Molecular Docking
3. Results
3.1. Bioactive Compounds in C. aeruginosa
3.2. Predicted Target Genes of C. aeruginosa and AGA
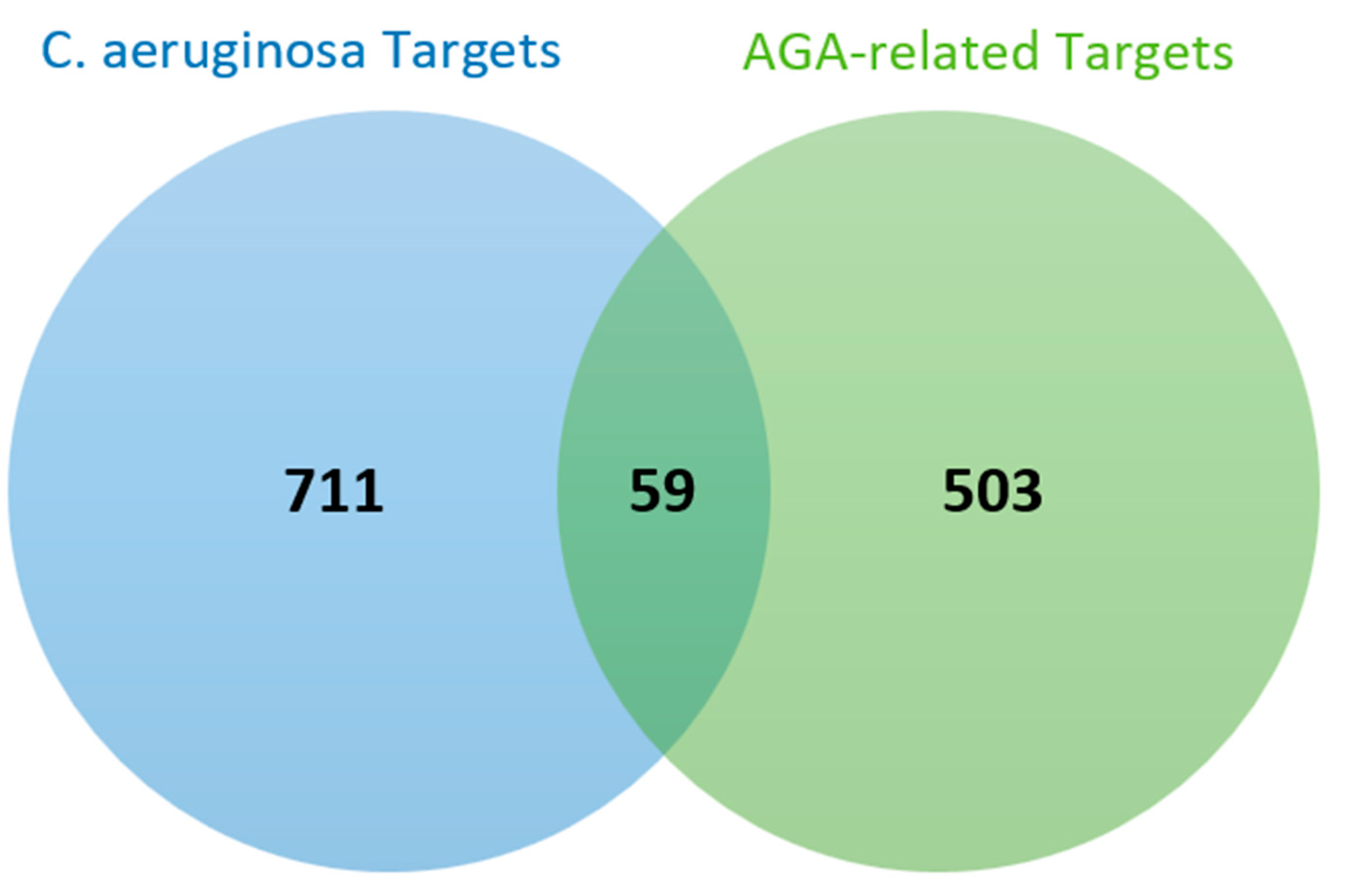
3.3. Common Targets of C. aeruginosa and AGA
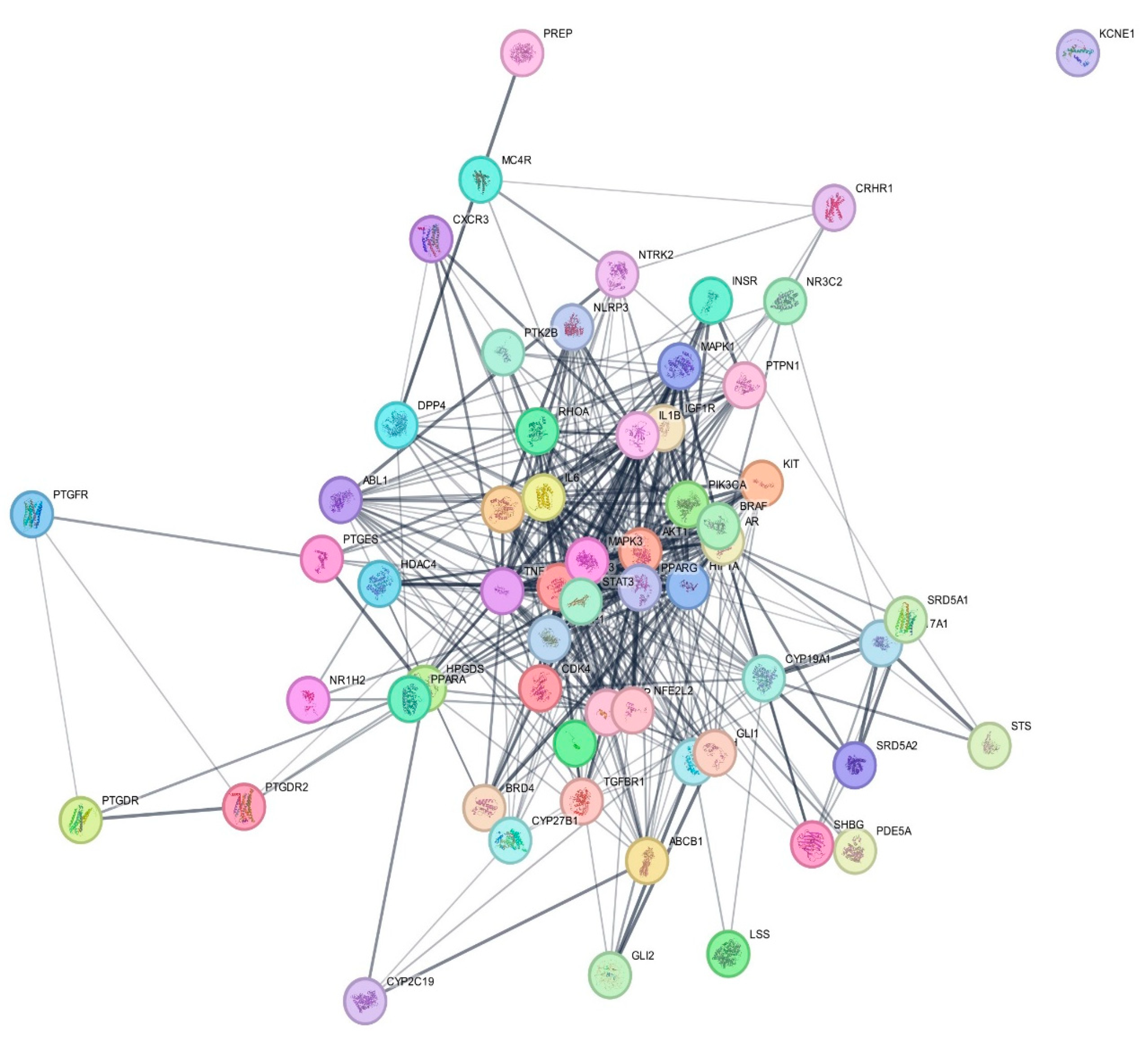
3.4. Protein–Protein Interaction (PPI) Network
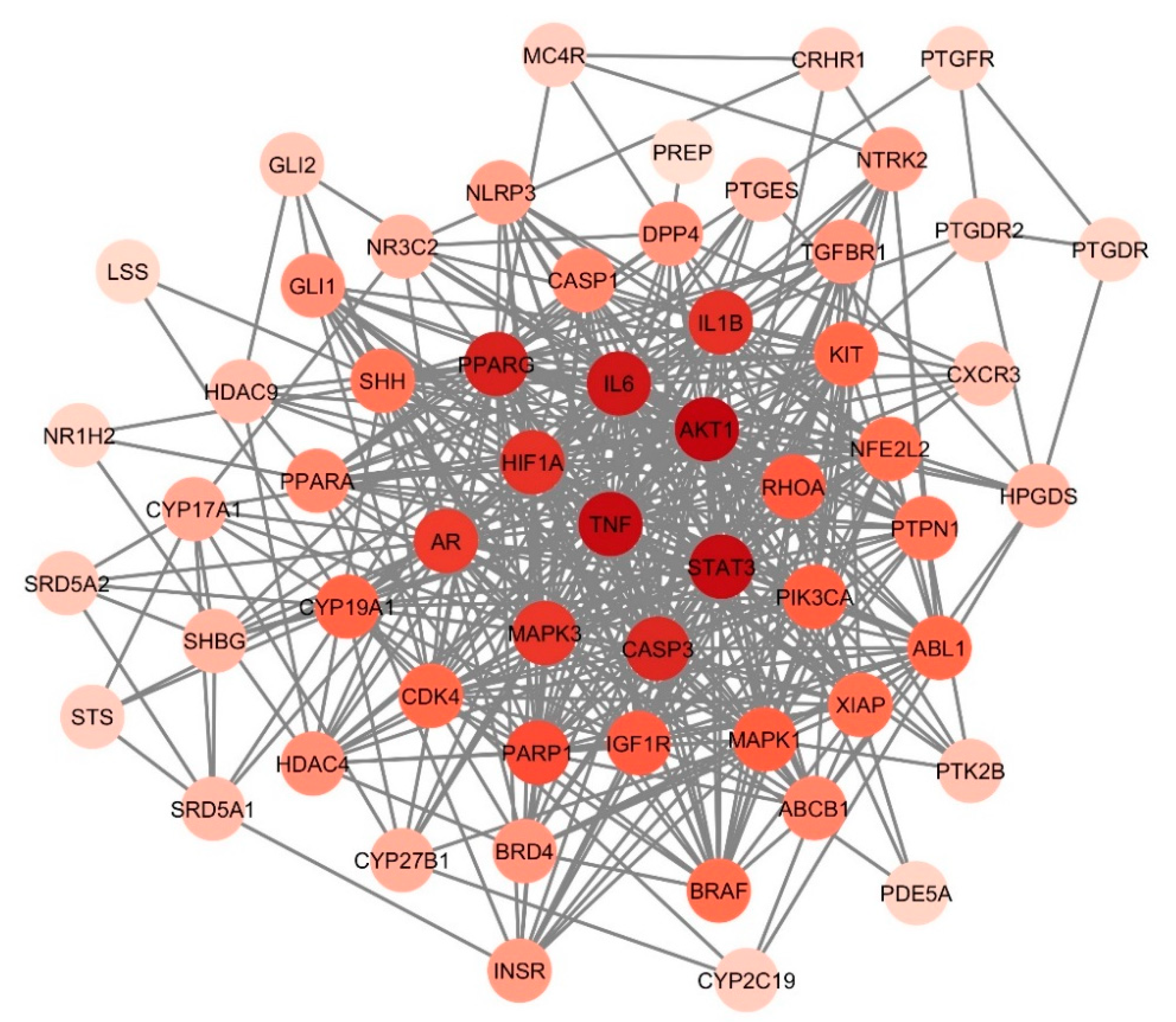
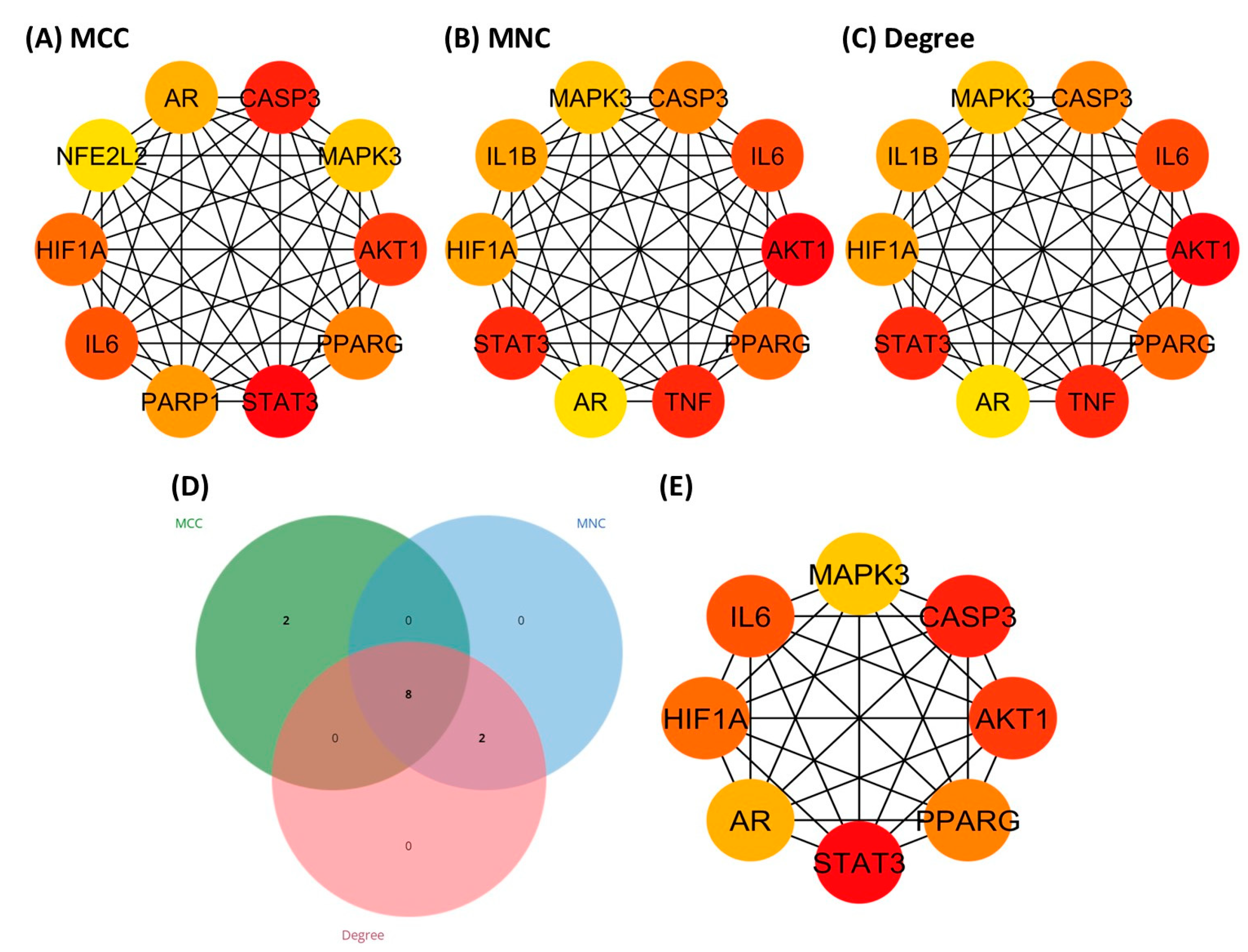
3.5. Identification of Hub Genes
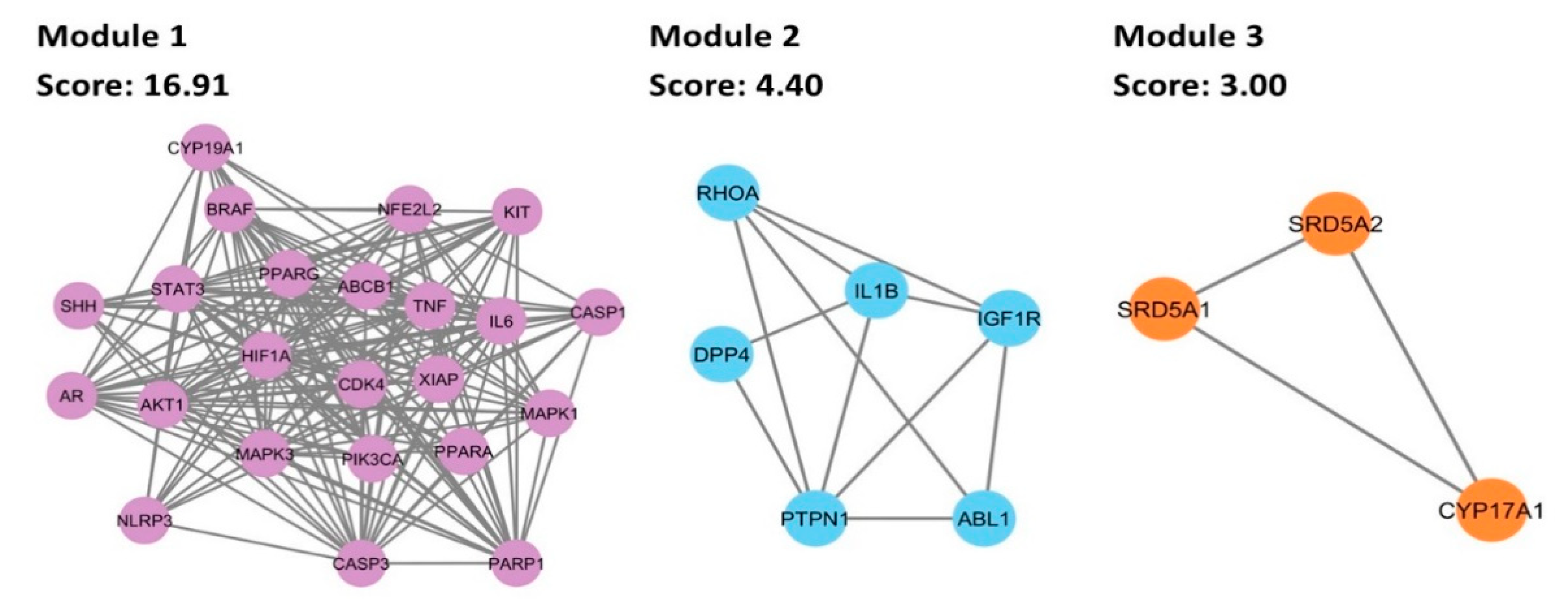
3.6. Cluster Analysis of PPI Network
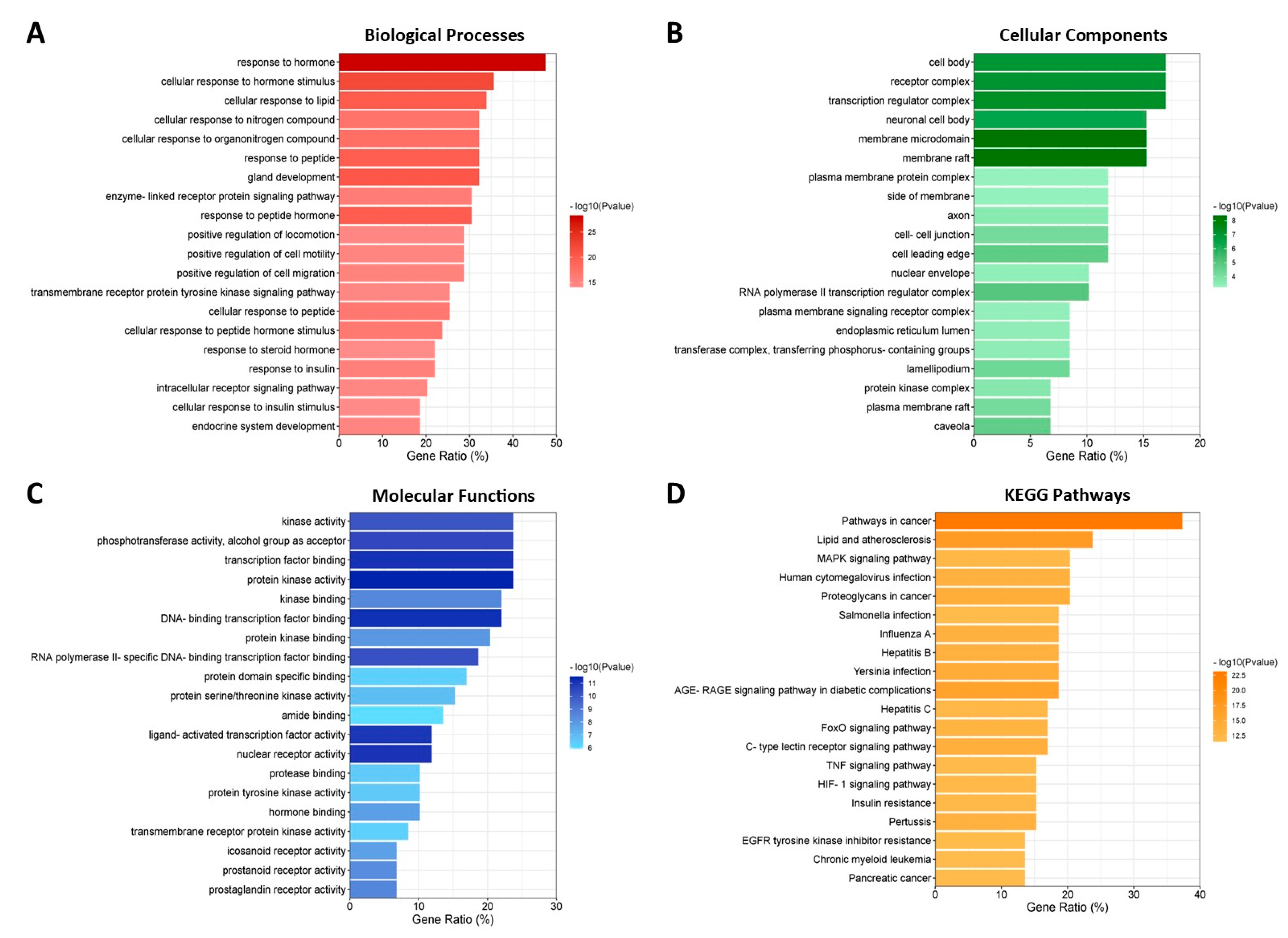
3.7. GO and KEGG Enrichment Analyses
3.8. Components–Targets–Pathways Network
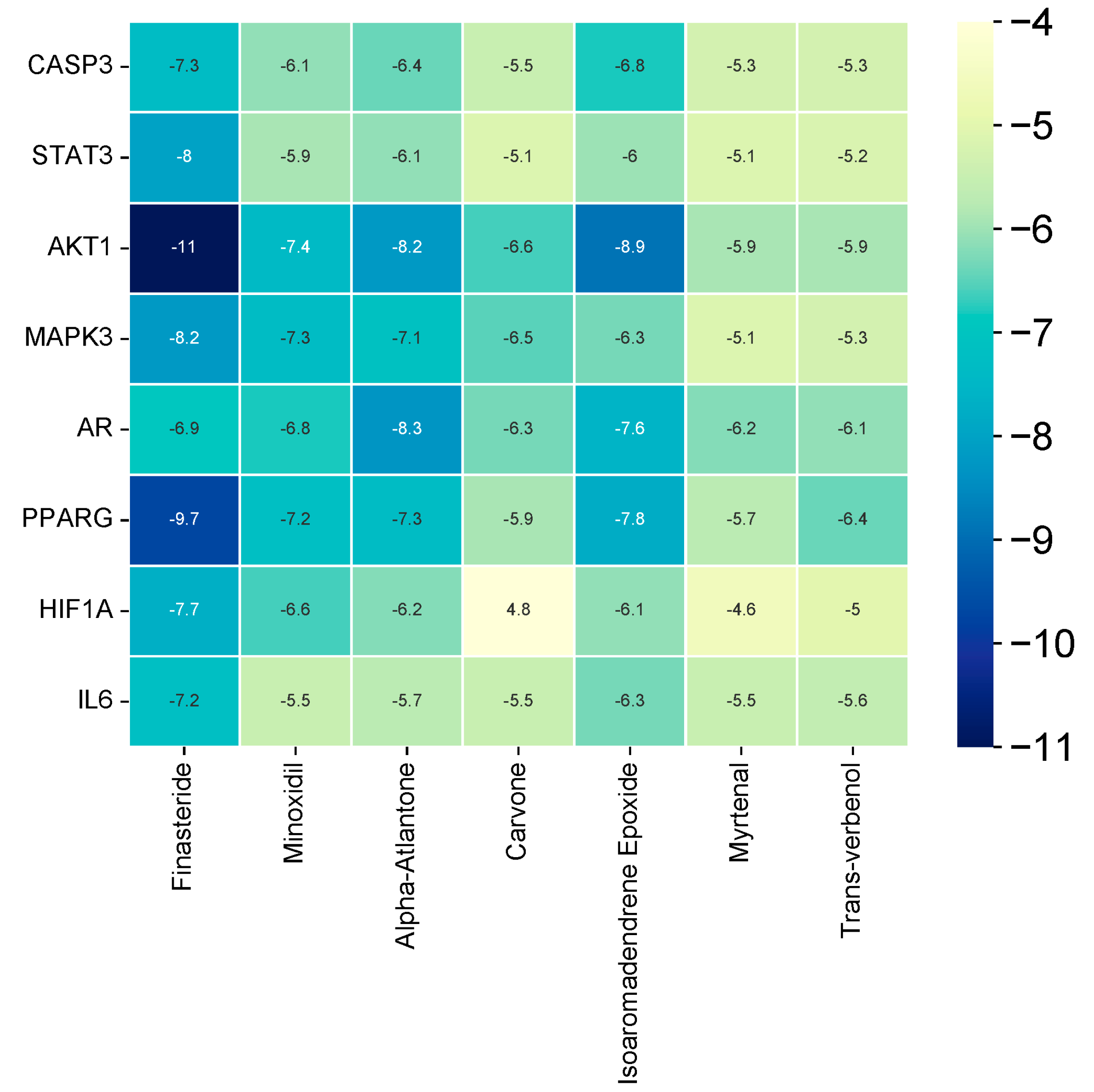
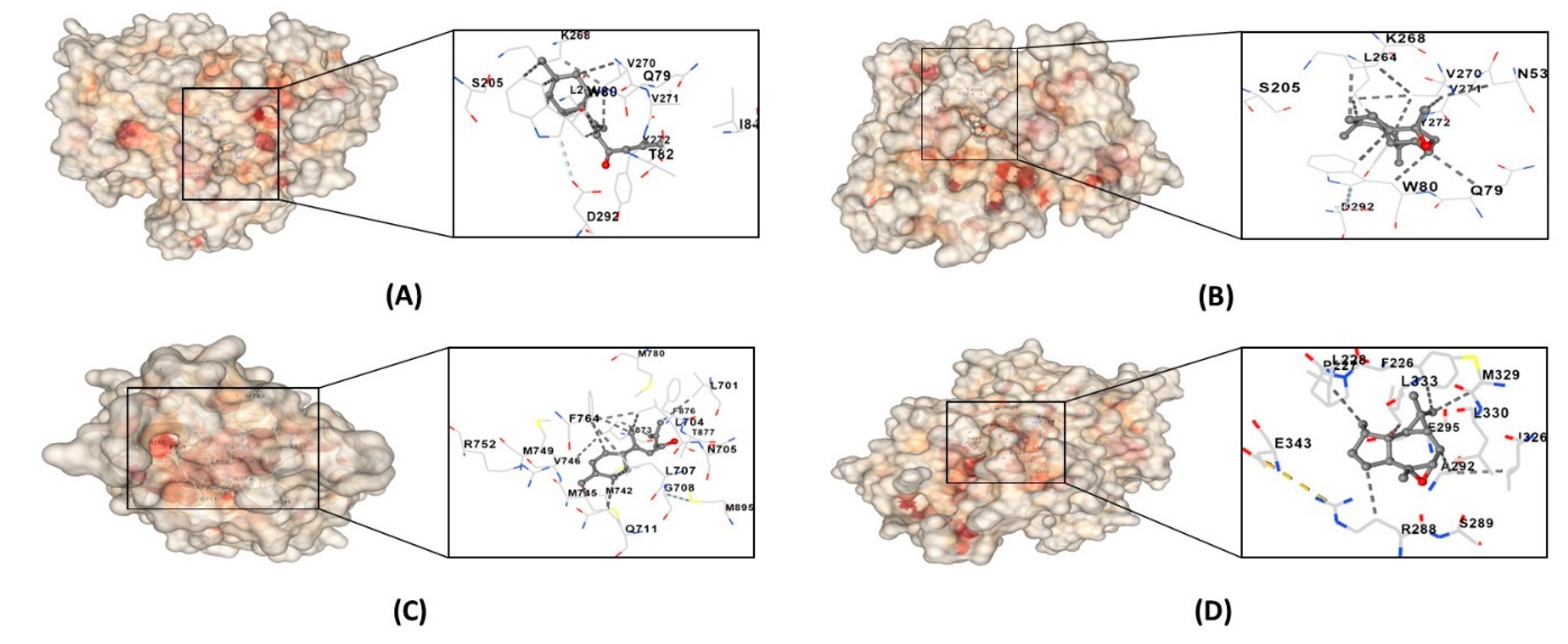
3.9. Molecular Docking
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| No. | Gene | Full Name |
|---|---|---|
| 1 | PIK3CA | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| 2 | CASP1 | caspase 1 |
| 3 | CYP2C19 | cytochrome P450 family 2 subfamily C member 19 |
| 4 | AR | androgen receptor |
| 5 | NR3C2 | nuclear receptor subfamily 3 group C member 2 |
| 6 | IGF1R | insulin-like growth factor 1 receptor |
| 7 | PARP1 | poly (ADP-ribose) polymerase 1 |
| 8 | KCNE1 | potassium voltage-gated channel subfamily E regulatory subunit 1 |
| 9 | VDR | vitamin D receptor |
| 10 | MAPK1 | mitogen-activated protein kinase 1 |
| 11 | CYP19A1 | cytochrome P450 family 19 subfamily A member 1 |
| 12 | PREP | prolyl endopeptidase |
| 13 | SRD5A1 | steroid 5 alpha-reductase 1 |
| 14 | HPGDS | hematopoietic prostaglandin D synthase |
| 15 | MC4R | melanocortin 4 receptor |
| 16 | CASP3 | caspase 3 |
| 17 | NLRP3 | NLR family pyrin domain containing 3 |
| 18 | BRD4 | bromodomain containing 4 |
| 19 | CYP17A1 | cytochrome P450 family 17 subfamily A member 1 |
| 20 | PTGES | prostaglandin E synthase |
| 21 | MAPK3 | mitogen-activated protein kinase 3 |
| 22 | SHBG | sex-hormone-binding globulin |
| 23 | SRD5A2 | steroid 5 alpha-reductase 2 |
| 24 | PPARG | peroxisome proliferator-activated receptor gamma |
| 25 | PPARA | peroxisome proliferator-activated receptor alpha |
| 26 | PTPN1 | protein tyrosine phosphatase non-receptor type 1 |
| 27 | IL6 | interleukin 6 |
| 28 | PTK2B | protein tyrosine kinase 2 beta |
| 29 | PDE5A | phosphodiesterase 5A |
| 30 | GLI2 | GLI family zinc finger 2 |
| 31 | GLI1 | GLI family zinc finger 1 |
| 32 | SHH | sonic hedgehog signaling molecule |
| 33 | DPP4 | dipeptidyl peptidase 4 |
| 34 | HIF1A | hypoxia-inducible factor 1 subunit alpha |
| 35 | NR1H2 | nuclear receptor subfamily 1 group H member 2 |
| 36 | PTGFR | prostaglandin F receptor |
| 37 | PTGDR | prostaglandin D2 receptor |
| 38 | TGFBR1 | transforming growth factor beta receptor 1 |
| 39 | IL1B | interleukin 1 beta |
| 40 | CRHR1 | corticotropin-releasing hormone receptor 1 |
| 41 | LSS | lanosterol synthase |
| 42 | TNF | tumor necrosis factor |
| 43 | CDK4 | cyclin-dependent kinase 4 |
| 44 | AKT1 | AKT serine/threonine kinase 1 |
| 45 | CXCR3 | C-X-C motif chemokine receptor 3 |
| 46 | KIT | KIT proto-oncogene receptor tyrosine kinase |
| 47 | RHOA | ras homolog family member A |
| 48 | ABCB1 | ATP-binding cassette subfamily B member 1 |
| 49 | BRAF | B-Raf proto-oncogene, serine/threonine kinase |
| 50 | STAT3 | signal transducer and activator of transcription 3 |
| 51 | PTGDR2 | prostaglandin D2 receptor 2 |
| 52 | ABL1 | ABL proto-oncogene 1 |
| 53 | STS | steroid sulfatase |
| 54 | NTRK2 | neurotrophic receptor tyrosine kinase 2 |
| 55 | XIAP | X-linked inhibitor of apoptosis |
| 56 | INSR | insulin receptor |
| 57 | NFE2L2 | NFE2-like bZIP transcription factor 2 |
| 58 | HDAC4 | histone deacetylase 4 |
| 59 | HDAC9 | histone deacetylase 9 |
| GO | Description | Count | % | Log (p-Value) |
|---|---|---|---|---|
| GO:0009725 | response to hormone | 28 | 47.46 | −28.2906 |
| GO:0032870 | cellular response to hormone stimulus | 21 | 35.59 | −22.1297 |
| GO:0048732 | gland development | 19 | 32.20 | −20.4392 |
| GO:0071396 | cellular response to lipid | 20 | 33.90 | −20.0435 |
| GO:0043434 | response to peptide hormone | 18 | 30.51 | −19.8568 |
| GO:1901652 | response to peptide | 19 | 32.20 | −19.7229 |
| GO:0071417 | cellular response to organonitrogen compound | 19 | 32.20 | −17.7248 |
| GO:1901699 | cellular response to nitrogen compound | 19 | 32.20 | −17.2401 |
| GO:1901653 | cellular response to peptide | 15 | 25.42 | −16.5512 |
| GO:0071375 | cellular response to peptide hormone stimulus | 14 | 23.73 | −16.4445 |
| GO:0007167 | enzyme-linked receptor protein signaling pathway | 18 | 30.51 | −16.0099 |
| GO:0032868 | response to insulin | 13 | 22.03 | −15.5597 |
| GO:0030335 | positive regulation of cell migration | 17 | 28.81 | −15.1975 |
| GO:0035270 | endocrine system development | 11 | 18.64 | −14.8955 |
| GO:2000147 | positive regulation of cell motility | 17 | 28.81 | −14.872 |
| GO:0030522 | intracellular receptor signaling pathway | 12 | 20.34 | −14.7579 |
| GO:0040017 | positive regulation of locomotion | 17 | 28.81 | −14.7092 |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 15 | 25.42 | −14.6503 |
| GO:0032869 | cellular response to insulin stimulus | 11 | 18.64 | −14.1934 |
| GO:0048545 | response to steroid hormone | 13 | 22.03 | −14.1231 |
| GO:0004672 | protein kinase activity | 14 | 23.72 | −11.5209 |
| GO:0140297 | DNA-binding transcription factor binding | 13 | 22.03 | −11.1747 |
| GO:0008134 | transcription factor binding | 14 | 23.73 | −11.1417 |
| GO:0004879 | nuclear receptor activity | 7 | 11.86 | −11.0367 |
| GO:0098531 | ligand-activated transcription factor activity | 7 | 11.86 | −10.9758 |
| GO:0016773 | phosphotransferase activity, alcohol group as acceptor | 14 | 23.73 | −10.5067 |
| GO:0061629 | RNA polymerase II-specific DNA-binding transcription factor binding | 11 | 18.64 | −10.2395 |
| GO:0016301 | kinase activity | 14 | 23.73 | −10.0562 |
| GO:0004955 | prostaglandin receptor activity | 4 | 6.78 | −8.56851 |
| GO:0019900 | kinase binding | 13 | 22.03 | −8.5368 |
| GO:0004954 | prostanoid receptor activity | 4 | 6.78 | −8.37285 |
| GO:0019901 | protein kinase binding | 12 | 20.34 | −8.04403 |
| GO:0042562 | hormone binding | 6 | 10.17 | −7.7874 |
| GO:0004953 | icosanoid receptor activity | 4 | 6.78 | −7.75875 |
| GO:0004674 | protein serine/threonine kinase activity | 9 | 15.25 | −6.86615 |
| GO:0004713 | protein tyrosine kinase activity | 6 | 10.17 | −6.52868 |
| GO:0002020 | protease binding | 6 | 10.17 | −6.49179 |
| GO:0019199 | transmembrane receptor protein kinase activity | 5 | 8.47 | −6.32271 |
| GO:0019904 | protein domain specific binding | 10 | 16.95 | −6.32105 |
| GO:0033218 | amide binding | 8 | 13.56 | −5.94186 |
| GO | Description | Count | % | Log (p-Value) |
|---|---|---|---|---|
| GO:0045121 | membrane raft | 9 | 15.25 | −8.37279 |
| GO:0098857 | membrane microdomain | 9 | 15.25 | −8.35955 |
| GO:0005667 | transcription regulator complex | 10 | 16.95 | −7.25901 |
| GO:0043235 | receptor complex | 10 | 16.95 | −7.04516 |
| GO:0044297 | cell body | 10 | 16.95 | −6.97898 |
| GO:0043025 | neuronal cell body | 9 | 15.25 | −6.39411 |
| GO:0090575 | RNA polymerase II transcription regulator complex | 6 | 10.17 | −4.98282 |
| GO:0031252 | cell leading edge | 7 | 11.86 | −4.73873 |
| GO:0005901 | caveola | 4 | 6.78 | −4.71441 |
| GO:0030027 | lamellipodium | 5 | 8.47 | −4.32896 |
| GO:0005911 | cell–cell junction | 7 | 11.86 | −4.20332 |
| GO:0044853 | plasma membrane raft | 4 | 6.78 | −4.16185 |
| GO:1902911 | protein kinase complex | 4 | 6.78 | −3.68335 |
| GO:0030424 | axon | 7 | 11.86 | −3.58421 |
| GO:0061695 | transferase complex, transferring phosphorus-containing groups | 5 | 8.47 | −3.47364 |
| GO:0005788 | endoplasmic reticulum lumen | 5 | 8.47 | −3.44166 |
| GO:0005635 | nuclear envelope | 6 | 10.17 | −3.39345 |
| GO:0098802 | plasma membrane signaling receptor complex | 5 | 8.47 | −3.36715 |
| GO:0098552 | side of membrane | 7 | 11.86 | −3.2816 |
| GO:0098797 | plasma membrane protein complex | 7 | 11.86 | −3.25701 |
| Pathway ID | Pathway Name | Log (p-Value) | Count | Gene Hits |
|---|---|---|---|---|
| hsa05200 | Pathways in cancer | −23.1337 | 22 | ABL1, AKT1, XIAP, AR, RHOA, BRAF, CASP3, CDK4, GLI1, GLI2, HIF1A, IGF1R, IL6, KIT, NFE2L2, PIK3CA, PPARG, MAPK1, MAPK3, SHH, STAT3, TGFBR1 |
| hsa05417 | Lipid and atherosclerosis | −17.2688 | 14 | AKT1, RHOA, CASP1, CASP3, IL1B, IL6, NFE2L2, PIK3CA, PPARG, MAPK1, MAPK3, STAT3, TNF, NLRP3 |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | −16.151 | 11 | AKT1, CASP3, CDK4, IL1B, IL6, PIK3CA, MAPK1, MAPK3, STAT3, TGFBR1, TNF |
| hsa05135 | Yersinia infection | −14.6017 | 11 | AKT1, RHOA, CASP1, PTK2B IL1B, IL6, PIK3CA, MAPK1 MAPK3, TNF, NLRP3 |
| hsa05205 | Proteoglycans in cancer | −14.248 | 12 | AKT1, RHOA, BRAF, CASP3, HIF1A, IGF1R, PIK3CA, MAPK1, MAPK3, SHH, STAT3, TNF |
| hsa04625 | C-type lectin receptor signaling pathway | −14.0983 | 10 | AKT1, RHOA, CASP1, IL1B, IL6, PIK3CA, MAPK1, MAPK3, TNF, NLRP3 |
| hsa05161 | Hepatitis B | −13.7886 | 11 | AKT1, BRAF, CASP3, PTK2B, IL6, PIK3CA, MAPK1, MAPK3, STAT3, TGFBR1, TNF |
| hsa05163 | Human cytomegalovirus infection | −13.7625 | 12 | AKT1, RHOA, CASP3, CDK4, PTK2B, IL1B, IL6, PIK3CA, MAPK1, MAPK3, STAT3, TNF |
| hsa05133 | Pertussis | −13.5622 | 9 | RHOA, CASP1, CASP3, IL1B, IL6, MAPK1, MAPK3, TNF, NLRP3 |
| hsa05164 | Influenza A | −13.5279 | 11 | AKT1, CASP1, CASP3, CDK4, IL1B, IL6, PIK3CA, MAPK1, MAPK3, TNF, NLRP3 |
| hsa04068 | FoxO signaling pathway | −13.0723 | 10 | AKT1, BRAF, IGF1R, IL6, INSR, PIK3CA, MAPK1, MAPK3, STAT3, TGFBR1 |
| hsa04010 | MAPK signaling pathway | −12.2603 | 12 | AKT1, BRAF, CASP3, IGF1R, IL1B, INSR, KIT NTRK2, MAPK1, MAPK3, TGFBR1, TNF |
| hsa05160 | Hepatitis C | −12.249 | 10 | AKT1, BRAF, CASP3, CDK4, PIK3CA, PPARA, MAPK1, MAPK3, STAT3, TNF |
| hsa04931 | Insulin resistance | −12.1443 | 9 | AKT1, IL6, INSR, PIK3CA, PPARA, PTPN1, STAT3, TNF, NR1H2 |
| hsa04066 | HIF-1 signaling pathway | −12.1075 | 9 | AKT1, HIF1A, IGF1R, IL6, INSR, PIK3CA, MAPK1 MAPK3, STAT3 |
| hsa04668 | TNF signaling pathway | −11.9288 | 9 | AKT1, XIAP, CASP3, IL1B, IL6, PIK3CA, MAPK1, MAPK3, TNF |
| hsa05132 | Salmonella infection | −11.7368 | 11 | AKT1, RHOA, CASP1 CASP3, IL1B, IL6, PIK3CA, MAPK1, MAPK3, TNF, NLRP3 |
| hsa05212 | Pancreatic cancer | −11.6678 | 8 | AKT1, BRAF, CDK4, PIK3CA, MAPK1, MAPK3, STAT3, TGFBR1 |
| hsa05220 | Chronic myeloid leukemia | −11.6678 | 8 | ABL1, AKT1, BRAF, CDK4, PIK3CA, MAPK1, MAPK3, TGFBR1 |
| hsa01521 | EGFR tyrosine kinase inhibitor resistance | −11.5287 | 8 | AKT1, BRAF, IGF1R, IL6, PIK3CA, MAPK1, MAPK3, STAT3 |
References
- Schwartzenfeld, D.M.; Karamikian, J. Hair Transplantation. In Plastic Surgery Secrets Plus; Elsevier: Amsterdam, The Netherlands, 2010; pp. 123–127. [Google Scholar]
- Frith, H.; Jankowski, G.S. Psychosocial Impact of Androgenetic Alopecia on Men: A Systematic Review and Meta-Analysis. Psychol. Health Med. 2023, 29, 822–842. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.S.; Karna, K.K.; Choi, B.R.; Park, J.K. Finasteride and Erectile Dysfunction in Patients with Benign Prostatic Hyperplasia or Male Androgenetic Alopecia. World J. Men’s Health 2019, 37, 157. [Google Scholar] [CrossRef]
- BinJadeed, H.; Almudimeegh, A.M.; Alomran, S.A.; Alshathry, A.H. A Case of Contact Allergic Dermatitis to Topical Minoxidil. Cureus 2021, 13, e12510. [Google Scholar] [CrossRef]
- Herman, A.; Herman, A.P. Mechanism of Action of Herbs and Their Active Constituents Used in Hair Loss Treatment. Fitoterapia 2016, 114, 18–25. [Google Scholar] [CrossRef]
- Nurcholis, W.; Priosoeryanto, B.P.; Purwakusumah, E.D.; Katayama, T.; Suzuki, T. Antioxidant, Cytotoxic Activities and Total Phenolic Content of Four Indonesian Medicinal Plants. J. Kim. Val. 2012, 2, 4. [Google Scholar] [CrossRef]
- Zohmachhuana, A.; Malsawmdawngliana; Lalnunmawia, F.; Mathipi, V.; Lalrinzuali, K.; Kumar, N.S. Curcuma aeruginosa Roxb. Exhibits Cytotoxicity in A-549 and HeLa Cells by Inducing Apoptosis through Caspase-Dependent Pathways. Biomed. Pharmacother. 2022, 150, 113039. [Google Scholar] [CrossRef]
- Akarchariya, N.; Sirilun, S.; Julsrigival, J.; Chansakaowa, S. Chemical Profiling and Antimicrobial Activity of Essential Oil from Curcuma aeruginosa Roxb., Curcuma Glans K. Larsen & J. Mood and Curcuma Cf. Xanthorrhiza Roxb. Collected in Thailand. Asian Pac. J. Trop. Biomed. 2017, 7, 881–885. [Google Scholar] [CrossRef]
- Sillapachaiyaporn, C.; Rangsinth, P.; Nilkhet, S.; Moungkote, N.; Chuchawankul, S. HIV-1 Protease and Reverse Transcriptase Inhibitory Activities of Curcuma aeruginosa Roxb. Rhizome Extracts and the Phytochemical Profile Analysis: In Vitro and In Silico Screening. Pharmaceuticals 2021, 14, 1115. [Google Scholar] [CrossRef]
- Thaina, P.; Tungcharoen, P.; Wongnawa, M.; Reanmongkol, W.; Subhadhirasakul, S. Uterine Relaxant Effects of Curcuma aeruginosa Roxb. Rhizome Extracts. J. Ethnopharmacol. 2009, 121, 433–443. [Google Scholar] [CrossRef]
- Suphrom, N.; Pumthong, G.; Khorana, N.; Waranuch, N.; Limpeanchob, N.; Ingkaninan, K. Anti-Androgenic Effect of Sesquiterpenes Isolated from the Rhizomes of Curcuma aeruginosa Roxb. Fitoterapia 2012, 83, 864–871. [Google Scholar] [CrossRef]
- Pumthong, G.; Asawanonda, P.; Varothai, S.; Jariyasethavong, V.; Triwongwaranat, D.; Suthipinittharm, P.; Ingkaninan, K.; Leelapornpisit, P.; Waranuch, N. Curcuma aeruginosa, a Novel Botanically Derived 5α-Reductase Inhibitor in the Treatment of Male-Pattern Baldness: A Multicenter, Randomized, Double-Blind, Placebo-Controlled Study. J. Dermatol. Treat. 2012, 23, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Srivilai, J.; Waranuch, N.; Tangsumranjit, A.; Khorana, N.; Ingkaninan, K. Germacrone and Sesquiterpene-Enriched Extracts from Curcuma aeruginosa Roxb. Increase Skin Penetration of Minoxidil, a Hair Growth Promoter. Drug Deliv. Transl. Res. 2018, 8, 140–149. [Google Scholar] [CrossRef]
- Li, S.; Zhang, B. Traditional Chinese Medicine Network Pharmacology: Theory, Methodology and Application. Chin. J. Nat. Med. 2013, 11, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Liu, L.; Li, J.; Zhang, Y.; Meng, D.-L. Virtual Screening of Active Compounds from Jasminum Lanceolarium and Potential Targets against Primary Dysmenorrhea Based on Network Pharmacology. Nat. Prod. Res. 2021, 35, 5853–5856. [Google Scholar] [CrossRef] [PubMed]
- Vivek-Ananth, R.P.; Mohanraj, K.; Sahoo, A.K.; Samal, A. IMPPAT 2.0: An Enhanced and Expanded Phytochemical Atlas of Indian Medicinal Plants. ACS Omega 2023, 8, 8827–8845. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated Data and New Features for Efficient Prediction of Protein Targets of Small Molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- Wang, J.-H.; Zhao, L.-F.; Wang, H.-F.; Wen, Y.-T.; Jiang, K.-K.; Mao, X.-M.; Zhou, Z.-Y.; Yao, K.-T.; Geng, Q.-S.; Guo, D.; et al. GenCLiP 3: Mining Human Genes’ Functions and Regulatory Networks from PubMed Based on Co-Occurrences and Natural Language Processing. Bioinformatics 2020, 36, 1973–1975. [Google Scholar] [CrossRef]
- Piñero, J.; Bravo, À.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A Comprehensive Platform Integrating Information on Human Disease-Associated Genes and Variants. Nucleic Acids Res. 2017, 45, D833–D839. [Google Scholar] [CrossRef]
- Amberger, J.S.; Bocchini, C.A.; Schiettecatte, F.; Scott, A.F.; Hamosh, A. OMIM.Org: Online Mendelian Inheritance in Man (OMIM®), an Online Catalog of Human Genes and Genetic Disorders. Nucleic Acids Res. 2015, 43, D789–D798. [Google Scholar] [CrossRef]
- Safran, M.; Dalah, I.; Alexander, J.; Rosen, N.; Iny Stein, T.; Shmoish, M.; Nativ, N.; Bahir, I.; Doniger, T.; Krug, H.; et al. GeneCards Version 3: The Human Gene Integrator. Database 2010, 2010, baq020. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING Database in 2011: Functional Interaction Networks of Proteins, Globally Integrated and Scored. Nucleic Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z.-X.; Cao, Y. CB-Dock2: Improved Protein-Ligand Blind Docking by Integrating Cavity Detection, Docking and Homologous Template Fitting. Nucleic Acids Res. 2022, 50, W159–W164. [Google Scholar] [CrossRef]
- Sari, A.P.; Supratman, U. Phytochemistry and Biological Activities of Curcuma aeruginosa (Roxb.). Indones. J. Chem. 2022, 22, 576–598. [Google Scholar] [CrossRef]
- Martin, Y.C. A Bioavailability Score. J. Med. Chem. 2005, 48, 3164–3170. [Google Scholar] [CrossRef] [PubMed]
- Saad, S.; Cavelier-Balloy, B.; Smadja, J.; Assouly, P.; Reygagne, P. Inflammatory Complications after Hair Transplantation: Report of 10 Cases. J. Cosmet. Dermatol. 2022, 21, 5938–5941. [Google Scholar] [CrossRef]
- Almohanna, H.M.; Perper, M.; Tosti, A. Safety Concerns When Using Novel Medications to Treat Alopecia. Expert Opin. Drug Saf. 2018, 17, 1115–1128. [Google Scholar] [CrossRef]
- Dou, J.; Zhang, Z.; Xu, X.; Zhang, X. Exploring the Effects of Chinese Herbal Ingredients on the Signaling Pathway of Alopecia and the Screening of Effective Chinese Herbal Compounds. J. Ethnopharmacol. 2022, 294, 115320. [Google Scholar] [CrossRef]
- Fan, X.; Chen, J.; Zhang, Y.; Wang, S.; Zhong, W.; Yuan, H.; Wu, X.; Wang, C.; Zheng, Y.; Wei, Y.; et al. Alpinetin Promotes Hair Regeneration via Activating Hair Follicle Stem Cells. Chin. Med. 2022, 17, 63. [Google Scholar] [CrossRef]
- Martinez-Jacobo, L.; Ancer-Arellano, C.I.; Ortiz-Lopez, R.; Salinas-Santander, M.; Villarreal-Villarreal, C.D.; Ancer-Rodriguez, J.; Camacho-Zamora, B.; Zomosa-Signoret, V.; Medina-De la Garza, C.E.; Ocampo-Candiani, J.; et al. Evaluation of the Expression of Genes Associated with Inflammation and Apoptosis in Androgenetic Alopecia by Targeted RNA-Seq. Skin Appendage Disord. 2018, 4, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wang, Z.; Zhang, J.; Shen, Y.; Hou, X.; Su, L.; Chen, W.; Chen, J.; Guo, X.; Song, H. Treatment of Androgenetic Alopecia by Exosomes Secreted from Hair Papilla Cells and the Intervention Effect of LTF. J. Cosmet. Dermatol. 2023, 22, 2996–3007. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Fan, Z.; Wang, X.; Mo, M.; Zeng, S.B.; Xu, R.-H.; Wang, X.; Wu, Y. PI3K/Akt Signaling Pathway Is Essential for de Novo Hair Follicle Regeneration. Stem Cell Res. Ther. 2020, 11, 144. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Yang, L.; Feldman, R.I.; Sun, X.; Bhalla, K.N.; Jove, R.; Nicosia, S.V.; Cheng, J.Q. Activation of Phosphatidylinositol 3-Kinase/Akt Pathway by Androgen through Interaction of P85α, Androgen Receptor, and Src. J. Biol. Chem. 2003, 278, 42992–43000. [Google Scholar] [CrossRef]
- Hibberts, N.; Howell, A.; Randall, V. Balding Hair Follicle Dermal Papilla Cells Contain Higher Levels of Androgen Receptors than Those from Non-Balding Scalp. J. Endocrinol. 1998, 156, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.A.; Stebbing, M.; Harrap, S.B. Polymorphism of the Androgen Receptor Gene Is Associated with Male Pattern Baldness. J. Investig. Dermatol. 2001, 116, 452–455. [Google Scholar] [CrossRef] [PubMed]
- Sawaya, M.E.; Price, V.H. Different Levels of 5α-Reductase Type I and II, Aromatase, and Androgen Receptor in Hair Follicles of Women and Men with Androgenetic Alopecia. J. Investig. Dermatol. 1997, 109, 296–300. [Google Scholar] [CrossRef] [PubMed]
- Hodgins, M.B.; Choudhry, R.; Parker, G.; Oliver, R.F.; Jahoda, C.A.B.; Withers, A.P.; Brinkmann, A.O.; Van Der Kwast, T.H.; Boersma, W.J.A.; Lammers, K.M.; et al. Androgen Receptors in Dermal Papilla Cells of Scalp Hair Follicles in Male Pattern Baldness. Ann. N. Y. Acad. Sci. 1991, 642, 448–451. [Google Scholar] [CrossRef]
- Mahmoud, E.A.; Elgarhy, L.H.; Hasby, E.A.; Mohammad, L. Dickkopf-1 Expression in Androgenetic Alopecia and Alopecia Areata in Male Patients. Am. J. Dermatopathol. 2019, 41, 122–127. [Google Scholar] [CrossRef]
- Tsuji, Y.; Denda, S.; Soma, T.; Raftery, L.; Momoi, T.; Hibino, T. A Potential Suppressor of TGF-β Delays Catagen Progression in Hair Follicles. J. Investig. Dermatol. Symp. Proc. 2003, 8, 65–68. [Google Scholar] [CrossRef]
- Kwack, M.H.; Ahn, J.S.; Kim, M.K.; Kim, J.C.; Sung, Y.K. Dihydrotestosterone-Inducible IL-6 Inhibits Elongation of Human Hair Shafts by Suppressing Matrix Cell Proliferation and Promotes Regression of Hair Follicles in Mice. J. Investig. Dermatol. 2012, 132, 43–49. [Google Scholar] [CrossRef]
- Ramot, Y.; Alam, M.; Oláh, A.; Bíró, T.; Ponce, L.; Chéret, J.; Bertolini, M.; Paus, R. Peroxisome Proliferator–Activated Receptor-Γ−Mediated Signaling Regulates Mitochondrial Energy Metabolism in Human Hair Follicle Epithelium. J. Investig. Dermatol. 2018, 138, 1656–1659. [Google Scholar] [CrossRef]
- Kwack, M.H.; Sung, Y.K.; Chung, E.J.; Im, S.U.; Ahn, J.S.; Kim, M.K.; Kim, J.C. Dihydrotestosterone-Inducible Dickkopf 1 from Balding Dermal Papilla Cells Causes Apoptosis in Follicular Keratinocytes. J. Investig. Dermatol. 2008, 128, 262–269. [Google Scholar] [CrossRef]
- Winiarska, A.; Mandt, N.; Kamp, H.; Hossini, A.; Seltmann, H.; Zouboulis, C.C.; Blume-Peytavi, U. Effect of 5α-Dihydrotestosterone and Testosterone on Apoptosis in Human Dermal Papilla Cells. Skin Pharmacol. Physiol. 2006, 19, 311–321. [Google Scholar] [CrossRef]
- Sano, S.; Kira, M.; Takagi, S.; Yoshikawa, K.; Takeda, J.; Itami, S. Two Distinct Signaling Pathways in Hair Cycle Induction: Stat3-Dependent and -Independent Pathways. Proc. Natl. Acad. Sci. USA 2000, 97, 13824–13829. [Google Scholar] [CrossRef]
- Kim, D.J.; Kataoka, K.; Rao, D.; Kiguchi, K.; Cotsarelis, G.; DiGiovanni, J. Targeted Disruption of Stat3 Reveals a Major Role for Follicular Stem Cells in Skin Tumor Initiation. Cancer Res. 2009, 69, 7587–7594. [Google Scholar] [CrossRef]
- Rao, D.; Macias, E.; Carbajal, S.; Kiguchi, K.; DiGiovanni, J. Constitutive Stat3 Activation Alters Behavior of Hair Follicle Stem and Progenitor Cell Populations. Mol. Carcinog. 2015, 54, 121–133. [Google Scholar] [CrossRef]
- Seo, J.; Yan, L.; Kageyama, T.; Nanmo, A.; Chun, Y.-S.; Fukuda, J. Hypoxia Inducible Factor-1α Promotes Trichogenic Gene Expression in Human Dermal Papilla Cells. Sci. Rep. 2023, 13, 1478. [Google Scholar] [CrossRef]
- Bukowiecki, J.; Pförringer, D.; Thor, D.; Duscher, D.; Brett, E. HIF-1α Stimulators Function Equally to Leading Hair Loss Agents in Enhancing Dermal Papilla Growth. Skin Pharmacol. Physiol. 2020, 33, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Akilli Öztürk, Ö.; Pakula, H.; Chmielowiec, J.; Qi, J.; Stein, S.; Lan, L.; Sasaki, Y.; Rajewsky, K.; Birchmeier, W. Gab1 and Mapk Signaling Are Essential in the Hair Cycle and Hair Follicle Stem Cell Quiescence. Cell Rep. 2015, 13, 561–572. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.; Choi, S.-J.; Jang, S.; Choi, H.-I.; Kang, B.-M.; Hwang, S.T.; Kwon, O. Shikimic Acid, a Mannose Bioisostere, Promotes Hair Growth with the Induction of Anagen Hair Cycle. Sci. Rep. 2019, 9, 17008. [Google Scholar] [CrossRef] [PubMed]
- Stamatas, G.N.; Wu, J.; Pappas, A.; Mirmirani, P.; McCormick, T.S.; Cooper, K.D.; Consolo, M.; Schastnaya, J.; Ozerov, I.V.; Aliper, A.; et al. An Analysis of Gene Expression Data Involving Examination of Signaling Pathways Activation Reveals New Insights into the Mechanism of Action of Minoxidil Topical Foam in Men with Androgenetic Alopecia. Cell Cycle 2017, 16, 1578–1584. [Google Scholar] [CrossRef]
- Deng, Z.; Chen, M.; Liu, F.; Wang, Y.; Xu, S.; Sha, K.; Peng, Q.; Wu, Z.; Xiao, W.; Liu, T.; et al. Androgen Receptor–Mediated Paracrine Signaling Induces Regression of Blood Vessels in the Dermal Papilla in Androgenetic Alopecia. J. Investig. Dermatol. 2022, 142, 2088–2099.e9. [Google Scholar] [CrossRef]
- Garza, L.A.; Liu, Y.; Yang, Z.; Alagesan, B.; Lawson, J.A.; Norberg, S.M.; Loy, D.E.; Zhao, T.; Blatt, H.B.; Stanton, D.C.; et al. Prostaglandin D 2 Inhibits Hair Growth and Is Elevated in Bald Scalp of Men with Androgenetic Alopecia. Sci. Transl. Med. 2012, 4, 126ra34. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.H.; Chae, C.W.; Choi, G.E.; Shin, H.C.; Lim, J.R.; Chang, H.S.; Park, J.; Cho, J.H.; Park, M.R.; Lee, H.J.; et al. Cyanidin 3-O-Arabinoside Suppresses DHT-Induced Dermal Papilla Cell Senescence by Modulating P38-Dependent ER-Mitochondria Contacts. J. Biomed. Sci. 2022, 29, 17. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.S.; Smart, R.C. An Estrogen Receptor Pathway Regulates the Telogen-Anagen Hair Follicle Transition and Influences Epidermal Cell Proliferation. Proc. Natl. Acad. Sci. USA 1996, 93, 12525–12530. [Google Scholar] [CrossRef] [PubMed]
- Liedert, A.; Nemitz, C.; Haffner-Luntzer, M.; Schick, F.; Jakob, F.; Ignatius, A. Effects of Estrogen Receptor and Wnt Signaling Activation on Mechanically Induced Bone Formation in a Mouse Model of Postmenopausal Bone Loss. Int. J. Mol. Sci. 2020, 21, 8301. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.Y. Targeting Wnt/β-Catenin Pathway for Developing Therapies for Hair Loss. Int. J. Mol. Sci. 2020, 21, 4915. [Google Scholar] [CrossRef] [PubMed]
- Bellanti, F.; Matteo, M.; Rollo, T.; De Rosario, F.; Greco, P.; Vendemiale, G.; Serviddio, G. Sex Hormones Modulate Circulating Antioxidant Enzymes: Impact of Estrogen Therapy. Redox Biol. 2013, 1, 340–346. [Google Scholar] [CrossRef]
- Choi, S.; Zhang, B.; Ma, S.; Gonzalez-Celeiro, M.; Stein, D.; Jin, X.; Kim, S.T.; Kang, Y.-L.; Besnard, A.; Rezza, A.; et al. Corticosterone Inhibits GAS6 to Govern Hair Follicle Stem-Cell Quiescence. Nature 2021, 592, 428–432. [Google Scholar] [CrossRef]
- Raman, M.; Chen, W.; Cobb, M.H. Differential Regulation and Properties of MAPKs. Oncogene 2007, 26, 3100–3112. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, S.R.; Choi, Y.-H.; Shin, J.Y.; Kim, C.D.; Kang, N.-G.; Park, B.C.; Lee, S. Quercitrin Stimulates Hair Growth with Enhanced Expression of Growth Factors via Activation of MAPK/CREB Signaling Pathway. Molecules 2020, 25, 4004. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Y.; He, J.; Wang, J.; Chen, X.; Yang, R. Regulation of signaling pathways in hair follicle stem cells. Burns Trauma 2022, 10, tkac022. [Google Scholar] [CrossRef]
- Huang, H.-C.; Lin, H.; Huang, M.-C. Lactoferrin Promotes Hair Growth in Mice and Increases Dermal Papilla Cell Proliferation through Erk/Akt and Wnt Signaling Pathways. Arch. Dermatol. Res. 2019, 311, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.K.; Kang, J.-I.; Hyun, J.W.; Koh, Y.S.; Kang, J.-H.; Hyun, C.-G.; Yoon, K.-S.; Lee, K.S.; Lee, C.M.; Kim, T.Y.; et al. Myristoleic Acid Promotes Anagen Signaling by Autophagy through Activating Wnt/β-Catenin and ERK Pathways in Dermal Papilla Cells. Biomol. Ther. 2021, 29, 211–219. [Google Scholar] [CrossRef]
- Bikkavilli, R.K.; Feigin, M.E.; Malbon, C.C. P38 Mitogen-Activated Protein Kinase Regulates Canonical Wnt–β-Catenin Signaling by Inactivation of GSK3β. J. Cell Sci. 2008, 121, 3598–3607. [Google Scholar] [CrossRef]
- He, Y.; Cai, C.; Sun, S.; Wang, X.; Li, W.; Li, H. Effect of JNK Inhibitor SP600125 on Hair Cell Regeneration in Zebrafish (Danio rerio) Larvae. Oncotarget 2016, 7, 51640–51650. [Google Scholar] [CrossRef] [PubMed]
- Yum, S.; Jeong, S.; Kim, D.; Lee, S.; Kim, W.; Yoo, J.-W.; Kim, J.-A.; Kwon, O.; Kim, D.-D.; Min, D.; et al. Minoxidil Induction of VEGF Is Mediated by Inhibition of HIF-Prolyl Hydroxylase. Int. J. Mol. Sci. 2017, 19, 53. [Google Scholar] [CrossRef]
- Imamura, Y.; Tomita, S.; Imanishi, M.; Kihira, Y.; Ikeda, Y.; Ishizawa, K.; Tsuchiya, K.; Tamaki, T. HIF-2α/ARNT Complex Regulates Hair Development via Induction of P21 Waf1/Cip1 and P27 Kip1. FASEB J. 2014, 28, 2517–2524. [Google Scholar] [CrossRef]
- Pagani, A.; Aitzetmüller, M.M.; Brett, E.A.; König, V.; Wenny, R.; Thor, D.; Radtke, C.; Huemer, G.M.; Machens, H.-G.; Duscher, D. Skin Rejuvenation through HIF-1α Modulation. Plast. Reconstr. Surg. 2018, 141, 600e–607e. [Google Scholar] [CrossRef]
- Thor, D.; Pagani, A.; Bukowiecki, J.; Houschyar, K.S.; Kølle, S.-F.T.; Wyles, S.P.; Duscher, D. A Novel Hair Restoration Technology Counteracts Androgenic Hair Loss and Promotes Hair Growth in A Blinded Clinical Trial. J. Clin. Med. 2023, 12, 470. [Google Scholar] [CrossRef] [PubMed]
- Ceruti, J.M.; Leirós, G.J.; Balañá, M.E. Androgens and Androgen Receptor Action in Skin and Hair Follicles. Mol. Cell. Endocrinol. 2018, 465, 122–133. [Google Scholar] [CrossRef] [PubMed]
- O’Shaughnessy, P.J.; Antignac, J.P.; Le Bizec, B.; Morvan, M.-L.; Svechnikov, K.; Söder, O.; Savchuk, I.; Monteiro, A.; Soffientini, U.; Johnston, Z.C.; et al. Alternative (Backdoor) Androgen Production and Masculinization in the Human Fetus. PLoS Biol. 2019, 17, e3000002. [Google Scholar] [CrossRef] [PubMed]
- Fukami, M.; Homma, K.; Hasegawa, T.; Ogata, T. Backdoor Pathway for Dihydrotestosterone Biosynthesis: Implications for Normal and Abnormal Human Sex Development. Dev. Dyn. 2013, 242, 320–329. [Google Scholar] [CrossRef]
- Shen, Y.; Zhu, Y.; Zhang, L.; Sun, J.; Xie, B.; Zhang, H.; Song, X. New Target for Minoxidil in the Treatment of Androgenetic Alopecia. Drug Des. Dev. Ther. 2023, 17, 2537–2547. [Google Scholar] [CrossRef]

| No. | Compound | MW | OB | GIA | Drug-Likeness | ||||
|---|---|---|---|---|---|---|---|---|---|
| Lipinski | Ghose | Veber | Muegge | Egan | |||||
| 1 | Zedoarol | 246.31 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 2 | Myrcene * | 136.24 | 0.55 | Low | Yes | Yes | No | Yes | Yes |
| 3 | Trans-Tagetone | 152.24 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 4 | Furanodienone | 230.31 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 5 | gamma-Terpinene * | 136.24 | 0.55 | Low | Yes | Yes | No | Yes | Yes |
| 6 | 1-Hexen-3-OL | 100.16 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 7 | p-Cymene * | 134.22 | 0.55 | Low | Yes | Yes | No | Yes | Yes |
| 8 | Curdione | 236.36 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 9 | Myrtenal | 150.22 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 10 | Germacrone | 218.34 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 11 | Isocurcumenol | 234.34 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 12 | 1-Hexanol | 102.18 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 13 | beta-Cubebene * | 204.36 | 0.55 | Low | Yes | Yes | Yes | Yes | Yes |
| 14 | Eucalyptol | 154.25 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 15 | beta-Elemene * | 204.36 | 0.55 | Low | No | Yes | Yes | Yes | Yes |
| 16 | Furanogermenone | 232.32 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 17 | Curzerene | 216.32 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 18 | (+)-Curcumenol | 234.34 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 19 | 4-Carvomenthenol | 154.25 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 20 | (1R)-2-methyl-5-propan-2-ylbicyclo[3.1.0]hex-2-ene * | 136.24 | 0.55 | Low | Yes | Yes | No | Yes | Yes |
| 21 | Curzerenone | 230.31 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 22 | alpha-Selinene * | 204.36 | 0.55 | Low | No | Yes | Yes | Yes | Yes |
| 23 | cis-3-Hexen-1-ol | 100.16 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 24 | d-Borneol | 154.25 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 25 | Terpinolene * | 136.24 | 0.55 | Low | Yes | Yes | No | Yes | Yes |
| 26 | beta-Farnesene * | 204.36 | 0.55 | Low | No | Yes | Yes | Yes | Yes |
| 27 | Curcumanolide B | 234.34 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 28 | Curcumanolides A | 234.34 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 29 | 2-Hexen-1-OL | 100.16 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 30 | Humulene * | 204.36 | 0.55 | Low | No | Yes | Yes | Yes | Yes |
| 31 | Pulegone | 152.24 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 32 | Thujone | 152.24 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 33 | (+)-delta-Cadinene * | 204.36 | 0.55 | Low | No | Yes | Yes | Yes | Yes |
| 34 | (-)-cis-Carveol | 152.24 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 35 | Camphor | 152.24 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 36 | Linalool | 154.25 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 37 | alpha-Pinene * | 136.24 | 0.55 | Low | Yes | Yes | No | Yes | Yes |
| 38 | Carvone | 150.22 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 39 | beta-Pinene * | 136.24 | 0.55 | Low | Yes | Yes | No | Yes | Yes |
| 40 | alpha-Fenchol | 154.25 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 41 | alpha-Terpineol | 154.25 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 42 | Sabinene * | 136.24 | 0.55 | Low | Yes | Yes | No | Yes | Yes |
| 43 | trans-Pinocarveol | 152.24 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 44 | Caryophyllene oxide | 220.36 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 45 | Phytol * | 296.54 | 0.55 | Low | No | Yes | No | No | No |
| 46 | (Z)-beta-Ocimene * | 136.24 | 0.55 | Low | Yes | Yes | No | Yes | Yes |
| 47 | gamma-Elemene * | 204.36 | 0.55 | Low | No | Yes | Yes | Yes | Yes |
| 48 | beta-Selinene * | 204.36 | 0.55 | Low | No | Yes | Yes | Yes | Yes |
| 49 | beta-Caryophyllene * | 204.36 | 0.55 | Low | No | Yes | Yes | Yes | Yes |
| 50 | (E)-beta-ocimene * | 136.24 | 0.55 | Low | Yes | Yes | No | Yes | Yes |
| 51 | Camphene * | 136.24 | 0.55 | Low | Yes | Yes | No | Yes | Yes |
| 52 | Limonene * | 136.24 | 0.55 | Low | Yes | Yes | No | Yes | Yes |
| 53 | trans-Verbenol | 152.24 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 54 | Allo-Aromadendrene * | 204.36 | 0.55 | Low | Yes | Yes | Yes | Yes | Yes |
| 55 | 2-Heptanol | 116.2 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 56 | Myrtenol | 152.24 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 57 | beta-Bisabolene * | 204.36 | 0.55 | Low | No | Yes | Yes | Yes | Yes |
| 58 | Curcumenone | 234.43 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 59 | 2-Undecanol | 172.31 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 60 | gamma-Terpineol | 154.24 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 61 | Humuladienone | 220.36 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 62 | (-)-beta-Curcumene * | 204.36 | 0.55 | Low | No | Yes | Yes | Yes | Yes |
| 63 | Dehydrocurdione | 234.43 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 64 | Tetradecanal | 212.38 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 65 | Bisacumol | 218.34 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 66 | Tricyclene * | 136.24 | 0.55 | Low | Yes | Yes | No | Yes | Yes |
| 67 | Linalyl acetate | 196.29 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 68 | 4′-Methylacetophenone | 134.18 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 69 | Xanthorrhizol | 218.34 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 70 | 1-Methyl-4-(prop-1-en-2-yl)benzene * | 132.21 | 0.55 | Low | Yes | Yes | No | Yes | Yes |
| 71 | Linalyl isobutyrate | 224.34 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 72 | alpha-Guaiene * | 204.36 | 0.55 | Low | No | Yes | Yes | Yes | Yes |
| 73 | 2-Nonanol | 144.26 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 74 | 2-Nonanone | 142.24 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 75 | 3,7(11)-Eudesmadiene * | 204.46 | 0.55 | Low | No | Yes | Yes | Yes | Yes |
| 76 | Camphene hydrate | 154.25 | 0.55 | High | Yes | Yes | No | Yes | Yes |
| 78 | Curcuphenol | 218.34 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 79 | Turmerol | 220.36 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 80 | 2-Undecanone | 170.3 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 81 | beta-Eudesmol | 222.37 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 82 | Farnesol | 222.37 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 83 | alpha-Terpinene * | 136.23 | 0.55 | Low | Yes | Yes | No | Yes | Yes |
| 84 | cis-beta-Farnesene * | 204.36 | 0.55 | Low | No | Yes | Yes | Yes | Yes |
| 85 | Zingiberene * | 204.36 | 0.55 | Low | No | Yes | Yes | Yes | Yes |
| 86 | (+)-beta-Phellandrene * | 136.23 | 0.55 | Low | Yes | Yes | No | Yes | Yes |
| 87 | alpha-Curcumene * | 202.34 | 0.55 | Low | No | Yes | Yes | Yes | Yes |
| 88 | ar-Turmerone | 216.32 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 89 | 3-(1,5-Dimethyl-4-hexenyl)-6-methylene-1-cyclohexene * | 204.36 | 0.55 | Low | Yes | Yes | No | Yes | Yes |
| 90 | Linalool oxide B | 170.25 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 91 | delta-Elemene * | 204.36 | 0.55 | Low | No | Yes | Yes | Yes | Yes |
| 92 | alpha-Atlantone | 218.34 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 93 | alpha-Phellandrene * | 136.24 | 0.55 | Low | Yes | Yes | No | Yes | Yes |
| 94 | Nerolidol | 222.37 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 95 | Flavone | 222.24 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 96 | Zedoalactone A | 266.36 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 97 | Zedoarondiol | 252.35 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 98 | Furanodiene | 216.32 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 99 | Zederone | 246.30 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 100 | Pyrocurzerenone | 212.29 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 101 | Dehydrochromolaenin | 210.27 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 102 | Isoaromadendrene epoxide | 220.35 | 0.55 | High | Yes | Yes | Yes | Yes | Yes |
| 103 | Demethoxycurcumin | 338.35 | 0.55 | High | No | Yes | Yes | Yes | Yes |
| Compound | Protein | Binding Sites |
|---|---|---|
| alpha-atlantone | AR | Leu701, Leu704, Asn705, Leu707, Gly708, Gln711, Trp741, Met742, Met745, Val746, Met749, Arg752, Phe764, Met780, Met787, Leu873, Phe876, THR877 Leu880, Phe891, Met895, Ile899 |
| AKT1 | Asn53, Asn54, Ala58, Gln59, Leu78, Gln79, Trp80, Thr82, Ile84, Asn199, Val201, Ser205, Leu210, Thr211, Leu264, Lys268, Val270, Val271, Tyr272, Ile290, Thr291, Asp292 | |
| isoaromandendrene epoxide | AKT1 | Glu17, Tyr18, Asn53, Asn54, Ser56, Ala58, Gln59, Gln79, Trp80, Thr81, Thr82, Ile84, Glu85, Arg86, Lys179, Val201, Ser205, Leu210, Thr211, Leu213, Tyr263, Leu264, Lys268, Val270, Val271, Tyr272, Arg273, Asp274, Asn279, Thr291, Asp292, Phe293, Gly294, Cys296, Lys297, Glu298, Tyr326 |
| PPARG | Phe226, Pro227, Leu228, Gly284, Cys285, Arg288, Ser289, Glu291, Ala292 Glu295, Ile296, Ile325, Ile326, Met329, Leu330, Leu333, Val339, Leu340, Ile341, Ser342, Glu343, Gly344, Met364 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sintos, A.M.L.; Cabrera, H.S. Network Pharmacology Reveals Curcuma aeruginosa Roxb. Regulates MAPK and HIF-1 Pathways to Treat Androgenetic Alopecia. Biology 2024, 13, 497. https://doi.org/10.3390/biology13070497
Sintos AML, Cabrera HS. Network Pharmacology Reveals Curcuma aeruginosa Roxb. Regulates MAPK and HIF-1 Pathways to Treat Androgenetic Alopecia. Biology. 2024; 13(7):497. https://doi.org/10.3390/biology13070497
Chicago/Turabian StyleSintos, Aaron Marbyn L., and Heherson S. Cabrera. 2024. "Network Pharmacology Reveals Curcuma aeruginosa Roxb. Regulates MAPK and HIF-1 Pathways to Treat Androgenetic Alopecia" Biology 13, no. 7: 497. https://doi.org/10.3390/biology13070497
APA StyleSintos, A. M. L., & Cabrera, H. S. (2024). Network Pharmacology Reveals Curcuma aeruginosa Roxb. Regulates MAPK and HIF-1 Pathways to Treat Androgenetic Alopecia. Biology, 13(7), 497. https://doi.org/10.3390/biology13070497






