The Utility of the Koala Scat: A Scoping Review
Abstract
Simple Summary
Abstract
1. Introduction
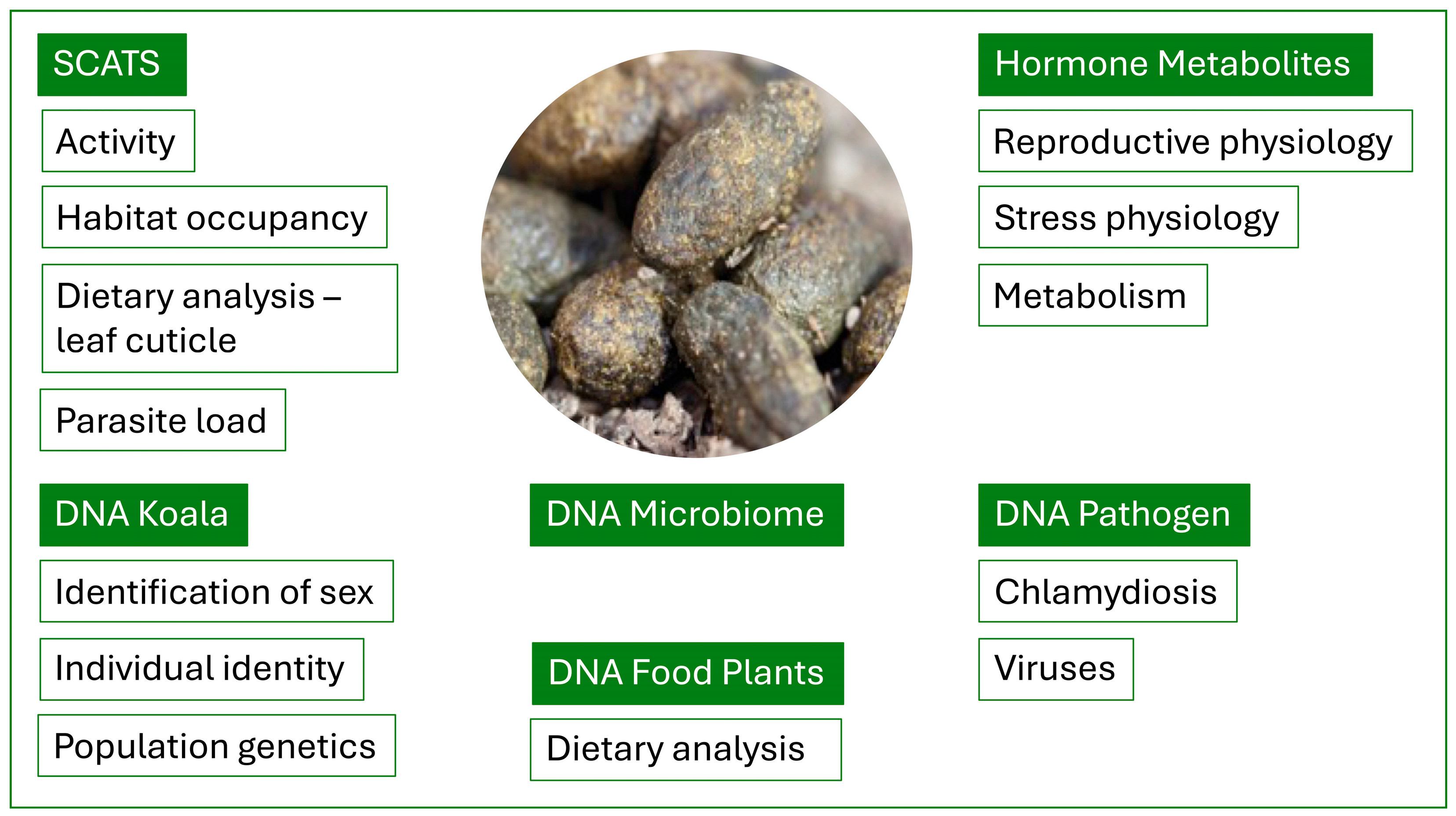
2. The Koala Scat
3. The Non-Invasive Sample
4. Habitat Occupancy and Activity
5. Dietary Analysis Using Cuticle Fragments
6. Koala DNA Extraction from Faeces
7. Genetic Analysis
8. Dietary Analysis Based on DNA
9. Microbiome
10. Chlamydiosis
11. Viruses
12. Koala Reproductive Hormones
13. Koala Glucocorticoid Hormones
14. Koala Metabolic Hormones
15. Implications: The Power of Koala Poo
Author Contributions
Funding
Conflicts of Interest
References
- Commonwealth of Australia, List of Threatened Species Amendment (Phascolarctos cinereus (combined populations of Queensland, New South Wales and the Australian Capital Territory) (280)) Instrument 2022. F2022L00131. C. 2022; Federal Register of Legislation. Available online: https://www.legislation.gov.au/Details/F2022L00131 (accessed on 21 June 2024).
- Queensland State Government Media Statement. Available online: https://statements.qld.gov.au/statements/98331 (accessed on 21 February 2024).
- Ghosal, R.; Edwards, K.L.; Chiarelli, T.L.; Fanson, K.V.; Ganswindt, A.; Keeley, T.; Koester, D.C.; Roberts, B.; Majelantle, T.; Wauters, J.; et al. Biomarkers of reproductive health in wildlife and techniques for their assessment. Theriogenology Wild 2023, 3, 100052. [Google Scholar] [CrossRef]
- Kersey, D.C.; Dehnhard, M. The use of non-invasive and minimally invasive methods in endocrinology for threatened mammalian species conservation. Gen. Comp. Endocrinol. 2014, 203, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Woosnam-Merchez, O.; Cristescu, R.; Dique, D.; Ellis, B.; Beeton, R.J.S.; Simmonds, J.; Carrick, F. What fecal pellet surveys can and can’t reveal about the ecology of koalas Phascolarctos cinereus. Aust. Zool. 2012, 36, 192–200. [Google Scholar] [CrossRef][Green Version]
- Jackson, S.; Reid, K.; Spittal, D.; Romer, L. Koalas. In Australian Mammals: Biology and Captive Management; Jackson, S., Ed.; CSIRO Publishing: Collingwood, VIC, Australia, 2003; pp. 145–181. [Google Scholar]
- Cristescu, R.H.; Foley, E.; Markula, A.; Jackson, G.; Jones, D.; Frère, C. Accuracy and efficiency of detection dogs: A powerful new tool for koala conservation and management. Sci. Rep. 2015, 5, 8349. [Google Scholar] [CrossRef] [PubMed]
- Youngentob, K.N.; Marsh, K.F.; Skewes, J. A Review of Koala Habitat Assessment Criteria and Methods; Report Prepared for the Department of Agriculture, Water and the Environment, Canberra, November 2021; CC BY 4.0. Available online: https://www.awe.gov.au/environment/epbc/publications (accessed on 21 June 2024).
- Palme, R. Measuring fecal steroids: Guidelines for practical application. Ann. N. Y Acad. Sci. 2005, 1046, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Palme, R. Non-invasive measurement of glucocorticoids: Advances and problems. Physiol. Behav. 2021, 199, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Melzer, A.; Carrick, F.; Menkhorst, P.; Lunney, D.; St John, B. Overview, critical assessment, and conservation implications of koala distribution and abundance. Conserv. Biol. 2000, 14, 619–628. [Google Scholar] [CrossRef]
- Phillips, S.; Callaghan, J. Spot assessment technique: A tool for determining localised levels of habitat use by koalas Phascolarctos cinereus. Aust. Zool. 2011, 35, 774–780. [Google Scholar] [CrossRef]
- Jiang, A.; Tribe, A.; Murray, P. The development of an improved scat survey method for koalas (Phascolarctos cinereus). Aust. J. Zool. 2019, 67, 125–133. [Google Scholar] [CrossRef]
- Cristescu, R.H.; Goethals, K.; Banks, P.B.; Carrick, F.N.; Free, C. Experimental Evaluation of Koala Scat Persistence and Detectability with Implications for Pellet-Based Fauna Census. Int. J. Zool. 2012, 631856. [Google Scholar] [CrossRef]
- Ellis, W.; Fitzgibbon, S.; Melzer, A.; Wilson, R.; Johnston, S.; Bercovitch, F.; Dique, D.; Carrick, F. Koala habitat use and population density: Using field data to test assumptions of ecological models. Aust. Mammal. 2013, 35, 160–165. [Google Scholar] [CrossRef]
- Marsh, K.J.; Blyton, M.D.J.; Foley, W.J.; Moore, B.D. Fundamental dietary specialisation explains differential use of resources within a koala population. Oecologia 2021, 196, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Moore, B.D.; Foley, W.J. A review of feeding and diet selection in koalas. Aust. J. Zool. 2000, 48, 317–333. [Google Scholar] [CrossRef]
- Carr, S.G.M.; Milkovits, L.; Carr, D.J. Eucalypt phytoglyphr: The microanatomical features of the epidermis in relation to taxonomy. Aust. J. Bot. 1971, 19, 173–190. [Google Scholar] [CrossRef]
- Tun, U.N. Diet Selection, Habitat Use and Reestablishment of Rehabilitated Koalas in Redland Shire, Queensland. Master’s Thesis, The University of Queensland, St Lucia, QLD, Australian, 1993. [Google Scholar]
- Hasegawa, M. Habitat utilisation by koalas (Phascolarctos cinereus) at Point Halloran. Master’s Thesis, The University of Queensland, Brisbane, QLD, Australia, 1995. [Google Scholar]
- Ellis, W.; Carrick, F.; Lundgren, P.; Veary, A.; Cohen, B. The use of faecal cuticle examination to determine the dietary composition of koalas. Aust. Zool. 1999, 31, 127–133. [Google Scholar] [CrossRef]
- Melzer, A.; Cristescu, R.; Ellis, W.; FitzGibbon, S.; Manno, G. The habitat and diet of 768 koalas (Phascolarctos cinereus) in Queensland. Aust. Mammal. 2014, 36, 189–199. [Google Scholar] [CrossRef]
- Wu, H.; McAlpine, C.; Seabrook, L. The dietary preferences of koalas, Phascolarctos cinereus, in southwest Queensland. Aust. Zool. 2012, 36, 93–102. [Google Scholar] [CrossRef]
- Blyton, M.D.J.; Brice, K.L.; Heller-Uszynska, K.; Pascoe, J.; Jaccoud, D.; Leigh, K.A.; Moore, B.D. A new genetic method for diet determination from faeces that provides species level resolution in the koala. bioRxiv 2023. [Google Scholar] [CrossRef]
- Wedrowicz, F.; Mosse, J.; Wright, W.; Hogan, F.E. Isolating DNA sourced non-invasive from koala scats: A comparison of four commercial DNA stool kits. Conserv. Genet. Resour. 2019, 11, 219–229. [Google Scholar] [CrossRef]
- Beja-Pereira, A.; Oliveira, R.; Alves, P.C.; Schwartz, M.K.; Luikart, G. Advancing ecological understandings through technological transformations in non-invasive genetics. Mol. Ecol. Resour. 2009, 9, 1279–1301. [Google Scholar] [CrossRef]
- Schultz, A.J.; Cristescu, R.H.; Littleford-Colquhoun, B.; Jaccoud, D.; Freer, C.H. Fresh is best: Accurate SMP genotyping form koala scats. Ecol. Evol. 2018, 8, 3139–3151. [Google Scholar] [CrossRef] [PubMed]
- Wedrowicz, F.; Karsa, M.; Mosse, J.; Hogan, F.E. Reliable genotyping of the koala (Phascolarctos cinereus) using DNA isolated from a single faecal pellet. Mol. Ecol. Resour. 2013, 13, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Chiou, K.L.; Bergey, C.M. Methylation-based enrichment facilitates low-cost, non-invasive genomic scale sequencing of populations from feces. Sci. Rep. 2018, 8, 1975. [Google Scholar] [CrossRef] [PubMed]
- Hogg, C.J.; Silver, L.; McLennan, E.A.; Belov, K. Koala Genome Survey: An Open Data Resource to Improve Conservation Planning. Genes 2023, 14, 546. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.N.; O’Meally, D.; Chen, Z.; Etherington, G.J.; Ho, S.Y.W.; Nash, W.J.; Grueber, C.E.; Cheng, Y.; Whittington, C.M.; Dennison, S.; et al. Adaptation and conservation insights from the koala genome. Nat. Genet. 2018, 50, 1102–1111. [Google Scholar] [CrossRef] [PubMed]
- Kjeldsen, S.R.; Zenger, K.R.; Leigh, K.; Ellis, W.; Tobey, J.; Phalen, D.; Melzer, A.; FitzGibbon, S.; Raadsma, H.W. Genome-wide SNP loci reveal novel insights into koala (Phascolarctos cinereus) population variability across its range. Conserv. Genet. 2016, 17, 337–353. [Google Scholar] [CrossRef]
- Kjeldsen, S.R.; Raadsma, H.W.; Leigh, K.A.; Tobey, J.R.; Phalen, D.; Krockenberger, A.; Ellis, W.A.; Hynes, E.; Higgins, D.P.; Zenger, K.R. Genomic comparisons reveal biogeographic and anthropogenic impacts in the koala (Phascolarctos cinereus): A dietary-specialist species distributed across heterogeneous environments. Heredity 2019, 122, 525–544. [Google Scholar] [CrossRef] [PubMed]
- Schultz, A.J.; Cristescu, R.H.; Hanger, J.; Loader, J.; de Villiers, D.; Frere, C.H. Inbreeding and disease avoidance in a free-ranging koala population. Mol. Ecol. 2020, 29, 2416–2430. [Google Scholar] [CrossRef]
- Barker, C.J.; Gillett, A.; Polkinghorne, A.; Timms, P. Investigation of the koala (Phascolarctos cinereus) hindgut microbiome via 16S pryosequencing. Vet. Microbial. 2013, 167, 554–564. [Google Scholar] [CrossRef]
- Alfano, N.; Courtiol, A.; Vielgrader, H.; Timms, P.; Roca, A.L.; Greenwood, A.D. Variation in koala microbiomes within and between individuals: Effect of body region and captivity status. Sci. Rep. 2015, 5, 10189. [Google Scholar] [CrossRef]
- Shiffman, M.E.; Soo, R.M.; Dennis, P.G.; Morrison, M.; Tyson, G.W.; Hugenholtz, P. Gene and genome-centric analyses of koala and wombat fecal microbiomes point to metabolic specialization for Eucalyptus digestion. PeerJ 2017, 5, e4075. [Google Scholar] [CrossRef]
- Brice, K.L.; Trivedi, P.; Jeffries, T.C.; Blyton, M.D.J.; Mitchell, C.; Singh, B.K.; Moore, B.D. The Koala (Phascolarctos cinereus) faecal microbiome differs with diet in a wild population. PeerJ 2019, 7, e6534. [Google Scholar] [CrossRef]
- Blyton, M.D.J.; Soo, R.M.; Whisson, D.; Marsh, K.J.; Pascoe, J.; Le Pla, M.; Foley, W.; Hugenholtz, P.; Moore, B. Faecal inoculations alter the gastrointestinal microbiome and allow dietary expansion in a wild specialist herbivore, the koala. Anim. Microbiome 2019, 1, 6. [Google Scholar] [CrossRef]
- Blyton, M.D.J.; Soo, R.M.; Hugenholtz, P.; Moore, B.D. Maternal inheritance of the koala gut microbiome and its compositional and functional maturation during juvenile development. Environ. Microbiol. 2022, 24, 475–493. [Google Scholar] [CrossRef]
- Blyton, M.D.J.; Soo, R.M.; Hugenholtz, P.; Moore, B.D. (Characterisation of juvenile koala gut microbiome across wild populations. Environ. Microbiol. 2022, 24, 4209–4219. [Google Scholar] [CrossRef]
- Gillett, A.; Hanger, J. Koala. In Current Therapy in Medicine of Australian Mammals; Vogelnest, L., Portas, T., Eds.; CSIRO Publishing: Clayton, VIC, Australia, 2019; pp. 463–486. [Google Scholar]
- Quigley, B.L.; Timms, P. Helping koalas battle disease—Recent advances on Chlamydia and koala retrovirus (KoRV) disease understanding and treatment in koalas. FEMS Microbiol. Rev. 2020, 44, 583–605. [Google Scholar] [CrossRef]
- Wedrowicz, F.; Saxton, T.; Moses, J.; Wright, W.; Hogan, F.E. A non-invasive tool for assessing pathogen prevalence in koala (Phascolarctos cinereus) populations: Detection of Chlamydia pecorum and koala retrovirus (KoRV) DNA in genetic material sourced from scats. Conserv. Genet. Resour. 2016, 8, 511–521. [Google Scholar] [CrossRef]
- Cristescu, R.; Miller, R.L.; Schultz, A.J.; Hulse, L.; Jaccoud, D.; Johnston, S.; Hanger, J.; Booth, R.; Frere, C.H. Developing non-invasive methodologies to assess koala population health through detecting Chlamydia from scats. Mol. Ecol. 2019, 19, 957–969. [Google Scholar] [CrossRef]
- Jelocnik, M.; Islam, M.M.; Madden, D.; Jenkins, C.; Branley, J.; Carver, S.; Polkinghorne, A. Development and evaluation of rapid novel isothermal amplification assays for important veterinary pathogens: Chlamydia psittaci and Chlamydia pecorum. PeerJ 2017, 5, e3799. [Google Scholar] [CrossRef]
- Hulse, L.S.; McDonald, S.; Johnston, S.D.; Beagley, K.W. Rapid point-of care diagnostics for the detection of Chlamydia pecorum in koalas (Phascolarctos cinereus) using loop-mediated isothermal amplification without nucleic acid purification. MicrobiologyOpen 2019, 8, e916. [Google Scholar] [CrossRef]
- Canfield, P.J.; Sabine, J.M.; Love, D.N. Virus particles associated with leukaemia in a koala. Aust. Vet. J. 1988, 65, 327–328. [Google Scholar] [CrossRef]
- Tarlinton, R.; Meers, J.; Hanger, J.; Young, P. Real-time reverse transcriptase PCR for the endogenous koala retrovirus reveals an association between plasma viral load and neoplastic disease in koalas. J. Gen. Virol. 2005, 86, 783–787. [Google Scholar] [CrossRef]
- Ito, N.; Yoshida, T.; Ichikawa, R.; Makino, E.; Akema, S.; Fukumori, J.; Takahashi, N.; Nakahara, J.; Yamashita, R.; Orihara, K.; et al. Clinical and pathological characteristics of acute myelogenous leukemia in a female koala with diabetes mellitus. J. Vet. Med. Sci. 2019, 81, 1229–1233. [Google Scholar] [CrossRef]
- Legione, A.R.; Patterson, J.L.; Whiteley, P.; Firestone, S.M.; Curnick, M.; Bodley, K.; Lynch, M.; Gilkerson, J.R.; Samsom, F.M.; Devlin, J.M. Koala retrovirus genotyping analyses reveal a low prevalence of KoRV-A in Victorian koalas and an association with clinical disease. J. Med. Microbiol. 2017, 66, 236–244. [Google Scholar] [CrossRef]
- Waugh, C.A.; Hanger, J.; Loader, J.; King, A.; Hobbs, M.; Johnson, R.; Timms, P. Infection with koala retrovirus subgroup B (KoRV-B), but not KoRV-A, is associated with chlamydial disease in free-ranging koalas (Phascolarctos cinereus). Sci. Rep. 2017, 7, 134. [Google Scholar] [CrossRef]
- Quigley, B.L.; Ong, V.A.; Hanger, J.; Timms, P. Molecular dynamics and mode of transmission of Koala Retrovirus (KoRV) as it invades and spreads through a wild Queensland koala population. J. Virol. 2018, 92, e01871–01817. [Google Scholar] [CrossRef]
- Xu, W.; Stadler, C.K.; Gorman, K.; Jensen, N.; Kim, D.; Zheng, H.; Tang, S.; Switzer, W.M.; Pye, G.W.; Eiden, M.V. An exogenous retrovirus isolated from koalas with malignant neoplasias in a US zoo. Proc. Natl. Acad. Sci. USA 2013, 110, 11547–11552. [Google Scholar] [CrossRef]
- Blyton, M.D.J.; Young, P.; Moore, B.; Chappell, K. Geographical patterns of koala retrovirus genetic diversity, endogenization and sub-type distributions. Proc. Natl. Acad. Sci. USA 2021, 119, e2122680119. [Google Scholar] [CrossRef]
- Wright, B.R.; Jelocnik, M.; Casteriano, A.; Muir, Y.S.S.; Legione, A.R.; Vaz, P.K.; Devlin, J.M.; Higgins, D.P. Development of diagnostic and point of care assays for a gammaherpesvirus infecting koalas. PLoS ONE 2023, 18, e0286407. [Google Scholar] [CrossRef]
- Schwarzenberger, F.; Most, E.; Palme, R.; Bamberg, E. Faecal steroid analysis for non-invasive monitoring of reproductive status in farm, wild and zoo animals. Anim. Reprod. Sci. 1996, 42, 515–526. [Google Scholar] [CrossRef]
- Schwarzenberger, F.; Brown, J. Hormone monitoring: An important tool for the breeding management of wildlife species. Wien. Tierärztli Monatsschr 2013, 100, 209–225. [Google Scholar]
- Kumar, V.; Umapathy, G. Non-invasive monitoring of steroid hormones in wildlife for conservation of endangered species: A review. Indian. J. Exp. Biol. 2019, 57, 307–314. [Google Scholar]
- Keeley, T.; Johnston, S. Assessment and management of reproduction in Australian monotremes and marsupials. In Current Therapy in Medicine of Australian Mammals; Vogelnest, L., Portas, T., Eds.; CSIRO Publishing: Clayton South, VIC, Australia, 2019; pp. 63–84. [Google Scholar]
- Kusuda, S.; Hashikawa, H.; Takeda, M.; Takeda, M.; Ito, H.; Ogata-Kobayashi, Y.; Hashimoto, M.; Ogata, M.; Morikaku, K.; Araki, S.; et al. Non-invasive monitoring of reproductive activity based on fecal progestogen profiles and sexual behaviour in koalas, Phascolarctos cinereus. Biol. Reprod. 2013, 81, 1033–1040. [Google Scholar] [CrossRef]
- Kusuda, S.; Hashikawa, H.; Takeda, M.; Ito, H.; Goto, A.; Oguchi, J.; Doi, O. Season- and age-related reproductive changes based on fecal androgen concentrations in male koalas, Phascolarctos cinereus. J. Reprod. Dev. 2013, 59, 308–313. [Google Scholar] [CrossRef]
- Keeley, T.; Barnes, M.; Mucci, A.; Seaton, J.; Johnston, S.D. Non-invasive hormone monitoring as a management tool for breeding koalas under human care. In Proceedings of the Australian Mammal Society Conference, Adelaide, SA, Australia, 18–22 September 2023. [Google Scholar]
- Fanson, K.V.; Best, E.C.; Bunch, A.; Fanson, B.G.; Hogan, L.A.; Keeley, T.; Narayan, E.J.; Palme, R.; Parrot, M.L.; Sharp, T.M.; et al. One size does not fit all: Monitoring faecal glucocorticoid metabolites in marsupials. Gen. Comp. Endocrinol. 2017, 244, 146–156. [Google Scholar] [CrossRef]
- Davies, N.; Gillett, A.; McAlpine, C.; Seabrook, L.; Baxter, G.; Lunney, D.; Bradley, A. The effect of ACTH upon faecal glucocorticoid excretion in the koala. J. Endocrinol. 2013, 219, 1–12. [Google Scholar] [CrossRef]
- Davies, N.A.; Gramotnev, G.; McAlpine, C.; Seabrook, L.; Baxter, G.; Lunny, D.; Rhodes, J.R.; Bradley, A. Physiological stress in koala populations near the arid edge of their distribution. PLoS ONE 2013, 8, e79136. [Google Scholar] [CrossRef]
- Narayan, E.; Webster, K.; Nicolson, V.; Mucci, A.; Hero, J. Non-invasive evaluation of physiological stress in an iconic Australian marsupial: The koala (Phascolarctos cinereus). Gen. Comp. Endocrinol. 2013, 187, 39–47. [Google Scholar] [CrossRef]
- Johnston, S.D.; Booth, R.A.; Pyne, M.; Keeley, T.; Mackie, J.T.; Hulse, L.; Ellis, W. Preliminary study of faecal cortisol and corticosterone as an index of acute cortisol secretion in the koala (Phascolarctos cinereus). Aust. Vet. J. 2013, 91, 534–537. [Google Scholar] [CrossRef]
- Webster, K.; Narayan, E.; de Los, N. Fecal glucocorticoid metabolite responses of captive koalas (Phascolarctos cinereus) to visitor encounters. Gen. Comp. Endocrinol. 2017, 244, 157–163. [Google Scholar] [CrossRef]
- Hogg, C.; Brandies, P.; Wright, B.; Grueber, C. Measuring the Impact of the Woolgoolga to Ballina Upgrade on Local Koala Populations: Faecal Cortisol Metabolite Concentrations before, during and after Phased Resource Reduction and during Clearing; Report by The Australian Wildlife Genomics Group, Faculty of Science, School of Environment and Life Sciences, The University of Sydney: Sydney, NSW, Australia, 2018. Available online: https://www.pacifichighway.nsw.gov.au/sites/default/files/media/documents/2018/Woolgoolga%20to%20Ballina%20upgrade%20Koala%20Cortisol%20Stress%20Study%20REPORT%20Mar%202018.pdf (accessed on 21 February 2024).
- Narayan, E. Physiological stress levels in wild koala sub-populations facing anthropogenic induced environmental trauma and disease. Sci. Rep. 2019, 9, 6031. [Google Scholar] [CrossRef]
- Narayan, E.; Vanderneut, T. Physiological stress in rescued wild koalas are influenced by habitat, demographics, environmental, stressors, and clinical intervention. Front. Endocrinol. 2019, 10, 18. [Google Scholar] [CrossRef]
- Santamaria, F.; Barlow, C.K.; Schlagloth, R.; Schittenhelm, R.B.; Palme, R.; Henning, J. Identification of koala (Phascolarctos cinereus) faecal cortisol metabolites using liquid chromatography-mass spectrometry and enzyme immunoassays. Metabolites 2021, 11, 393. [Google Scholar] [CrossRef]
- Santamaria, F.; Schlagloth, R.; Palme, R.; Henning, J. Over time decay of cortisol metabolites in faecal pellets of koalas in central Queensland. Animals 2021, 11, 3376. [Google Scholar] [CrossRef]
- Santamaria, F.; Palme, R.; Schlagloth, R.; Klobetz-Rassam, E.; Henning, J. Seasonal variations of faecal cortisol metabolites in koalas in South-east Queensland. Animals 2021, 11, 1622. [Google Scholar] [CrossRef]
- Charalambous, R.; Simonato, T.; Peel, M.; Narayan, E.J. Physiological stress in rescued wild koalas (Phascolarctos cinereus) being held in a rehabilitation sanctuary: A pilot study. Animals 2021, 11, 2864. [Google Scholar] [CrossRef]
- Beaman, J.E.; Mulligan, C.; Moore, C.; Mitchell, D.; Narayan, E.; da Silva, K.B. Resident wild koalas how resilience to large-scale translocation of bushfire-rescued koalas Conserv. Physiol. 2023, 11, coac088. [Google Scholar] [CrossRef]
- Samarawickrama, A. The Influence of Cultural Burns on the Density and Stress Response of Koalas on Minjerribah, North Stradbroke Island. Master’s Thesis, The University of Sunshine Coast, Sippy Downs, QLD, Australia, 2023. [Google Scholar]
- Santamaria, F.; Schlagloth, R.; Valenza, L.; Palme, R.; de Villiers, D.; Henning, J. The effect of disease and injury on faecal cortisol metabolites, as an indicator of stress in wild hospitalised koalas, endangered Australian marsupials. Vet. Sci. 2023, 10, 65. [Google Scholar] [CrossRef]
- Parker Fischer, C.; Romero, L.M. Chronic captivity stress in wild animals is highly species-specific. Conserv. Physiol. 2019, 7, coz093. [Google Scholar] [CrossRef]
- Dickens, M.J.; Romero, L.M. A consensus endocrine profile for chronically stressed wild animals does not exist. Gen. Comp. Endocrinol. 2013, 191, 177–189. [Google Scholar] [CrossRef]
- MacDougall-Shackleton, S.A.; Bonier, F.; Romero, L.M.; Moore, I.T. Glucocorticoids and “stress” are not synonymous. Integr. Org. Biol. 2019, 1, obz017. [Google Scholar] [CrossRef]
- Behringer, V.; Deimel, C.; Hohmann, G.; Negrey, J.; Schaebs, F.S.; Deschner, T. Applications for non-invasive thyroid hormone measurements in mammalian ecology, growth, and maintenance. Horm. Behav. 2018, 105, 66–85. [Google Scholar] [CrossRef]
- Wasser, S.K.; Azkarate, J.C.; Booth, R.K.; Hayward, L.; Hunt, K.; Ayres, K.; Vynne, C.; Gobush, K.; Canales-Espinosa, D.; Rodriguez-Luna, E. Non-invasive measurement of thyroid hormone in feces of a diverse array of avian and mammalian species. Gen. Comp. Endocrinol. 2010, 168, 1–7. [Google Scholar] [CrossRef]
| Metabolite | Type of Study | Assay | Reference |
|---|---|---|---|
| Cortisol | V—ACTH, Captive | EIA | [65] |
| Cortisol | A—Habitat (Aridity) (Semi-arid Zone), Wild | EIA | [66] |
| Cortisol | V—ACTH, A—Captive V Wild, Handling | EIA | [67] |
| Cortisol, CS | V—ACTH | EIA | [68] |
| Cortisol, CS, 72a, 37e | V—ACTH, Captive | EIA | [64] |
| Cortisol | A—Zoo visitor, Captive | EIA | [69] |
| Cortisol | A—Habitat clearing, Wild | EIA | [70] |
| Cortisol | A—Disease, Trauma, Hospital, Bushfire, Wild | EIA | [71] |
| Cortisol | A—Disease, Trauma, Hospital, Bushfire, Wild | EIA | [72] |
| Cortisol, 37e, 50c | V—Hydrocortisone, Captive | LCMS, EIA | [73] |
| Cortisol, 37e, 50c | V—Time decay, Water loss, Captive | EIA | [74] |
| Cortisol, 37e, 50c | A—Seasonality, Captive | EIA | [75] |
| Cortisol | A—Hospitalised, Rehabilitation | EIA | [76] |
| Cortisol | A—Translocation, Wild | EIA | [77] |
| Cortisol | A—Wild, Influence of cultural burns | EIA | [78] |
| Cortisol, 50c | A—Hospitalised | EIA | [79] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Johnston, S.D.; Hulse, L.; Keeley, T.; Mucci, A.; Seddon, J.; Maynard, S. The Utility of the Koala Scat: A Scoping Review. Biology 2024, 13, 523. https://doi.org/10.3390/biology13070523
Johnston SD, Hulse L, Keeley T, Mucci A, Seddon J, Maynard S. The Utility of the Koala Scat: A Scoping Review. Biology. 2024; 13(7):523. https://doi.org/10.3390/biology13070523
Chicago/Turabian StyleJohnston, Stephen D., Lyndal Hulse, Tamara Keeley, Albano Mucci, Jennifer Seddon, and Sam Maynard. 2024. "The Utility of the Koala Scat: A Scoping Review" Biology 13, no. 7: 523. https://doi.org/10.3390/biology13070523
APA StyleJohnston, S. D., Hulse, L., Keeley, T., Mucci, A., Seddon, J., & Maynard, S. (2024). The Utility of the Koala Scat: A Scoping Review. Biology, 13(7), 523. https://doi.org/10.3390/biology13070523






