ClpL Chaperone as a Possible Component of the Disaggregase Activity of Limosilactobacillus fermentum U-21
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Plasmids
- L. fermentum U-21 (collection number VKPM V-12075, NCBI Genome assembly ASM286982v2);
- E. coli SG20250 (ΔlacU169 araD flbB relA clpB+), and its insertion derivative SG22100 clpB::kan− (kindly provided by S. Gottesman) [21];
- E. coli XL1-Blue (recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F’proAB lacIqZΔM15 Tn10 (Tetr)]) (Stratagene, La Jolla, CA, USA);
- E. coli BL21-Gold (DE3) (F− ompT dcm+ TetR gal lon hsdSB(rB− mB−) λ(DE3 [lacI lacUV5-T7p07 ind1sam7 nin5]) used for the biosynthesis of luciferase LuxAB and NADH-FMN oxidoreductases LuxG (obtained from VKPM);
- E. coli Nico21(DE3) can::CBD fhuA2 [lon] ompT gal (λ DE3) [dcm] arnA::CBD slyD::CBD glmS6Ala ∆hsdS λ DE3 = λ sBamHIo ∆EcoRI-B int::(lacI::PlacUV5::T7 gene1) i21 ∆nin5 (NEB strain catalog no. C2529H);
- E. coli TG1 (thi relA supE44 hsdR17 hsdM Δ(lacproAB) [F’traD36 proAB lacIqZ ΔM15]) for plasmid preparation (obtained from VKPM).
2.2. Cultivation Conditions
2.3. DNA Manipulations
2.4. Expression of the ClpL Gene from L. fermentum U-21 Strain in E. coli
2.5. In Vivo Luminescence Measurement
2.6. Biosynthesis, Isolation, and Purification of P. luminescens Luciferase, V. aquamarinus LuxG, and L. fermentum U-21 ClpL
2.7. Spent L. fermentum U-21 Culture Medium
2.8. Measurement of P. luminescens Luciferase Activity In Vitro
2.9. Phylogenetic and Molecular Evolutionary Analysis
3. Results
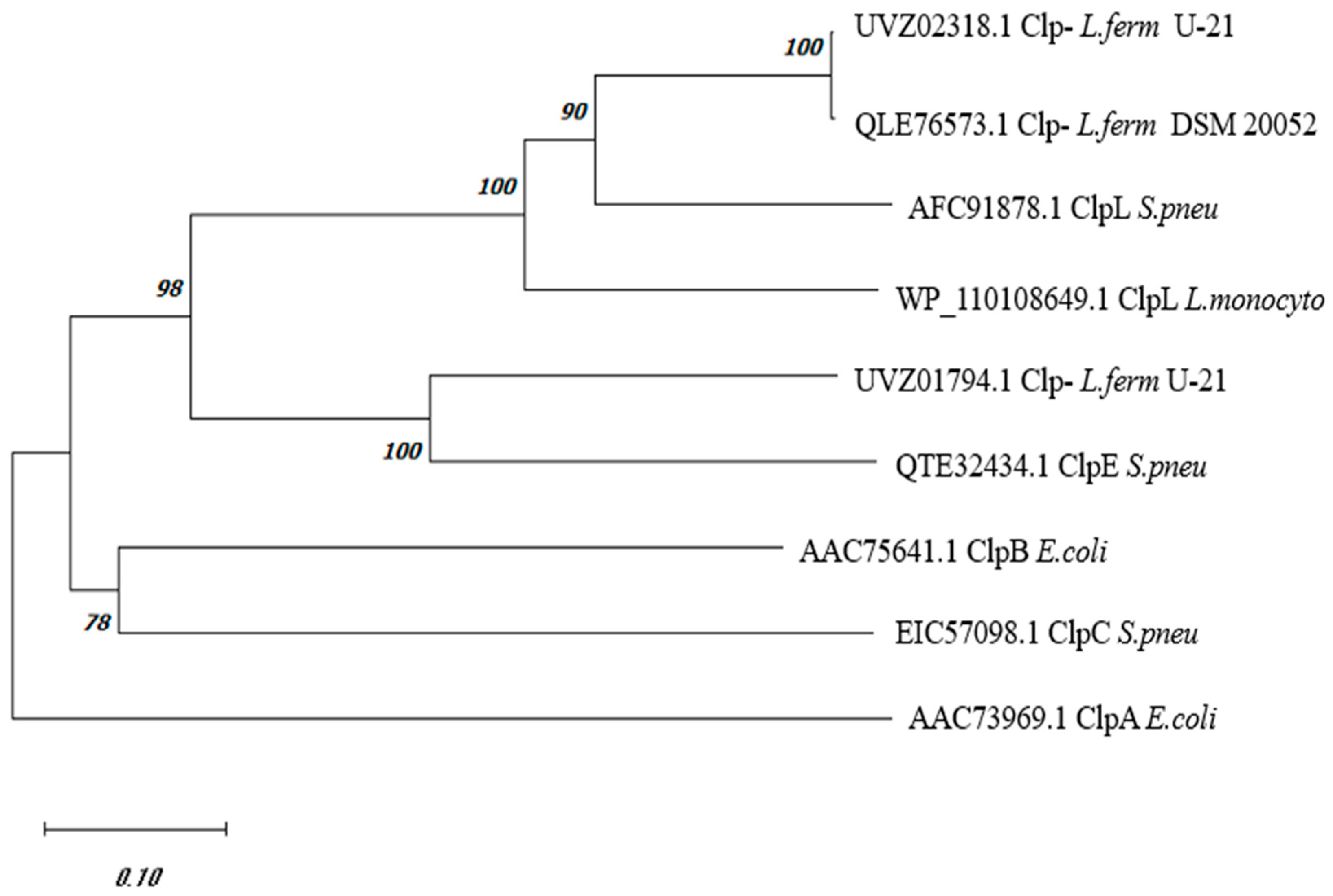
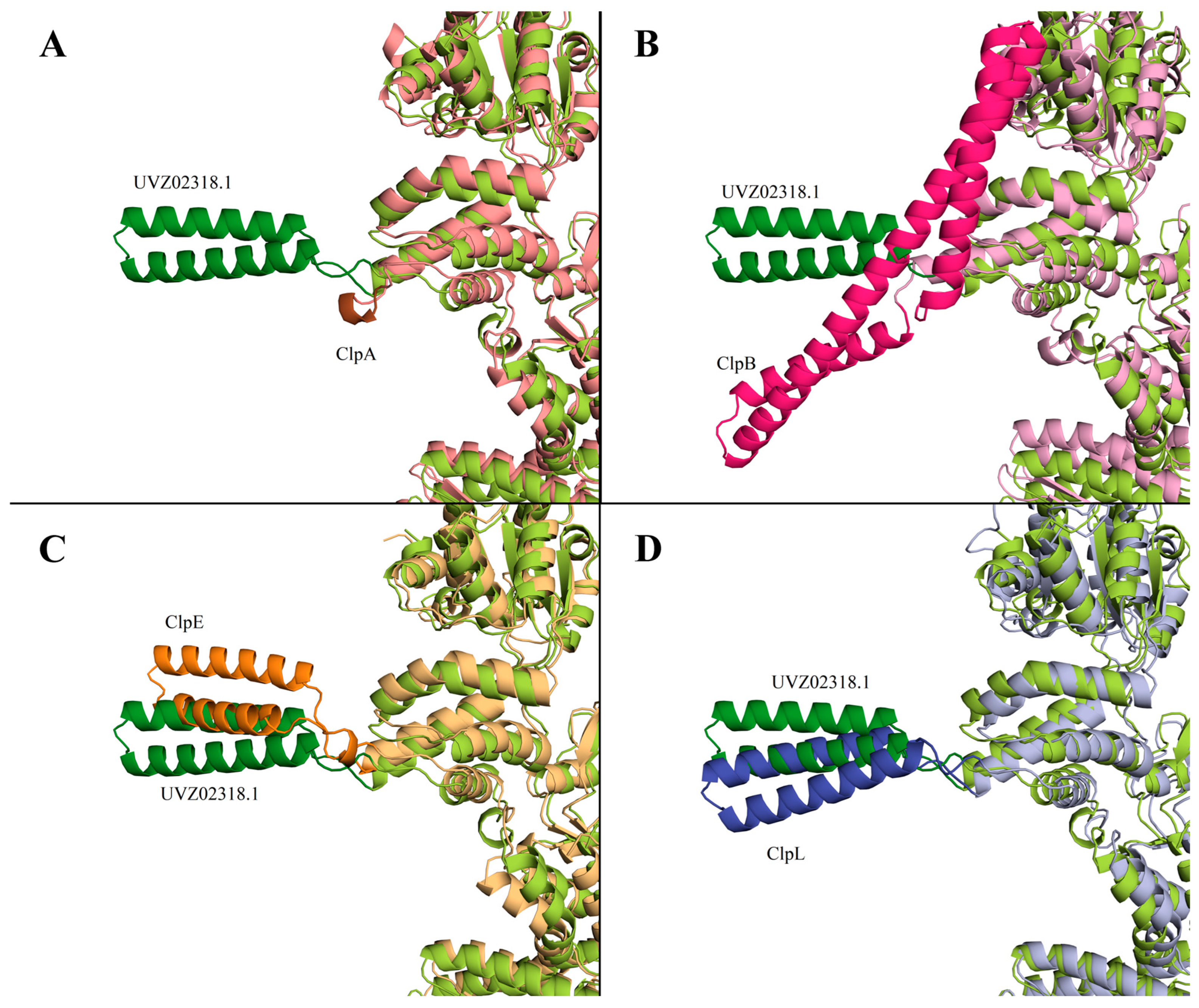
3.1. Phylogenetic Analysis of C0965_000195 L. fermentum U-21
3.2. Gene Cloning and Expression and Chaperone Activity Investigation of the L. fermentum U-21 ClpL Protein in E. coli Cells
3.3. Investigation of the Chaperone Activity of L. fermentum U-21 SCM In Vitro
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Katayama, Y.; Gottesman, S.; Pumphrey, J.; Rudikoff, S.; Clark, W.P.; Maurizi, M.R. The Two-Component, ATP-Dependent Clp Protease of Escherichia Coli. Purification, Cloning, and Mutational Analysis of the ATP-Binding Component. J. Biol. Chem. 1988, 263, 15226–15236. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, M.; Wada, C.; Yoshioka, S.; Yura, T. Expression of ClpB, an Analog of the ATP-Dependent Protease Regulatory Subunit in Escherichia coli, Is Controlled by a Heat Shock Sigma Factor (Sigma 32). J. Bacteriol. 1991, 173, 4247–4253. [Google Scholar] [CrossRef] [PubMed]
- Hoskins, J.R.; Sharma, S.; Sathyanarayana, B.K.; Wickner, S. Clp ATPases and Their Role in Protein Unfolding and Degradation. Adv. Protein Chem. 2001, 59, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Wawrzynow, A.; Wojtkowiak, D.; Marszalek, J.; Banecki, B.; Jonsen, M.; Graves, B.; Georgopoulos, C.; Zylicz, M. The ClpX Heat-Shock Protein of Escherichia coli, the ATP-Dependent Substrate Specificity Component of the ClpP-ClpX Protease, Is a Novel Molecular Chaperone. EMBO J. 1995, 14, 1867–1877. [Google Scholar] [CrossRef] [PubMed]
- Wickner, S.; Gottesman, S.; Skowyra, D.; Hoskins, J.; McKenney, K.; Maurizi, M.R. A Molecular Chaperone, ClpA, Functions like DnaK and DnaJ. Proc. Natl. Acad. Sci. USA 1994, 91, 12218. [Google Scholar] [CrossRef] [PubMed]
- Weber-Ban, E.U.; Reid, B.G.; Miranker, A.D.; Horwich, A.L. Global Unfolding of a Substrate Protein by the Hsp100 Chaperone ClpA. Nature 1999, 401, 90–93. [Google Scholar] [CrossRef] [PubMed]
- Hoskins, J.R.; Singh, S.K.; Maurizi, M.R.; Wickner, S. Protein Binding and Unfolding by the Chaperone ClpA and Degradation by the Protease ClpAP. Proc. Natl. Acad. Sci. USA 2000, 97, 8892–8897. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, S.; Wickner, S.; Maurizi, M.R. Protein Quality Control: Triage by Chaperones and Proteases. Genes Dev. 1997, 11, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, K.; Watanabe, Y.; Yohda, M.; Yoshida, M. Heat-Inactivated Proteins Are Rescued by the DnaK.J-GrpE Set and ClpB Chaperones. Proc. Natl. Acad. Sci. USA 1999, 96, 7184–7189. [Google Scholar] [CrossRef]
- Zavilgelsky, G.B.; Kotova, V.Y.; Mazhul’, M.M.; Manukhov, I.V. The Effect of Clp Proteins on DnaK-Dependent Refolding of Bacterial Luciferases. Mol. Biol. 2004, 38, 427–433. [Google Scholar] [CrossRef]
- Kim, G.; Lee, S.G.; Han, S.; Jung, J.; Jeong, H.S.; Hyun, J.k.; Rhee, D.K.; Kim, H.M.; Lee, S. ClpL Is a Functionally Active Tetradecameric AAA+ Chaperone, Distinct from Hexameric/Dodecameric Ones. FASEB J. 2020, 34, 14353–14370. [Google Scholar] [CrossRef] [PubMed]
- Namy, O.; Mock, M.; Fouet, A. Co-Existence of ClpB and ClpC in the Bacillaceae. FEMS Microbiol. Lett. 1999, 173, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Breton, J.; Legrand, R.; Akkermann, K.; Järv, A.; Harro, J.; Déchelotte, P.; Fetissov, S.O. Elevated Plasma Concentrations of Bacterial ClpB Protein in Patients with Eating Disorders. Int. J. Eat. Disord. 2016, 49, 805–808. [Google Scholar] [CrossRef] [PubMed]
- Dominique, M.; Breton, J.; Guérin, C.; Bole-Feysot, C.; Lambert, G.; Déchelotte, P.; Fetissov, S. Effects of Macronutrients on the In Vitro Production of ClpB, a Bacterial Mimetic Protein of α-MSH and Its Possible Role in Satiety Signaling. Nutrients 2019, 11, 2115. [Google Scholar] [CrossRef] [PubMed]
- Tennoune, N.; Chan, P.; Breton, J.; Legrand, R.; Chabane, Y.N.; Akkermann, K.; Järv, A.; Ouelaa, W.; Takagi, K.; Ghouzali, I.; et al. Bacterial ClpB Heat-Shock Protein, an Antigen-Mimetic of the Anorexigenic Peptide α-MSH, at the Origin of Eating Disorders. Transl. Psychiatry 2014, 4, e458. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Sojo, M.J.; Ruiz-Malagón, A.J.; Rodríguez-Cabezas, M.E.; Gálvez, J.; Rodríguez-Nogales, A. Limosilactobacillus fermentum CECT5716: Mechanisms and Therapeutic Insights. Nutrients 2021, 13, 1016. [Google Scholar] [CrossRef] [PubMed]
- De Luna Freire, M.O.; Cruz Neto, J.P.R.; de Albuquerque Lemos, D.E.; de Albuquerque, T.M.R.; Garcia, E.F.; de Souza, E.L.; de Brito Alves, J.L. Limosilactobacillus fermentum Strains as Novel Probiotic Candidates to Promote Host Health Benefits and Development of Biotherapeutics: A Comprehensive Review. Probiotics Antimicrob. Proteins 2024, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Marsova, M.; Poluektova, E.; Odorskaya, M.; Ambaryan, A.; Revishchin, A.; Pavlova, G.; Danilenko, V. Protective Effects of Lactobacillus fermentum U-21 against Paraquat-Induced Oxidative Stress in Caenorhabditis elegans and Mouse Models. World J. Microbiol. Biotechnol. 2020, 36, 104. [Google Scholar] [CrossRef]
- Danilenko, V.N.; Stavrovskaya, A.V.; Voronkov, D.N.; Gushchina, A.S.; Marsova, M.V.; Yamshchikova, N.G.; Ol’shansky, A.S.; Ivanov, M.V.; Illarioshkin, S.N. The Use of a Pharmabiotic Based on the Lactobacillus fermentum U-21 Strain to Modulate the Neurodegenerative Process in an Experimental Model of Parkinson Disease. Ann. Clin. Exp. Neurol. 2020, 14, 62–69. [Google Scholar] [CrossRef]
- Poluektova, E.U.; Mavletova, D.A.; Odorskaya, M.V.; Marsova, M.V.; Klimina, K.M.; Koshenko, T.A.; Yunes, R.A.; Danilenko, V.N. Comparative Genomic, Transcriptomic, and Proteomic Analysis of the Limosilactobacillus fermentum U-21 Strain Promising for the Creation of a Pharmabiotic. Russ. J. Genet. 2022, 58, 1079–1090. [Google Scholar] [CrossRef]
- Gottesman, S.; Roche, E.; Zhou, Y.N.; Sauer, R.T. The ClpXP and ClpAP Proteases Degrade Proteins with Carboxy-Terminal Peptide Tails Added by the SsrA-Tagging System. Genes Dev. 1998, 12, 1338–1347. [Google Scholar] [CrossRef]
- Bazhenov, S.; Novoyatlova, U.; Scheglova, E.; Fomin, V.; Khrulnova, S.; Melkina, O.; Chistyakov, V.; Manukhov, I. Influence of the luxR Regulatory Gene Dosage and Expression Level on the Sensitivity of the Whole-Cell Biosensor to Acyl-Homoserine Lactone. Biosensors 2021, 11, 166. [Google Scholar] [CrossRef] [PubMed]
- Zavil’gel’skiĭ, G.B.; Kotova, V.I.; Manukhov, I.V. A Kinetic Method of Determining the Frequency of Homologous Recombination of Plasmids in Escherichia coli Cells. Mol. Biol. (Mosk) 1994, 28, 1299–1307. [Google Scholar]
- Zavilgelsky, G.B.; Zarubina, A.P.; Manukhov, I.V. Sequencing and Comparative Analysis of the lux-Operon of Photorhabdus luminescens Strain ZM1: ERIC Elements as Putative Recombination Spots. Mol. Biol. 2002, 36, 792–804. [Google Scholar] [CrossRef]
- Joseph, S.; Maccallum, P.; William, R.D. Molecular Cloning a Laboratory Manual, Volume 1; CSHL Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic Assembly of DNA Molecules up to Several Hundred Kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Nazarov, P.A.; Khrulnova, S.A.; Kessenikh, A.G.; Novoyatlova, U.S.; Kuznetsova, S.B.; Bazhenov, S.V.; Sorochkina, A.I.; Karakozova, M.V.; Manukhov, I.V. Observation of Cytotoxicity of Phosphonium Derivatives Is Explained: Metabolism Inhibition and Adhesion Alteration. Antibiotics 2023, 12, 720. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Park, S.S.; Kwon, H.Y.; Tran, T.D.H.; Choi, M.H.; Jung, S.H.; Lee, S.; Briles, D.E.; Rhee, D.K. ClpL Is a Chaperone without Auxiliary Factors. FEBS J. 2015, 282, 1352–1367. [Google Scholar] [CrossRef] [PubMed]
- Bohl, V.; Hollmann, N.M.; Melzer, T.; Katikaridis, P.; Meins, L.; Simon, B.; Flemming, D.; Sinning, I.; Hennig, J.; Mogk, A. The Listeria Monocytogenes Persistence Factor ClpL Is a Potent Stand-Alone Disaggregase. eLife 2024, 12, RP92746. [Google Scholar] [CrossRef]
- Rotanova, T.V.; Andrianova, A.G.; Kudzhaev, A.M.; Li, M.; Botos, I.; Wlodawer, A.; Gustchina, A. New Insights into Structural and Functional Relationships between LonA Proteases and ClpB Chaperones. FEBS Open Bio 2019, 9, 1536. [Google Scholar] [CrossRef]
- Huang, D.C.; Huang, X.F.; Novel, G.; Novel, M. Two Genes Present on a Transposon-like Structure in Lactococcus Lactis Are Involved in a Clp-Family Proteolytic Activity. Mol. Microbiol. 1993, 7, 957–965. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.Y.; Kim, S.W.; Choi, M.H.; Ogunniyi, A.D.; Paton, J.C.; Park, S.H.; Pyo, S.N.; Rhee, D.K. Effect of Heat Shock and Mutations in ClpL and ClpP on Virulence Gene Expression in Streptococcus pneumoniae. Infect. Immun. 2003, 71, 3757–3765. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Biswas, I. ClpL Is Required for Folding of CtsR in Streptococcus Mutans. J. Bacteriol. 2013, 195, 576. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.N.; Khan, R.H. Protein Misfolding and Related Human Diseases: A Comprehensive Review of Toxicity, Proteins Involved, and Current Therapeutic Strategies. Int. J. Biol. Macromol. 2022, 223, 143–160. [Google Scholar] [CrossRef]
- Lo, A.C.; Callaerts-Vegh, Z.; Nunes, A.F.; Rodrigues, C.M.P.; D’Hooge, R. Tauroursodeoxycholic Acid (TUDCA) Supplementation Prevents Cognitive Impairment and Amyloid Deposition in APP/PS1 Mice. Neurobiol. Dis. 2013, 50, 21–29. [Google Scholar] [CrossRef]
- Almeida, Z.L.; Brito, R.M.M. Amyloid Disassembly: What Can We Learn from Chaperones? Biomedicines 2022, 10, 3276. [Google Scholar] [CrossRef]
- Auburger, G.; Key, J.; Gispert, S. The Bacterial ClpXP-ClpB Family Is Enriched with RNA-Binding Protein Complexes. Cells 2022, 11, 2370. [Google Scholar] [CrossRef]
- Odorskaya, M.V.; Mavletova, D.A.; Nesterov, A.A.; Tikhonova, O.V.; Soloveva, N.A.; Reznikova, D.A.; Galanova, O.O.; Vatlin, A.A.; Slynko, N.M.; Vasilieva, A.R.; et al. The Use of Omics Technologies in Creating LBP and Postbiotics Based on the Limosilactobacillus fermentum U-21. Front. Microbiol. 2024, 15, 1416688. [Google Scholar] [CrossRef]
- Stavrovskaya, A.V.; Voronkov, D.N.; Marsova, M.V.; Olshansky, A.S.; Gushchina, A.S.; Danilenko, V.N.; Illarioshkin, S.N. Effects of the Pharmabiotic U-21 in a Combined Neuroinflammatory Model of Parkinson’s Disease in Rats. Bull. Exp. Biol. Med. 2024, 177, 193–199. [Google Scholar] [CrossRef]
- Ojha, S.; Patil, N.; Jain, M.; Kole, C.; Kaushik, P. Probiotics for Neurodegenerative Diseases: A Systemic Review. Microorganisms 2023, 11, 1083. [Google Scholar] [CrossRef]
- Dhyani, P.; Goyal, C.; Dhull, S.B.; Chauhan, A.K.; Singh Saharan, B.; Harshita; Duhan, J.S.; Goksen, G. Psychobiotics for Mitigation of Neuro-Degenerative Diseases: Recent Advancements. Mol. Nutr. Food Res. 2023, 68, 2300461. [Google Scholar] [CrossRef]
- Peña-Díaz, S.; García-Pardo, J.; Ventura, S. Development of Small Molecules Targeting α-Synuclein Aggregation: A Promising Strategy to Treat Parkinson’s Disease. Pharmaceutics 2023, 15, 839. [Google Scholar] [CrossRef]
- Chu, C.; Yu, L.; Li, Y.; Guo, H.; Zhai, Q.; Chen, W.; Tian, F. Meta-Analysis of Randomized Controlled Trials of the Effects of Probiotics in Parkinson’s Disease. Food Funct. 2023, 14, 3406–3422. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant Computational Platform for Mass Spectrometry-Based 41 Shotgun Proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]


| Name | Description | Source |
|---|---|---|
| p15Tc-lac | Expression vector. Ptac-MCS, lacI, ori-p15A, and Tcr | [22] |
| pF6 | pACYC184 vector with luxABE from A. fischeri under Plac promoter, Cmr | [23] |
| p15FisAB | p15Tc-lac with luxAB from A. fischeri under Ptac promoter, Tcr | This study |
| pXen7 | pUC18 with luxCDABE operon from P. luminescens under control of its own promoter, Apr | [24] |
| p15XenAB | p15Tc-lac with luxAB from P. luminescens under Ptac promoter, Tcr | This study |
| pUC19:clpL | pUC19 vector (Sigma-Aldrich Inc., USA) with clpL from L. fermentum U-21 under Plac promoter, Apr | This study |
| pABX-T7 | pET15b with luxAB from P. luminescens under T7 promoter, Apr | This study |
| pLuxG-T7 | pET15b with luxG from V. aquamarinus under T7 promoter, Apr | This study |
| pET16b.clpL | pET16b vector (Novagen, Madison, WI, USA) with clpL from L. fermentum U-21 under T7 promoter, Apr | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al Ebrahim, R.N.; Alekseeva, M.G.; Bazhenov, S.V.; Fomin, V.V.; Mavletova, D.A.; Nesterov, A.A.; Poluektova, E.U.; Danilenko, V.N.; Manukhov, I.V. ClpL Chaperone as a Possible Component of the Disaggregase Activity of Limosilactobacillus fermentum U-21. Biology 2024, 13, 592. https://doi.org/10.3390/biology13080592
Al Ebrahim RN, Alekseeva MG, Bazhenov SV, Fomin VV, Mavletova DA, Nesterov AA, Poluektova EU, Danilenko VN, Manukhov IV. ClpL Chaperone as a Possible Component of the Disaggregase Activity of Limosilactobacillus fermentum U-21. Biology. 2024; 13(8):592. https://doi.org/10.3390/biology13080592
Chicago/Turabian StyleAl Ebrahim, Rahaf N., Maria G. Alekseeva, Sergey V. Bazhenov, Vadim V. Fomin, Dilara A. Mavletova, Andrey A. Nesterov, Elena U. Poluektova, Valeriy N. Danilenko, and Ilya V. Manukhov. 2024. "ClpL Chaperone as a Possible Component of the Disaggregase Activity of Limosilactobacillus fermentum U-21" Biology 13, no. 8: 592. https://doi.org/10.3390/biology13080592
APA StyleAl Ebrahim, R. N., Alekseeva, M. G., Bazhenov, S. V., Fomin, V. V., Mavletova, D. A., Nesterov, A. A., Poluektova, E. U., Danilenko, V. N., & Manukhov, I. V. (2024). ClpL Chaperone as a Possible Component of the Disaggregase Activity of Limosilactobacillus fermentum U-21. Biology, 13(8), 592. https://doi.org/10.3390/biology13080592







