Paternal Mitochondrial DNA Leakage in Natural Populations of Large-Scale Loach, Paramisgurnus dabryanus
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
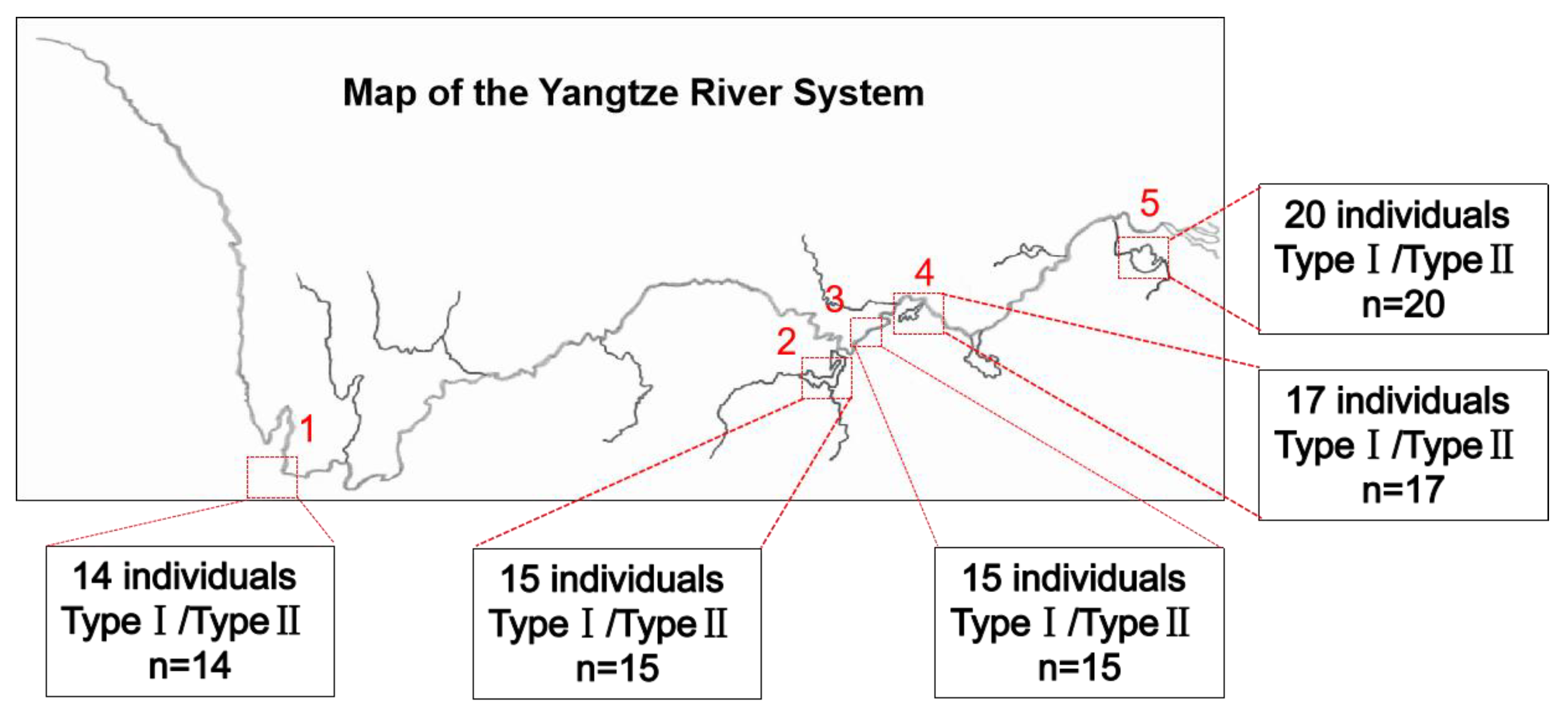
2.1. Experimental Animals
2.2. RNA Extraction and cDNA Synthesis
2.3. Polymerase Chain Reaction (PCR) Amplification and Sequencing
2.4. Gene Annotation and Sequence Analysis
2.5. Phylogenetic Analyses
2.6. Statistical Analysis
3. Results
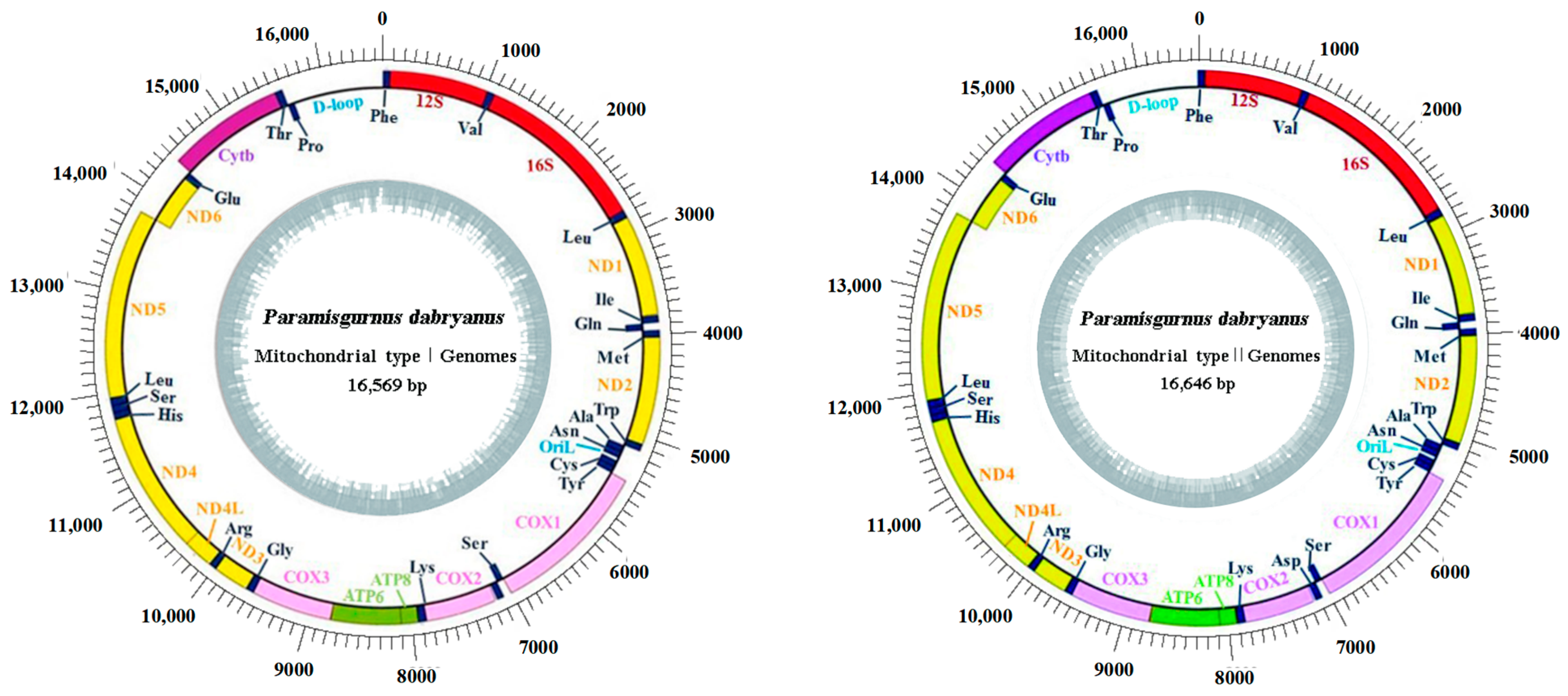
3.1. Mitogenome Organization and Composition
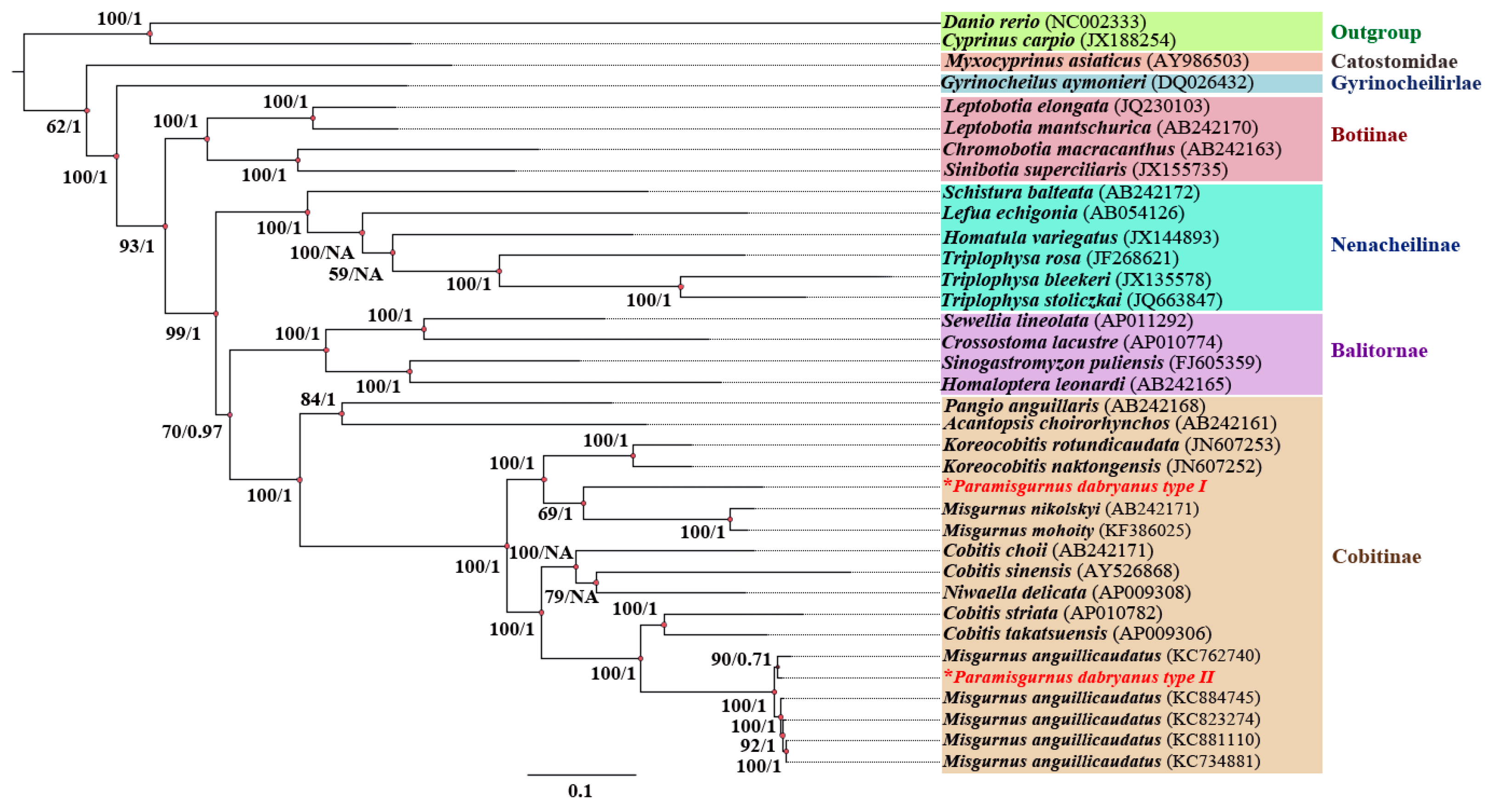
3.2. Phylogenetic Relationships
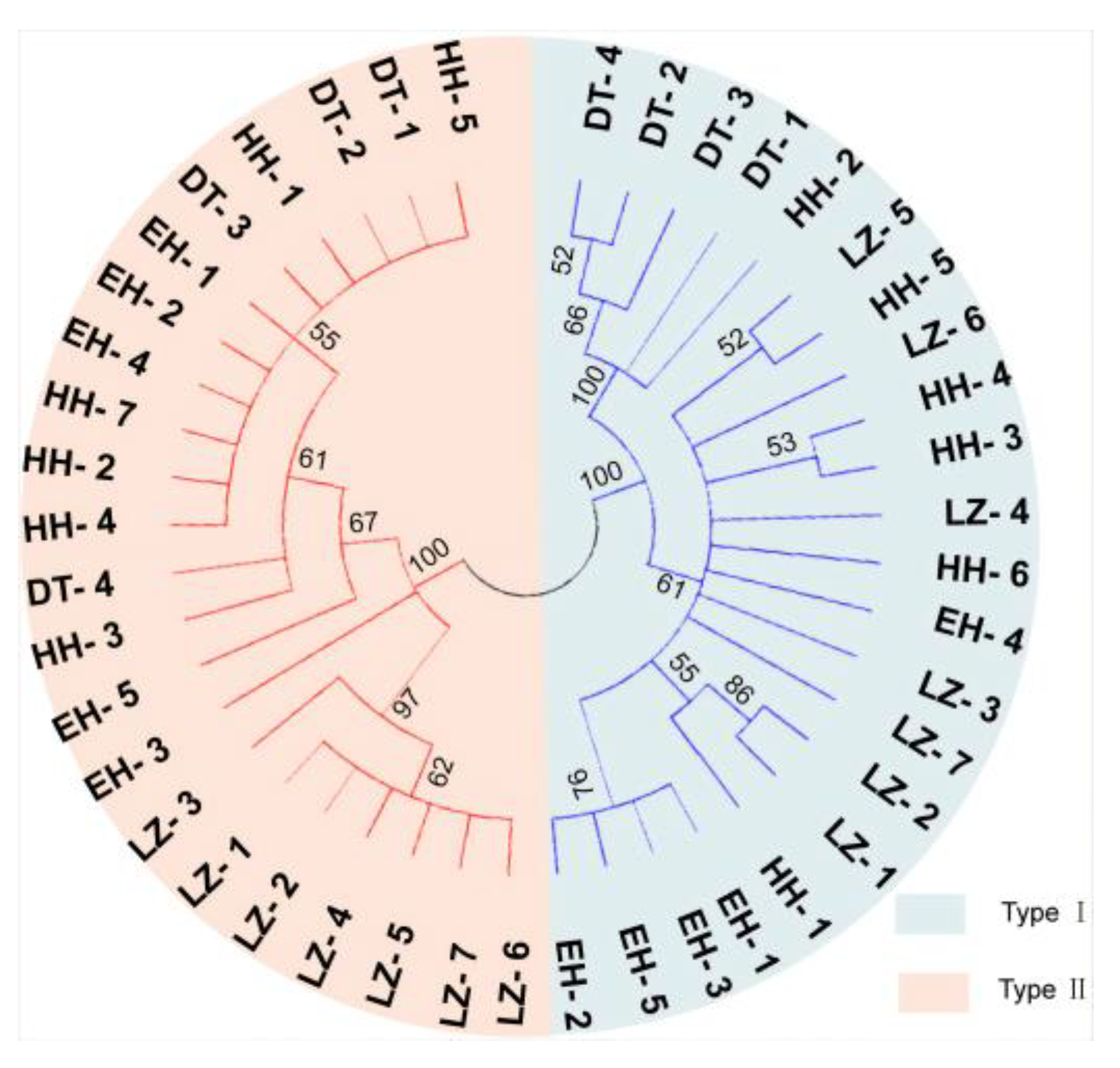
3.3. Investigation of Mitochondrial Heterogeneity in P. dabryanus from Different Regions
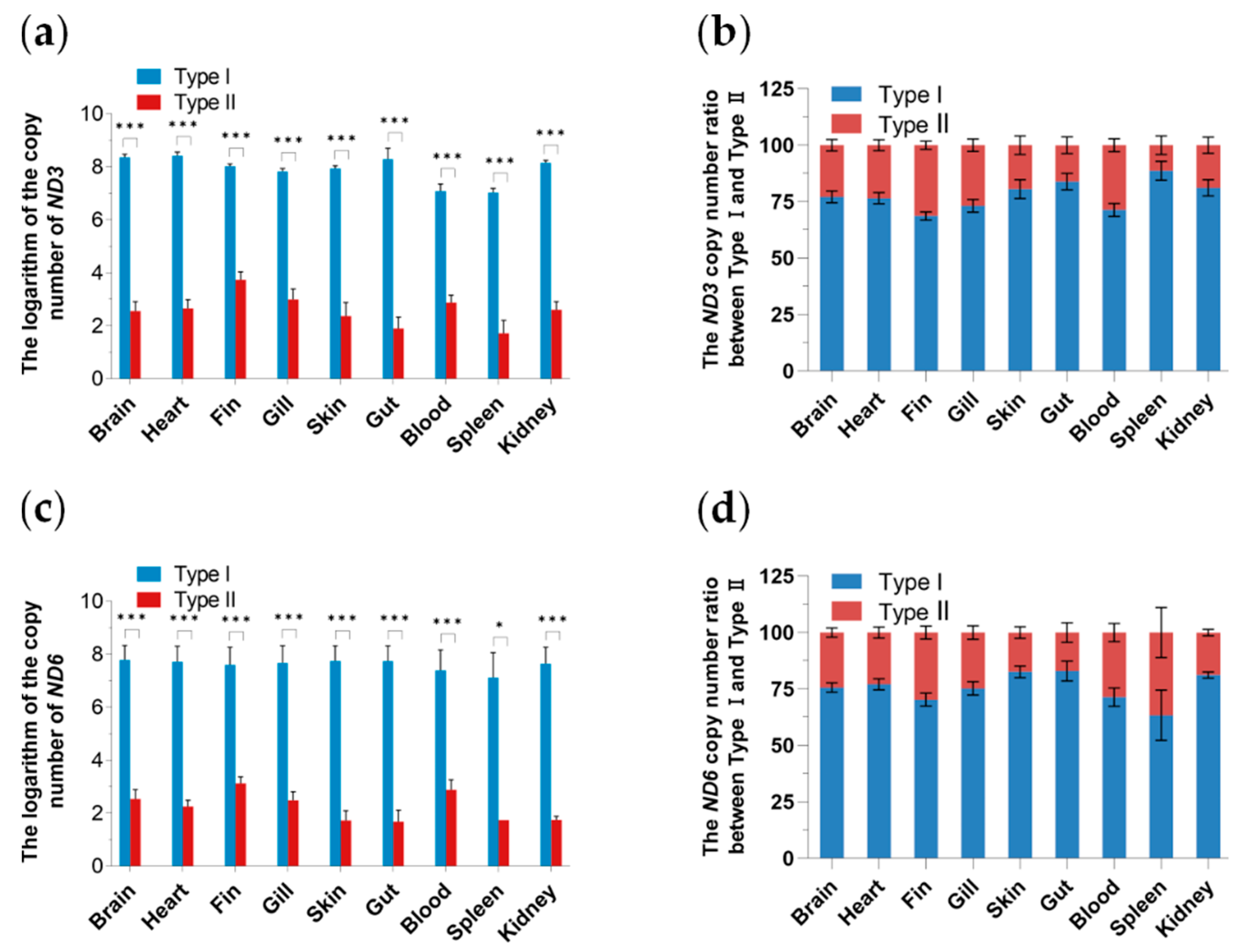
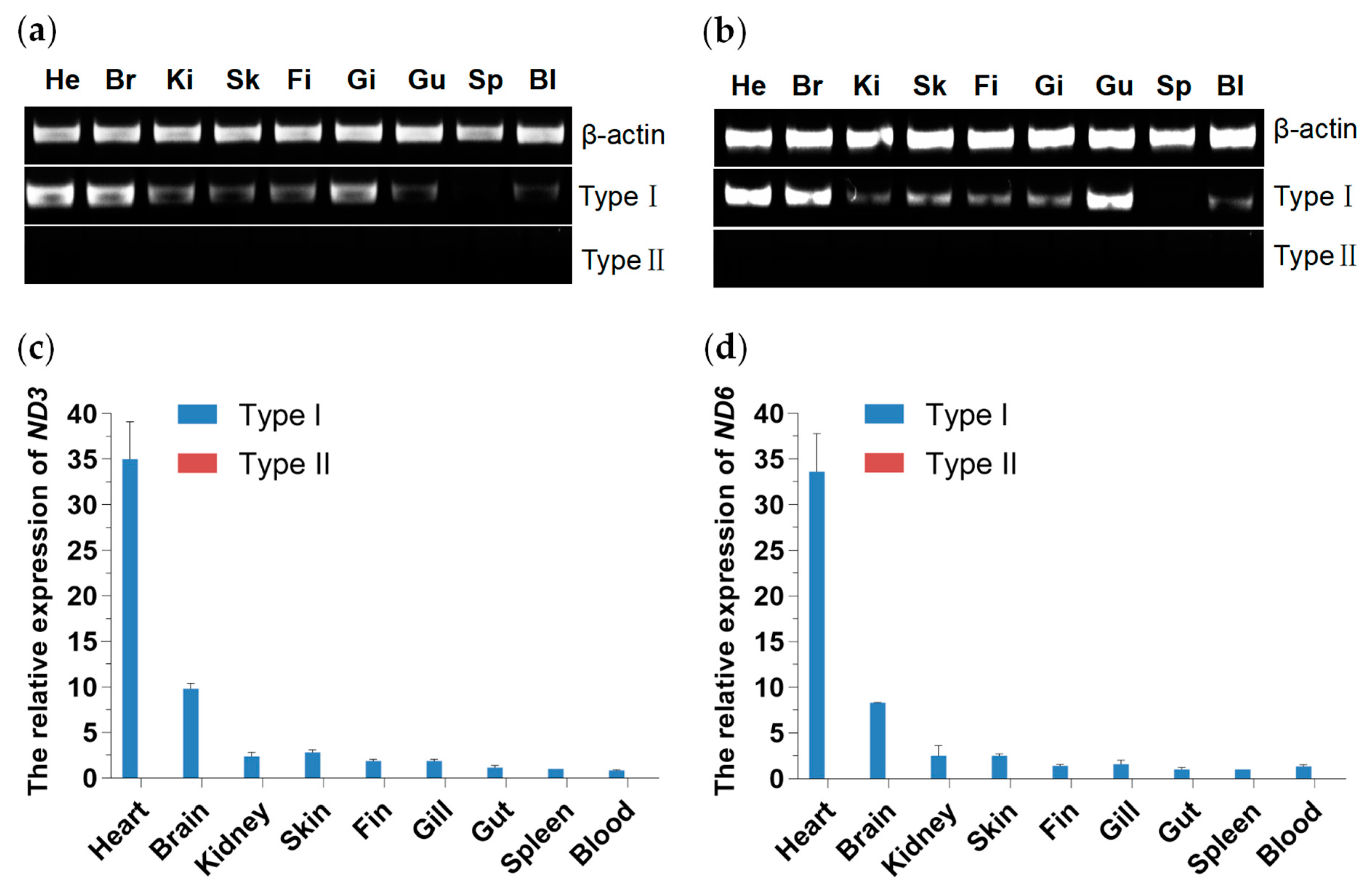
3.4. Tissue Distribution and Expression Analysis of Two Mitochondrial DNA Haplotypes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ma, X.; Niu, M.; Ni, H.M.; Ding, W.X. Mitochondrial dynamics, quality control, and mtDNA in alcohol-associated liver disease and liver cancer. Hepatology 2024. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, C.M.; Falkenberg, M.; Larsson, N.-G. Maintenance and Expression of Mammalian Mitochondrial DNA. Annu. Rev. Biochem. 2016, 85, 133–160. [Google Scholar] [CrossRef] [PubMed]
- Bi, C.; Wang, L.; Fan, Y.; Yuan, B.; Alsolami, S.; Zhang, Y.; Zhang, P.Y.; Huang, Y.; Yu, Y.; Izpisua Belmonte, J.C.; et al. Quantitative haplotype-resolved analysis of mitochondrial DNA heteroplasmy in Human single oocytes, blastoids, and pluripotent stem cells. Nucleic Acids Res. 2023, 51, 3793–3805. [Google Scholar] [CrossRef] [PubMed]
- Catanese, G.; Infante, C.; Manchado, M. Complete mitochondrial DNA sequences of the frigate tuna Auxis thazard and the bullet tuna Auxis rochei. DNA Seq. J. DNA Seq. Mapp. 2008, 19, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Yue, G.H.; Liew, W.C.; Orban, L. The complete mitochondrial genome of a basal teleost, the Asian arowana (Scleropages formosus, Osteoglossidae). BMC Genom. 2006, 7, 242. [Google Scholar] [CrossRef] [PubMed]
- Hoolahan, A.H.; Blok, V.C.; Gibson, T.; Dowton, M. Paternal leakage of mitochondrial DNA in experimental crosses of populations of the potato cyst nematode Globodera pallida. Genetica 2011, 139, 1509–1519. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.B.; Chinnery, P.F. Extreme heterogeneity of human mitochondrial DNA from organelles to populations. Nat. Rev. Genet. 2021, 22, 106–118. [Google Scholar] [CrossRef]
- Lee, J.; Willett, C.S.; Lopez, J. Frequent Paternal Mitochondrial Inheritance and Rapid Haplotype Frequency Shifts in Copepod Hybrids. J. Hered. 2022, 113, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.B.; Chinnery, P.F. The dynamics of mitochondrial DNA heteroplasmy: Implications for human health and disease. Nat. Rev. Genet. 2015, 16, 530–542. [Google Scholar] [CrossRef]
- Lunt, D.H.; Whipple, L.E.; Hyman, B.C. Mitochondrial DNA variable number tandem repeats (VNTRs): Utility and problems in molecular ecology. Mol. Ecol. 1998, 7, 1441–1455. [Google Scholar] [CrossRef]
- Moum, T.; Bakke, I. Mitochondrial control region structure and single site heteroplasmy in the razorbill (Alca torda; Aves). Curr. Genet. 2001, 39, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Densmore, L.D.; Wright, J.W.; Brown, W.M. Length variation and heteroplasmy are frequent in mitochondrial DNA from parthenogenetic and bisexual lizards (genus Cnemidophorus). Genetics 1985, 110, 689–707. [Google Scholar] [CrossRef] [PubMed]
- Kvist, L.; Martens, J.; Nazarenko, A.A.; Orell, M. Paternal leakage of mitochondrial DNA in the great tit (Parus major). Mol. Biol. Evol. 2003, 20, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.; Ho, S.Y.; Molak, M.; Barnett, R.; Carlborg, Ö.; Dorshorst, B.; Honaker, C.; Besnier, F.; Wahlberg, P.; Dobney, K.; et al. Mitogenomic analysis of a 50-generation chicken pedigree reveals a rapid rate of mitochondrial evolution and evidence for paternal mtDNA inheritance. Biol. Lett. 2015, 11, 20150561. [Google Scholar] [CrossRef] [PubMed]
- Gyllensten, U.; Wharton, D.; Josefsson, A.; Wilson, A.C. Paternal inheritance of mitochondrial DNA in mice. Nature 1991, 352, 255–257. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, N.; Guo, W.; Hu, X.; Liu, Z.; Gong, G.; Wang, A.; Feng, J.; Wu, C. Further evidence for paternal inheritance of mitochondrial DNA in the sheep (Ovis aries). Heredity 2004, 93, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Valencia, C.A.; Zhang, J.; Lee, N.C.; Slone, J.; Gui, B.; Wang, X.; Li, Z.; Dell, S.; Brown, J.; et al. Biparental Inheritance of Mitochondrial DNA in Humans. Proc. Natl. Acad. Sci. USA 2018, 115, 13039–13044. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.N. A105 family decoded: Discovery of genome-wide fingerprints for personalized genomic medicine. In Proceedings of the International Congress on Up Close & Personalized, Florence, Italy, 2–5 February 2012. [Google Scholar]
- Passamonti, M.; Ghiselli, F. Doubly uniparental inheritance: Two mitochondrial genomes, one precious model for organelle DNA inheritance and evolution. DNA Cell Biol. 2009, 28, 79–89. [Google Scholar] [CrossRef]
- Wolff, J.N.; Nafisinia, M.; Sutovsky, P.; Ballard, J.W. Paternal transmission of mitochondrial DNA as an integral part of mitochondrial inheritance in metapopulations of Drosophila simulans. Heredity 2013, 110, 57–62. [Google Scholar] [CrossRef]
- Meusel, M.S.; Moritz, R.F. Transfer of paternal mitochondrial DNA during fertilization of honeybee (Apis mellifera L.) eggs. Curr. Genet. 1993, 24, 539–543. [Google Scholar] [CrossRef]
- Fontaine, K.M.; Cooley, J.R.; Simon, C. Evidence for paternal leakage in hybrid periodical cicadas (Hemiptera: Magicicada spp.). PLoS ONE 2007, 2, e892. [Google Scholar] [CrossRef] [PubMed]
- Vilaça, S.T.; Maroso, F.; Lara, P.; de Thoisy, B.; Chevallier, D.; Arantes, L.S.; Santos, F.R.; Bertorelle, G.; Mazzoni, C.J. Evidence of backcross inviability and mitochondrial DNA paternal leakage in sea turtle hybrids. Mol. Ecol. 2023, 32, 628–643. [Google Scholar] [CrossRef] [PubMed]
- Gandolfi, A.; Crestanello, B.; Fagotti, A.; Simoncelli, F.; Chiesa, S.; Girardi, M.; Giovagnoli, E.; Marangoni, C.; Di Rosa, I.; Lucentini, L. New Evidences of Mitochondrial DNA Heteroplasmy by Putative Paternal Leakage between the Rock Partridge (Alectoris graeca) and the Chukar Partridge (Alectoris chukar). PLoS ONE 2017, 12, e0170507. [Google Scholar] [CrossRef] [PubMed]
- Polovina, E.S.; Parakatselaki, M.E.; Ladoukakis, E.D. Paternal leakage of mitochondrial DNA and maternal inheritance of heteroplasmy in Drosophila hybrids. Sci. Rep. 2020, 10, 2599. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, W.; Gu, Q.; Yao, J.; Tan, H.; Huang, X.; Qin, Q.; Tao, M.; Zhang, C.; Liu, S. Variations in the Mitochondrial Genome of a Goldfish-Like Hybrid [Koi Carp (♀) × Blunt Snout Bream (♂)] Indicate Paternal Leakage. Front. Genet. 2021, 11, 613520. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Jiao, N.; Zhao, L.; Zhang, M.; Zhou, P.; Huang, X.; Hu, F.; Yang, C.; Shu, Y.; Li, W.; et al. Evidence for the paternal mitochondrial DNA in the crucian carp-like fish lineage with hybrid origin. Sci. China Life Sci. 2020, 63, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Z.S. The subfamily classification of the Cobitidae fishes and their phylogenetic relationships. Acta Zootaxon. Sin. 1984, 9, 201–207. [Google Scholar]
- Miya, M.; Nishida, M. Use of mitogenomic information in teleostean molecular phylogenetics: A tree-based exploration under the maximum-parsimony optimality criterion. Mol. Phylogenet. Evol. 2000, 17, 437–455. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Nalbant, T.T.; Bianco, P.G. The loaches of Iran and adjacent regions with description of six new species (Cobitoidea). Ital. J. Zool. 1998, 65, 109–123. [Google Scholar] [CrossRef]
- Tang, Q.; Liu, H.; Mayden, R.; Xiong, B. Comparison of evolutionary rates in the mitochondrial DNA cytochrome b gene and control region and their implications for phylogeny of the Cobitoidea (Teleostei: Cypriniformes). Mol. Phylogenet. Evol. 2006, 39, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, W.; Song, W.; Huang, S.; Cao, X. The complete mitochondrial genome of natural Paramisgurnus dabryanus (Cypriniformes: Cobitidae). Mitochondrial DNA 2015, 26, 937–938. [Google Scholar] [CrossRef]
- Dai, L.; Guo, B.; Chu, Z.; Wang, Y.; Wang, X.; Huang, T. Complete mitochondrial genome of Paramisgurnus dabryanus. Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2016, 27, 1257–1259. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Shen, K.; Feng, S.; Wang, C.; Xu, S. The complete mitochondrial genome of Platycephalus sp.1 (Teleostei, Platycephalidae) obtained by whole genome sequencing. Mitochondrial DNA. Part B Resour. 2021, 6, 1941–1943. [Google Scholar] [CrossRef]
- Bi, S.; Song, Y.; Liu, L.; Wan, J.; Zhou, Y.; Zhu, Q.; Liu, J. Complete Mitochondrial Genome of Piophila casei (Diptera: Piophilidae): Genome Description and Phylogenetic Implications. Genes 2023, 14, 883. [Google Scholar] [CrossRef]
- Kim, J.; Jeong, J.; Lee, S.; Nguyen, P.T.; Han, D.W.; Hettiarachchi, S.A.; Jo, E.; Kang, S.; Kim, I.C.; Kim, J.H. The complete mitochondrial genome of Gobionotothen gibberifrons (Perciformes, Nototheniidae). Mitochondrial DNA. Part B Resour. 2024, 9, 398–402. [Google Scholar] [CrossRef] [PubMed]
- Moritz, C.; Dowling, T.E.; Brown, W.M. Evolution of Animal Mitochondrial DNA: Relevance for Population Biology and Systematics. Annu. Rev. Ecol. Syst. 1987, 18, 269–292. [Google Scholar] [CrossRef]
- Hoeh, W.R.; Blakley, K.H.; Brown, W.M. Heteroplasmy suggests limited biparental inheritance of Mytilus mitochondrial DNA. Science 1991, 251, 1488–1490. [Google Scholar] [CrossRef]
- Ballard, J.W. Sequential evolution of a symbiont inferred from the host: Wolbachia and Drosophila simulans. Mol. Biol. Evol. 2004, 21, 428–442. [Google Scholar] [CrossRef]
- Mizi, A.; Zouros, E.; Moschonas, N.; Rodakis, G.C. The complete maternal and paternal mitochondrial genomes of the Mediterranean mussel Mytilus galloprovincialis: Implications for the doubly uniparental inheritance mode of mtDNA. Mol. Biol. Evol. 2005, 22, 952–967. [Google Scholar] [CrossRef]
- Saville, B.J.; Kohli, Y.; Anderson, J.B. mtDNA recombination in a natural population. Proc. Natl. Acad. Sci. USA 1998, 95, 1331–1335. [Google Scholar] [CrossRef] [PubMed]
- Sutovsky, P.; Moreno, R.D.; Ramalho-Santos, J.; Dominko, T.; Simerly, C.; Schatten, G. Ubiquitinated sperm mitochondria, selective proteolysis, and the regulation of mitochondrial inheritance in mammalian embryos. Biol. Reprod. 2000, 63, 582–590. [Google Scholar] [CrossRef]
- Sherengul, W.; Kondo, R.; Matsuura, E.T. Analysis of paternal transmission of mitochondrial DNA in Drosophila. Genes Genet. Syst. 2006, 81, 399–404. [Google Scholar] [CrossRef] [PubMed][Green Version]
- You, C.; Yu, X.; Tong, J. Detection of hybridization between two loach species (Paramisgurnus dabryanus and Misgurnus anguillicaudatus) in wild populations. Environ. Biol. Fishes 2009, 86, 65–71. [Google Scholar] [CrossRef]
- Kim, D.S.; Nam, Y.K.; Park, I.-S. Survival and karyological analysis of reciprocal diploid and triploid hybrids between mud loach (Misgurnus mizolepis) and pond loach (Misgurnus anguillicaudatus). Aquaculture 1995, 135, 257–265. [Google Scholar] [CrossRef]
- Nam, Y.K.; Park, I.-S.; Kim, D.S. Triploid hybridization of fast-growing transgenic mud loach Misgurnus mizolepis male to cyprinid loach Misgurnus anguillicaudatus female: The first performance study on growth and reproduction of transgenic polyploid hybrid fish. Aquaculture 2004, 231, 559–572. [Google Scholar] [CrossRef]
- Wallace, D.C.; Chalkia, D. Mitochondrial DNA Genetics and the Heteroplasmy Conundrum in Evolution and Disease. Cold Spring Harb. Perspect. Biol. 2013, 5, a021220. [Google Scholar] [CrossRef] [PubMed]
- Mastrantonio, V.; Latrofa, M.S.; Porretta, D.; Lia, R.P.; Parisi, A.; Iatta, R.; Dantas-Torres, F.; Otranto, D.; Urbanelli, S. Paternal leakage and mtDNA heteroplasmy in Rhipicephalus spp. ticks. Sci. Rep. 2019, 9, 1460. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Kalinowski, P.; Kim, B.; Pauls, A.D.; Poburko, D. Emerging methods for and novel insights gained by absolute quantification of mitochondrial DNA copy number and its clinical applications. Pharmacol. Ther. 2022, 232, 107995. [Google Scholar] [CrossRef]
- Wang, Z.-R.; Li, S.-Y.; Zhang, Y.-Z.; Li, Y.-A.; Huo, H.-H.; Yu, C.-Q.; Zhou, Q.-B. Metabolomic and transcriptomic profiling reveals the effect of dietary protein and lipid levels on growth performance in loach (Paramisgurnus dabryanus). Front. Immunol. 2023, 14, 1236812. [Google Scholar] [CrossRef]

| Forward | Sequence (5′–3′) | Reverse | Sequence (5′–3′) |
|---|---|---|---|
| Long PCR Primers | |||
| S-LA-16S-H | TGCACCATTRGGATGTCCTGATCCAACATC | S-LA-16S-L | CGATTAAAGTCCTACGTGATCTGAGTTCAG |
| L12321-Leu | GGTCTTAGGAACCAAAAACTCTTGGTGCAA | H15149-CYB | GGTGGCKCCTCAGAAGGACATTTGKCCTCA |
| P13-L | CCAGCAATAATACTTCCTCAG | ||
| Internal Primers | |||
| P1F | CACTGAAGATGCTAAGATGG | P1R | TCTCTGCCTGTTGTATGC |
| P2F | TTACACCGAGAAGACATCC | P2R | CTCGTCTTGTAGGTGTATGC |
| P3F | CAGTGACCACAAGTTCAAC | P3R | GCATATTCAGCCAGGAAGA |
| P4F | TCTCTAGCCTTGCCGTATA | P4R | GTTGATAGGATTAGACCTGTTG |
| P5F | ATAGCACAGCAGCATCAC | P5R | GCAGTTCCAACCATTCCA |
| P6F | AACTTAGACCAAGAGCCTTC | P6R | CACGAGTATCAACATCTATTCC |
| P7F | ACTGCCGTTCTTCTTCTAC | P7R | AGCCTAAGTCCTCATAGTCA |
| P8F | ATAACCAYTCTGCCAYTTC | P8R | CGATTRATTAGTCAGCCTTG |
| P9F | CGTTCCACTTGAGCACTT | P9R | ATGGTCAGAAGAAGCAGAAT |
| P10F | GTCTATTCATTCGTCCATTAGC | P10R | GCATTGTAGGAGATTGAGGTT |
| P11F | AAGACCGTGGTTCAACTC | P11R | TGTGTTCGCTCGTAAGTG |
| P12F | TGTTCACCTCTGACTACCTA | P12R | CATCTGCTCGTCCGTATC |
| P13F | CATCCTGATAYATRCACTCTGAC | P13R | CCAGCAAYAATACTTCCTCAG |
| P14F | CTGGCATTCCTTCACATCT | P14R | TTCTTCAAGTCACTGGTCTC |
| P15F | CTGCCACCACTAATCCTAA | P15R | GCTCACTCTAATGCCTTGT |
| P16F | GCCACTATTCTACATCTACTCT | P16R | GCATAAGTCAACACCTACTG |
| P17F | AACAAGGCATTAGAGTGAG | P17R | GTATGACAGCCAAGAGGT |
| P18F | GATACCAGTAGAACATCCAT | P18R | GCCCTCTTATCCCTAACTA |
| Forward | Sequence (5′–3′) | Reverse | Sequence (5′–3′) |
|---|---|---|---|
| Two Types of Mitochondrial Detection Primers | |||
| Test1-F | ACCCGTACTTATACTAAAACC | Test1-R | CGATGCCAATAGAACAGC |
| Test2-F | CCGTACTTATACTAAAACCG | Test2-R | CAAGAAGATTATTAGGGAGG |
| Test3-F | GCTAGAAATAGCAACTATGG | Test3-R | GGGAGGTGAGTAGTAGGG |
| Test4-F | CGTTCCACTTGAGCACTT | Test4-R | ATGGTCAGAAGAAGCAGAAT |
| Test5-F | TATCGTCGCCATAGTCTCCA | Test5-R | GTTTTAGTATAAGTACGGGTTTT |
| Test6-F | CAACTATGGAAGAAGTTACA | Test6-R | CAAGCAGGGTTAATAGGAAT |
| Test7-F | TACTTTACATTATCGTCGCC | Test7-R | GTTTTAGTATAAGTACGGGTTTT |
| Test8-F | TACTTTACATTATCGTCGCC | Test8-R | CTTCCATAGTTGCTATTTCT |
| Quantitative Real-Time PCR Primers | |||
| ND3-1F | CCTTGGATCTGCTCGTTT | ND3-1R | CCCTGAGCCCACTCGTAT |
| ND3-2F | GATCTGCCCGATTACCAT | ND3-2R | CATTCATAAACCAAGCCTAA |
| ND6-1F | TGGTTGCTGTGGCTTCTA | ND6-1R | GCCAGAGCTGCCGAATAG |
| ND6-2F | CTGGCCGCTGAACCATTT | ND6-2R | CTCCTCGTAACATACTAAATTCC |
| Name of Gene | Location | Size (bp) | Start Codon | Stop Codon | Intergenic Nucleotides | Stand | Nucleotide Divergence | |
|---|---|---|---|---|---|---|---|---|
| I | II | I/II | I/II | I/II | I/II | I/II | I to II | |
| tRNAPhe | 1–69 | 1–69 | 69 | 0 | H | 0.05 | ||
| 12S rRNA | 70–1023 | 70–1022 | 954/953 | 0 | H | 0.06 | ||
| tRNAVal | 1024–1095 | 1023–1094 | 72 | 0 | H | 0.03 | ||
| 16S rRNA | 1096–2773 | 1095–2773 | 1678/1679 | 0 | H | 0.1 | ||
| tRNALeu(UUR) | 2774–2848 | 2774–2848 | 75 | 1 | H | 0.01 | ||
| ND1 | 2850–3824 | 2850–3824 | 975 | ATG | TAA | 7/6 | H | 0.17 |
| tRNAIle | 3830–3901 | 3831–3902 | 72 | −2 | H | 0.04 | ||
| tRNAGln | 3900–3970 | 3901–3971 | 71 | 1 | L | 0 | ||
| tRNAMet | 3972–4040 | 3973–4041 | 69 | 0 | H | 0.03 | ||
| ND2 | 4041–5087 | 4042–5086 | 1047/1045 | ATG | TAA/T-- | −2/0 | H | 0.17 |
| tRNATrp | 5086–5155 | 5087–5156 | 70 | 2/1 | H | 0.08 | ||
| tRNAAla | 5158–5226 | 5158–5226 | 69 | 1 | L | 0.04 | ||
| tRNAAsn | 5228–5300 | 5228–5300 | 73 | 0 | L | 0.01 | ||
| OriL | 5301–5330 | 5301–5330 | 30 | 0 | L | 0.1 | ||
| tRNACys | 5331–5396 | 5331–5396 | 66 | 0 | L | 0.08 | ||
| tRNATyr | 5397–5465 | 5397–5465 | 69 | 1 | L | 0.03 | ||
| COX1 | 5467–7017 | 5467–7017 | 1551 | GTG | TAA | 1 | H | 0.14 |
| tRNASer(UCN) | 7019–7089 | 7019–7089 | 71 | 0/2 | L | 0.06 | ||
| tRNAAsp | 7090–7161 | 7092–7163 | 72 | 3/13 | H | 0.07 | ||
| COX2 | 7175–7865 | 7177–7911 | 691/735 | ATG | T--/TAA | 0/27 | H | 0.13 |
| tRNALys | 7866–7941 | 7939–8014 | 76 | 1 | H | 0.03 | ||
| ATPase8 | 7943–8110 | 8016–8183 | 168 | ATG | TAA | 0/−10 | H | 0.12 |
| ATPase6 | 8101–8784 | 8174–8857 | 684 | ATG | TAA | −1 | H | 0.15 |
| COX3 | 8784–9567 | 8857–9640 | 784 | ATG | T-- | 0 | H | 0.13 |
| tRNAGly | 9568–9640 | 9641–9713 | 73 | 0 | H | 0.03 | ||
| ND3 | 9641–9991 | 9714–10,062 | 351/349 | ATG | TAA/T-- | −1/0 | H | 0.17 |
| tRNAArg | 9991–10,060 | 10,063–10,132 | 70 | 0 | H | 0.06 | ||
| ND4L | 10,061–10,357 | 10,133–10,429 | 297 | ATG | TAA | −7 | H | 0.14 |
| ND4 | 10,351–11,733 | 10,423–11,804 | 1383/1382 | ATG | TAG/TA- | −1/0 | H | 0.17 |
| tRNAHis | 11,733–11,801 | 11,805–11,873 | 69 | 0 | H | 0 | ||
| tRNASer(AGY) | 11,802–11,869 | 11,874–11,941 | 68 | 0/1 | H | 0.04 | ||
| tRNALeu(CUN) | 11,871–11,943 | 11,943–12,015 | 73 | 0 | H | 0.01 | ||
| ND5 | 11,944–13,782 | 12,016–13,854 | 1839 | ATG | TAA/TAG | −4 | H | 0.18 |
| ND6 | 13,779–14,300 | 13,851–14,372 | 522 | ATG | TAA | 0 | L | 0.17 |
| tRNAGlu | 14,301–14,369 | 14,373–14,441 | 69 | 7/6 | L | 0 | ||
| Cytb | 14,377–15,517 | 14,448–15,588 | 1141 | ATG | T-- | 0 | H | 0.15 |
| tRNAThr | 15,518–15,589 | 15,589–15,660 | 72 | −2 | H | 0.06 | ||
| tRNAPro | 15,588–15,657 | 15,659–15,728 | 70 | 0 | L | 0.03 | ||
| D-loop | 15,658–16,569 | 15,729–16,646 | 912/918 | H | 0.14 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, Z.; Shi, J.; Yu, Y.; Yin, G.; Zhou, X.; Yu, Y. Paternal Mitochondrial DNA Leakage in Natural Populations of Large-Scale Loach, Paramisgurnus dabryanus. Biology 2024, 13, 604. https://doi.org/10.3390/biology13080604
Qi Z, Shi J, Yu Y, Yin G, Zhou X, Yu Y. Paternal Mitochondrial DNA Leakage in Natural Populations of Large-Scale Loach, Paramisgurnus dabryanus. Biology. 2024; 13(8):604. https://doi.org/10.3390/biology13080604
Chicago/Turabian StyleQi, Zixin, Jiaoxu Shi, Yue Yu, Guangmei Yin, Xiaoyun Zhou, and Yongyao Yu. 2024. "Paternal Mitochondrial DNA Leakage in Natural Populations of Large-Scale Loach, Paramisgurnus dabryanus" Biology 13, no. 8: 604. https://doi.org/10.3390/biology13080604
APA StyleQi, Z., Shi, J., Yu, Y., Yin, G., Zhou, X., & Yu, Y. (2024). Paternal Mitochondrial DNA Leakage in Natural Populations of Large-Scale Loach, Paramisgurnus dabryanus. Biology, 13(8), 604. https://doi.org/10.3390/biology13080604







