A Comparative Analysis of the Immunoglobulin Repertoire in Leukemia Cells and B Cells in Chinese Acute Myeloid Leukemia by High-Throughput Sequencing
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. AML Patient Samples
2.2. Isolation and Preparation of Bone Marrow Mononuclear Cells
2.3. Fluorescence-Activated Cell Sorting
2.4. Flow Cytometry Analysis
2.5. RNA Extraction and cDNA Synthesis by 5′-RACE
2.6. Nested PCR
2.7. PacBio Sequencing and Data Analysis
2.8. Protein Extraction and Western Blot Analysis
2.9. Statistical Analysis
3. Results
3.1. Characteristics of AML Patients and the Strategy of Ig Sequencing
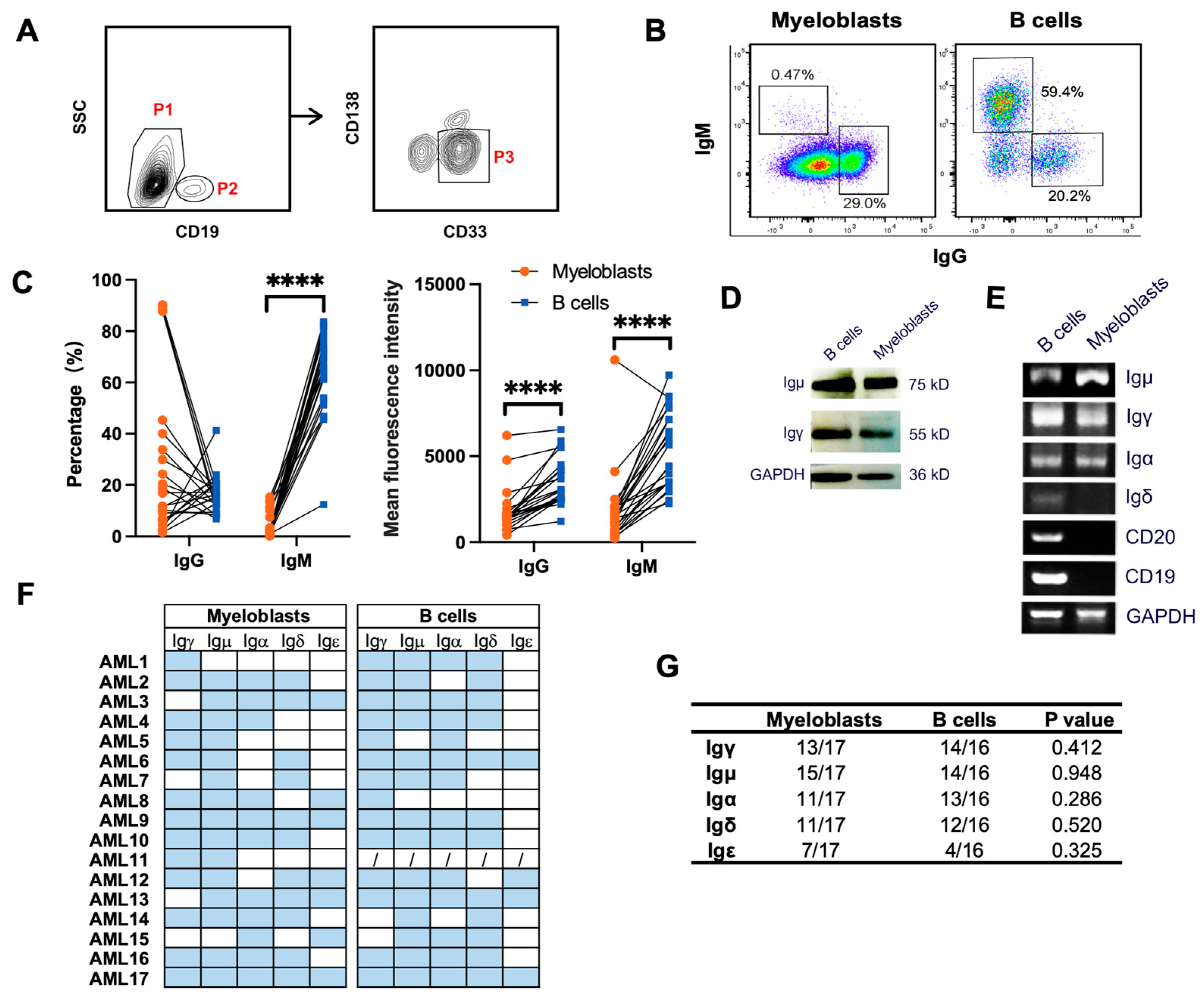
3.2. Primary Myeloblasts Express Different Types of Ig Heavy Chains
3.3. The Biased Usages of V(D)J Rearrangement Patterns in Myeloblasts Are Different from That of B Cells from the Same Patient
3.4. IGHV Mutations with Relatively High Frequency Occur at Unique Sites in Myeloblasts
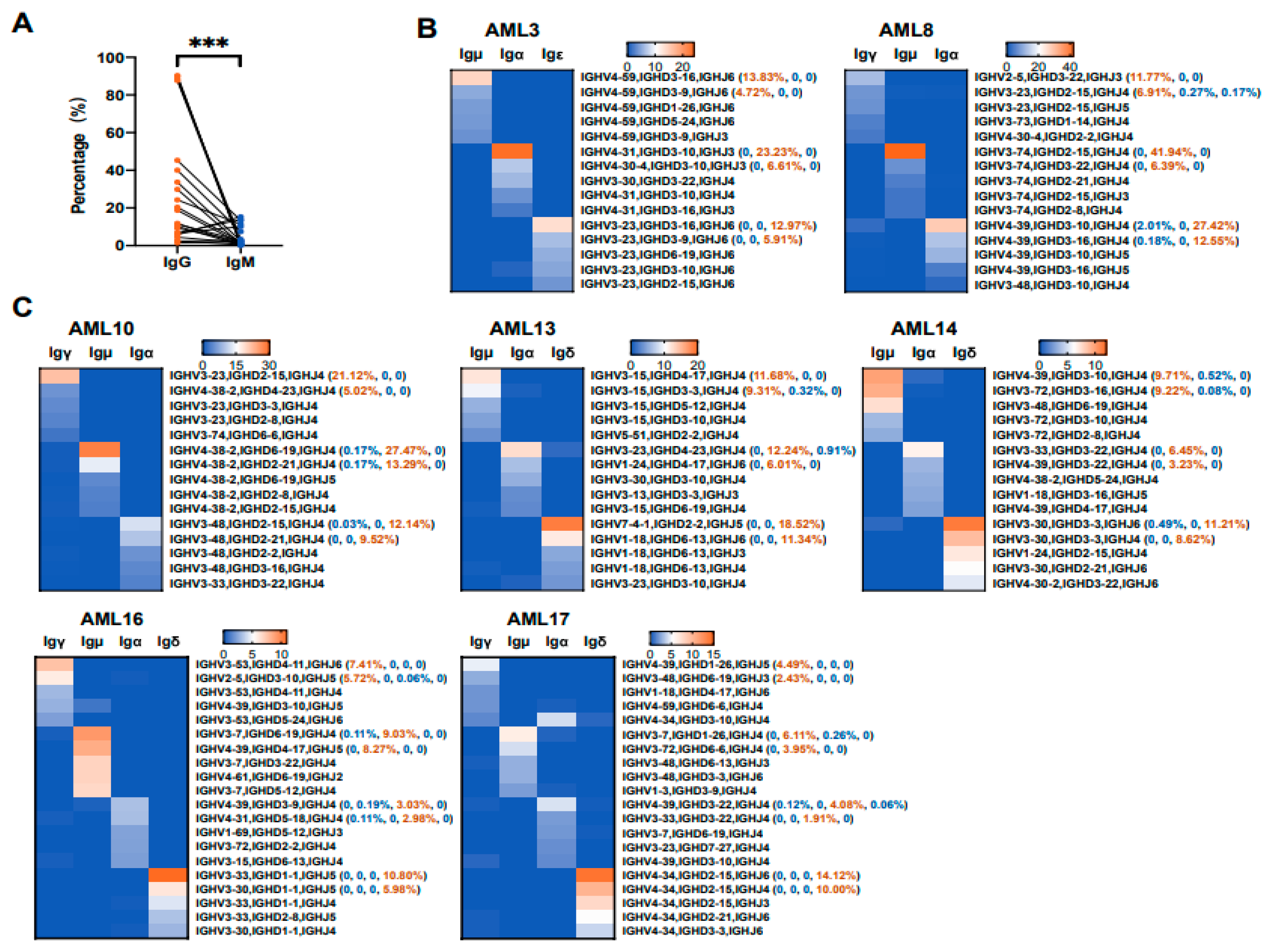
3.5. Various Ig Classes of AML-Ig from the Same Patient Tend to Use Different VHDJH Rearrangement Patterns
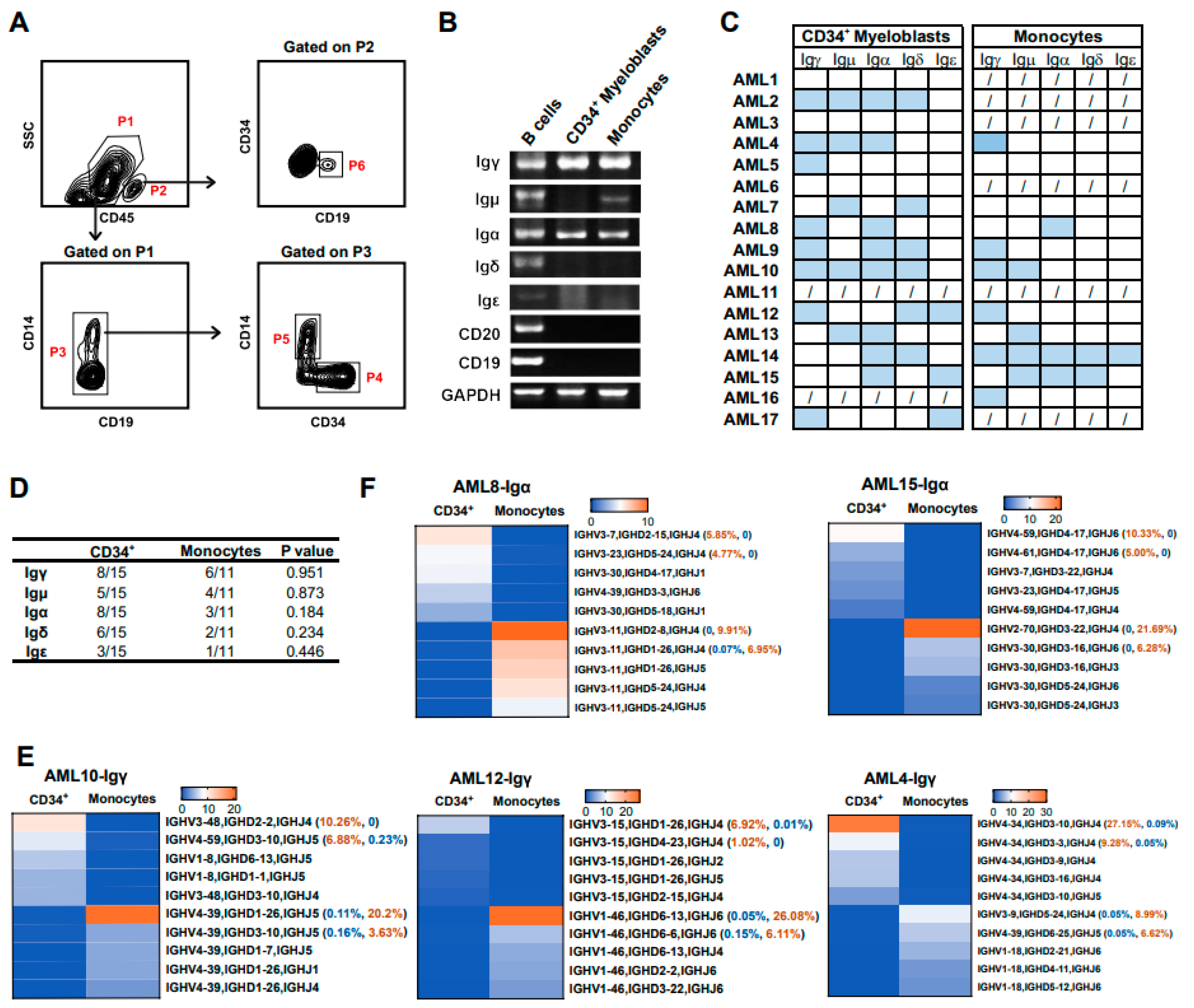
3.6. Myeloid Cells at Different Differentiation Stages Express Different Ig Rearrangement Patterns
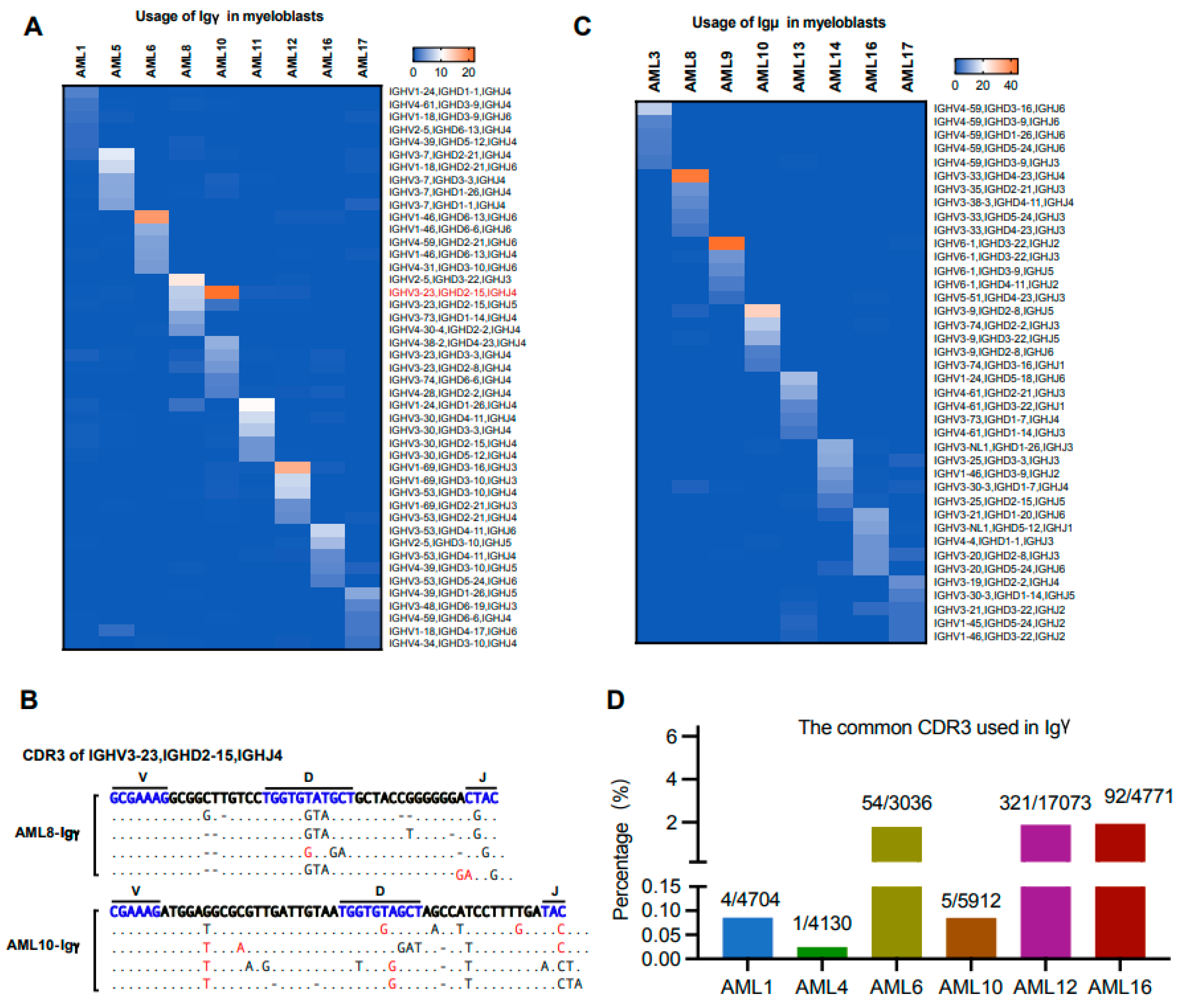
3.7. The Biased Usage of V(D)J Rearrangement Patterns Are Different in the Same Ig Class of Myeloblasts from Different Individuals
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qiu, X.; Zhu, X.; Zhang, L.; Mao, Y.; Zhang, J.; Hao, P.; Li, G.; Lv, P.; Li, Z.; Sun, X.; et al. Human epithelial cancers secrete immunoglobulin g with unidentified specificity to promote growth and survival of tumor cells. Cancer Res. 2003, 63, 6488–6495. [Google Scholar] [PubMed]
- Huang, X.; Zhang, S.; Tang, J.; Tian, T.; Pan, Y.; Wu, L.; Zhang, J.; Liu, Y.; Huang, J.; Dai, H.; et al. A Self-Propagating c-Met-SOX2 Axis Drives Cancer-Derived IgG Signaling That Promotes Lung Cancer Cell Stemness. Cancer Res. 2023, 83, 1866–1882. [Google Scholar] [CrossRef]
- Ma, J.; Jiang, D.; Gong, X.; Shao, W.; Zhu, Z.; Xu, W.; Qiu, X. Free immunoglobulin light chain (FLC) promotes murine colitis and colitis-associated colon carcinogenesis by activating the inflammasome. Sci. Rep. 2017, 7, 5165. [Google Scholar] [CrossRef]
- Huang, J.; Sun, X.; Gong, X.; He, Z.; Chen, L.; Qiu, X.; Yin, C.C. Rearrangement and expression of the immunoglobulin mu-chain gene in human myeloid cells. Cell Mol. Immunol. 2014, 11, 94–104. [Google Scholar] [CrossRef]
- Qiu, X.; Sun, X.; He, Z.; Huang, J.; Hu, F.; Chen, L.; Lin, P.; You, M.J.; Medeiros, L.J.; Yin, C.C. Immunoglobulin gamma heavy chain gene with somatic hypermutation is frequently expressed in acute myeloid leukemia. Leukemia 2013, 27, 92–99. [Google Scholar] [CrossRef]
- Fuchs, T.; Hahn, M.; Ries, L.; Giesler, S.; Busch, S.; Wang, C.; Han, J.; Schulze, T.J.; Puellmann, K.; Beham, A.W.; et al. Expression of combinatorial immunoglobulins in macrophages in the tumor microenvironment. PLoS ONE 2018, 13, e0204108. [Google Scholar] [CrossRef] [PubMed]
- Schmetzer, H.M.; Gerhartz, H.H. Acute myeloid leukemia (AML) can be oligoclonal. Leukemia 1993, 7, 1965–1970. [Google Scholar] [PubMed]
- Schmetzer, H.M.; Braun, S.; Wiesner, D.; Duell, T.; Gerhartz, H.H.; Mittermueller, J. Gene rearrangements in bone marrow cells of patients with acute myelogenous leukemia. Acta Haematol. 2000, 103, 125–134. [Google Scholar] [CrossRef]
- Yang, B.; Ma, C.; Chen, Z.; Yi, W.; McNutt, M.A.; Wang, Y.; Korteweg, C.; Gu, J. Correlation of immunoglobulin G expression and histological subtype and stage in breast cancer. PLoS ONE 2013, 8, e58706. [Google Scholar] [CrossRef]
- Tang, J.; Zhang, J.; Liu, Y.; Liao, Q.; Huang, J.; Geng, Z.; Xu, W.; Sheng, Z.; Lee, G.; Zhang, Y.; et al. Lung squamous cell carcinoma cells express non-canonically glycosylated IgG that activates integrin-FAK signaling. Cancer Lett. 2018, 430, 148–159. [Google Scholar] [CrossRef]
- Jiang, C.; Huang, T.; Wang, Y.; Huang, G.; Wan, X.; Gu, J. Immunoglobulin G expression in lung cancer and its effects on metastasis. PLoS ONE 2014, 9, e97359. [Google Scholar] [CrossRef] [PubMed]
- Chu, J.; Li, Y.; Deng, Z.; Zhang, Z.; Xie, Q.; Zhang, H.; Zhong, W.; Pan, B. IGHG1 Regulates Prostate Cancer Growth via the MEK/ERK/c-Myc Pathway. Biomed. Res. Int. 2019, 2019, 7201562. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hu, S.; Korteweg, C.; Chen, Z.; Qiu, Y.; Su, M.; Gu, J. Expression of immunoglobulin G in esophageal squamous cell carcinomas and its association with tumor grade and Ki67. Hum. Pathol. 2012, 43, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wang, J.; Zhang, Q.; Zhang, J.; Lou, Y.; Yang, J.; Chen, Y.; Wei, T.; Zhang, J.; Fu, Q.; et al. Tumour cell-derived debris and IgG synergistically promote metastasis of pancreatic cancer by inducing inflammation via tumour-associated macrophages. Br. J. Cancer 2019, 121, 786–795. [Google Scholar] [CrossRef]
- Chen, Z.; Huang, X.; Ye, J.; Pan, P.; Cao, Q.; Yang, B.; Li, Z.; Su, M.; Huang, C.; Gu, J. Immunoglobulin G is present in a wide variety of soft tissue tumors and correlates well with proliferation markers and tumor grades. Cancer 2010, 116, 1953–1963. [Google Scholar] [CrossRef]
- Fan, T.; Liao, Q.; Zhao, Y.; Dai, H.; Song, S.; He, T.; Wang, Z.; Huang, J.; Zeng, Z.; Guo, H.; et al. Sialylated IgG in epithelial cancers inhibits antitumor function of T cells via Siglec-7. Cancer Sci. 2023, 114, 370–383. [Google Scholar] [CrossRef]
- Wu, L.; Xia, M.; Sun, X.; Han, X.; Zu, Y.; Jabbour, E.J.; You, M.J.; Lin, P.; Li, S.; Xu, J.; et al. High levels of immunoglobulin expression predict shorter overall survival in patients with acute myeloid leukemia. Eur. J. Haematol. 2020, 105, 449–459. [Google Scholar] [CrossRef]
- Bagg, A.; Braziel, R.M.; Arber, D.A.; Bijwaard, K.E.; Chu, A.Y. Immunoglobulin heavy chain gene analysis in lymphomas: A multi-center study demonstrating the heterogeneity of performance of polymerase chain reaction assays. J. Mol. Diagn. 2002, 4, 81–89. [Google Scholar] [CrossRef]
- Xochelli, A.; Baliakas, P.; Kavakiotis, I.; Agathangelidis, A.; Sutton, L.A.; Minga, E.; Ntoufa, S.; Tausch, E.; Yan, X.J.; Shanafelt, T.; et al. Chronic Lymphocytic Leukemia with Mutated IGHV4-34 Receptors: Shared and Distinct Immunogenetic Features and Clinical Outcomes. Clin. Cancer Res. 2017, 23, 5292–5301. [Google Scholar] [CrossRef]
- Hussmann, D.; Starnawska, A.; Kristensen, L.; Daugaard, I.; Thomsen, A.; Kjeldsen, T.E.; Hansen, C.S.; Bybjerg-Grauholm, J.; Johansen, K.D.; Ludvigsen, M.; et al. IGHV-associated methylation signatures more accurately predict clinical outcomes of chronic lymphocytic leukemia patients than IGHV mutation load. Haematologica 2022, 107, 877–886. [Google Scholar] [CrossRef]
- Visentin, A.; Bonaldi, L.; Rigolin, G.M.; Mauro, F.R.; Martines, A.; Frezzato, F.; Imbergamo, S.; Scomazzon, E.; Pravato, S.; Bardi, M.A.; et al. The combination of complex karyotype subtypes and IGHV mutational status identifies new prognostic and predictive groups in chronic lymphocytic leukaemia. Br. J. Cancer 2019, 121, 150–156. [Google Scholar] [CrossRef]
- Wierda, W.G.; Byrd, J.C.; Abramson, J.S.; Bhat, S.; Bociek, G.; Brander, D.; Brown, J.; Chanan-Khan, A.; Coutre, S.E.; Davis, R.S.; et al. Hairy Cell Leukemia, Version 2.2018, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc Netw. 2017, 15, 1414–1427. [Google Scholar] [CrossRef]
- Arons, E.; Roth, L.; Sapolsky, J.; Suntum, T.; Stetler-Stevenson, M.; Kreitman, R.J. Evidence of canonical somatic hypermutation in hairy cell leukemia. Blood 2011, 117, 4844–4851. [Google Scholar] [CrossRef] [PubMed]
- Forconi, F.; Sozzi, E.; Cencini, E.; Zaja, F.; Intermesoli, T.; Stelitano, C.; Rigacci, L.; Gherlinzoni, F.; Cantaffa, R.; Baraldi, A.; et al. Hairy cell leukemias with unmutated IGHV genes define the minor subset refractory to single-agent cladribine and with more aggressive behavior. Blood 2009, 114, 4696–4702. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xia, M.; Sun, X.; He, Z.; Hu, F.; Chen, L.; Bueso-Ramos, C.E.; Qiu, X.; Yin, C.C. IGK with conserved IGKV/IGKJ repertoire is expressed in acute myeloid leukemia and promotes leukemic cell migration. Oncotarget 2015, 6, 39062–39072. [Google Scholar] [CrossRef]
- Xia, M.; Wu, L.; Sun, X.; Han, X.; Yan, H.; Huang, J.; Zhang, Y.; Hu, Z.; Zu, Y.; Yin, C.C.; et al. Next-Generation Sequencing Revealed a Distinct Immunoglobulin Repertoire with Specific Mutation Hotspots in Acute Myeloid Leukemia. Biology 2022, 11, 161. [Google Scholar] [CrossRef] [PubMed]
- Doenecke, A.; Winnacker, E.L.; Hallek, M. Rapid amplification of cDNA ends (RACE) improves the PCR-based isolation of immunoglobulin variable region genes from murine and human lymphoma cells and cell lines. Leukemia 1997, 11, 1787–1792. [Google Scholar] [CrossRef] [PubMed]
- Alamyar, E.; Duroux, P.; Lefranc, M.P.; Giudicelli, V. IMGT((R)) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol. Biol. 2012, 882, 569–604. [Google Scholar] [CrossRef]
- Miqueu, P.; Guillet, M.; Degauque, N.; Dore, J.C.; Soulillou, J.P.; Brouard, S. Statistical analysis of CDR3 length distributions for the assessment of T and B cell repertoire biases. Mol. Immunol. 2007, 44, 1057–1064. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N.; Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, A.; Hoadley, K.; Triche, T.J., Jr.; Laird, P.W.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef]
- Patel, J.P.; Gonen, M.; Figueroa, M.E.; Fernandez, H.; Sun, Z.; Racevskis, J.; Van Vlierberghe, P.; Dolgalev, I.; Thomas, S.; Aminova, O.; et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 2012, 366, 1079–1089. [Google Scholar] [CrossRef]
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef] [PubMed]
- DiNardo, C.D.; Cortes, J.E. Mutations in AML: Prognostic and therapeutic implications. Hematol. Am. Soc. Hematol. Educ. Program. 2016, 2016, 348–355. [Google Scholar] [CrossRef]
- Kanyavuz, A.; Marey-Jarossay, A.; Lacroix-Desmazes, S.; Dimitrov, J.D. Breaking the law: Unconventional strategies for antibody diversification. Nat. Rev. Immunol. 2019, 19, 355–368. [Google Scholar] [CrossRef]
- van de Bovenkamp, F.S.; Hafkenscheid, L.; Rispens, T.; Rombouts, Y. The Emerging Importance of IgG Fab Glycosylation in Immunity. J. Immunol. 2016, 196, 1435–1441. [Google Scholar] [CrossRef] [PubMed]
- Hamza, N.; Hershberg, U.; Kallenberg, C.G.; Vissink, A.; Spijkervet, F.K.; Bootsma, H.; Kroese, F.G.; Bos, N.A. Ig gene analysis reveals altered selective pressures on Ig-producing cells in parotid glands of primary Sjogren’s syndrome patients. J. Immunol. 2015, 194, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Rombouts, Y.; Willemze, A.; van Beers, J.J.; Shi, J.; Kerkman, P.F.; van Toorn, L.; Janssen, G.M.; Zaldumbide, A.; Hoeben, R.C.; Pruijn, G.J.; et al. Extensive glycosylation of ACPA-IgG variable domains modulates binding to citrullinated antigens in rheumatoid arthritis. Ann. Rheum. Dis. 2016, 75, 578–585. [Google Scholar] [CrossRef]
- Choe, H.; Li, W.; Wright, P.L.; Vasilieva, N.; Venturi, M.; Huang, C.C.; Grundner, C.; Dorfman, T.; Zwick, M.B.; Wang, L.; et al. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell 2003, 114, 161–170. [Google Scholar] [CrossRef]
- Huang, C.C.; Venturi, M.; Majeed, S.; Moore, M.J.; Phogat, S.; Zhang, M.Y.; Dimitrov, D.S.; Hendrickson, W.A.; Robinson, J.; Sodroski, J.; et al. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc. Natl. Acad. Sci. USA 2004, 101, 2706–2711. [Google Scholar] [CrossRef] [PubMed]
- Stavnezer, J.; Schrader, C.E. IgH chain class switch recombination: Mechanism and regulation. J. Immunol. 2014, 193, 5370–5378. [Google Scholar] [CrossRef]
- Xu, Z.; Zan, H.; Pone, E.J.; Mai, T.; Casali, P. Immunoglobulin class-switch DNA recombination: Induction, targeting and beyond. Nat. Rev. Immunol. 2012, 12, 517–531. [Google Scholar] [CrossRef] [PubMed]
- Matthews, A.J.; Zheng, S.; DiMenna, L.J.; Chaudhuri, J. Regulation of immunoglobulin class-switch recombination: Choreography of noncoding transcription, targeted DNA deamination, and long-range DNA repair. Adv. Immunol. 2014, 122, 1–57. [Google Scholar] [CrossRef]
- Qiao, Q.; Wang, L.; Meng, F.L.; Hwang, J.K.; Alt, F.W.; Wu, H. AID recognizes structured DNA for class switch recombination. Mol. Cell 2017, 67, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Siriwardena, S.U.; Chen, K.; Bhagwat, A.S. Functions and malfunctions of mammalian DNA-cytosine deaminases. Chem. Rev. 2016, 116, 12688–12710. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Huang, J.; Mao, Y.; Liu, S.; Sun, X.; Zhu, X.; Ma, T.; Zhang, L.; Ji, J.; Zhang, Y.; et al. Immunoglobulin gene transcripts have distinct VHDJH recombination characteristics in human epithelial cancer cells. J. Biol. Chem. 2009, 284, 13610–13619. [Google Scholar] [CrossRef]
- Zan, H.; Cerutti, A.; Dramitinos, P.; Schaffer, A.; Li, Z.; Casali, P. Induction of Ig somatic hypermutation and class switching in a human monoclonal IgM+ IgD+ B cell line in vitro: Definition of the requirements and modalities of hypermutation. J. Immunol. 1999, 162, 3437–3447. [Google Scholar] [CrossRef]
- Budeus, B.; Kibler, A.; Kuppers, R. Human IgM-expressing memory B cells. Front. Immunol. 2023, 14, 1308378. [Google Scholar] [CrossRef] [PubMed]
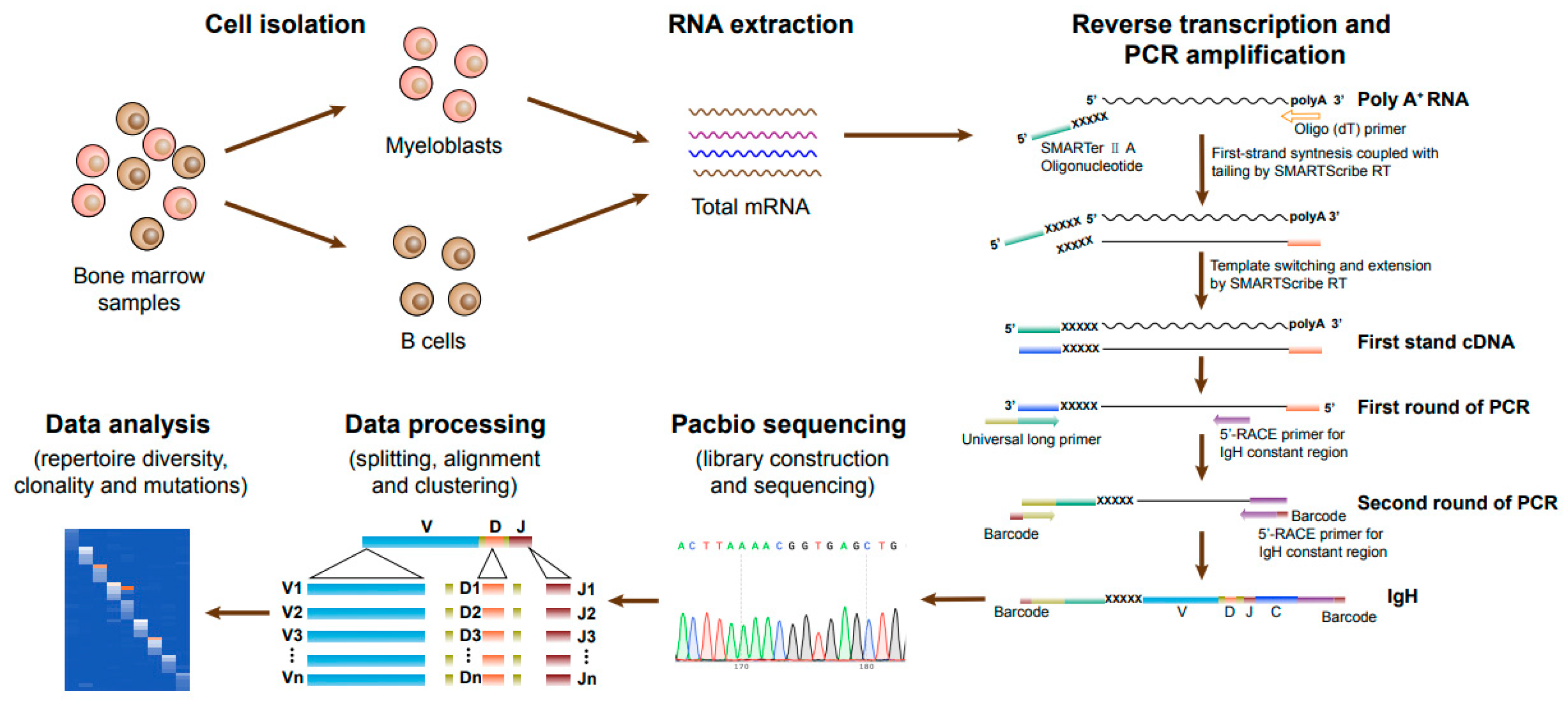



| Patient ID | Sex | Age (year) | 2022 WHO Classification | BM Blasts (%) | FLT3-ITD Mutation | NPM1 Mutation | EVI1 Overexpression | MLL-PTD | WT1 Mutation |
|---|---|---|---|---|---|---|---|---|---|
| AML1 | F | 40 | AML with NPM1 mutation | 49 | + | + | - | - | - |
| AML2 | F | 35 | AML with maturation | 55 | - | - | - | - | - |
| AML3 | F | 58 | AML with maturation | 52 | - | - | - | - | - |
| AML4 | F | 45 | AML with RUNX1::RUNX1T1 | 33 | - | - | - | - | + |
| AML5 | F | 51 | AML with maturation | 79 | - | - | + | + | - |
| AML6 | M | 23 | AML with RUNX1::RUNX1T1 | 25 | - | - | - | - | - |
| AML7 | M | 61 | AML with maturation | 53 | - | - | - | + | + |
| AML8 | M | 51 | AML with maturation | 72 | - | - | - | - | + |
| AML9 | M | 37 | AML with maturation | 86 | - | - | - | - | + |
| AML10 | M | 31 | AML with maturation | 63 | + | - | - | - | + |
| AML11 | M | 61 | AML with maturation | 41 | - | - | - | - | + |
| AML12 | F | 65 | AML with maturation | 57 | - | - | - | + | + |
| AML13 | M | 31 | AML with maturation | 58 | - | - | - | - | + |
| AML14 | M | 18 | Acute monocytic leukemia | 71 | + | - | - | + | + |
| AML15 | F | 31 | AML with MECOM rearrangement | 59 | - | - | + | - | + |
| AML16 | F | 38 | AML with NPM1 mutation | 53 | - | + | - | - | + |
| AML17 | F | 62 | Acute myelomonocytic leukemia | 85 | - | - | - | - | - |
| AML18 | F | 60 | Acute myelomonocytic leukemia | 44 | - | - | - | - | - |
| AML19 | M | 36 | AML with maturation | 41 | - | - | + | + | - |
| AML20 | M | 67 | AML with maturation | NA | - | - | + | + | - |
| AML21 | F | 54 | AML with KMT2A rearrangement | NA | - | - | - | - | - |
| AML22 | M | 83 | AML with maturation | 86 | + | + | - | - | - |
| AML23 | F | 60 | AML with maturation | 54 | - | + | - | - | + |
| AML24 | M | 52 | AML with NPM1 mutation | 41 | - | + | - | - | + |
| AML25 | F | 50 | APL with PML::RARA | 66 | - | - | - | - | + |
| AML26 | M | 54 | AML with maturation | 27 | - | - | - | + | + |
| AML27 | F | 35 | APL with PML::RARA | 79 | - | - | - | - | - |
| Patient ID | Sorted Cells | Igγ | Igμ | Igα | Igδ | Igε |
|---|---|---|---|---|---|---|
| AML1 | Myeloblasts | 4704 | NA | NA | NA | NA |
| B cells | 1356 | 2030 | 1148 | 2833 | NA | |
| AML2 | B cells | 1725 | 911 | NA | 2316 | NA |
| AML3 | Myeloblasts | NA | 1018 | 2680 | NA | 1058 |
| B cells | 3991 | 2416 | 3113 | 1481 | NA | |
| AML4 | B cells | 814 | 1401 | 317 | 1155 | NA |
| AML5 | Myeloblasts | 12,377 | NA | NA | NA | NA |
| B cells | 1877 | NA | 893 | NA | NA | |
| AML6 | Myeloblasts | 3036 | NA | NA | NA | NA |
| B cells | 2820 | 1343 | 4236 | 4545 | 244 | |
| AML7 | Myeloblasts | NA | NA | NA | NA | NA |
| B cells | 4425 | 1226 | 3202 | NA | NA | |
| AML8 | Myeloblasts | 3709 | 523 | 4365 | NA | NA |
| B cells | 9586 | NA | NA | NA | NA | |
| AML9 | Myeloblasts | NA | 5583 | 5848 | NA | NA |
| B cells | 457 | 1027 | 1737 | 3463 | NA | |
| AML10 | Myeloblasts | 4853 | 1355 | 2625 | NA | NA |
| B cells | 2179 | 1127 | 3120 | 2095 | NA | |
| AML11 | Myeloblasts | 3724 | NA | NA | NA | NA |
| AML12 | Myeloblasts | 7250 | NA | NA | NA | NA |
| B cells | 5628 | 2218 | 3104 | NA | 666 | |
| AML13 | Myeloblasts | NA | 880 | 1550 | 1651 | NA |
| B cells | NA | 1576 | 894 | 2123 | 1499 | |
| AML14 | Myeloblasts | NA | 293 | 2155 | 295 | NA |
| B cells | NA | 1402 | NA | 1685 | NA | |
| AML15 | Myeloblasts | NA | NA | 1085 | NA | NA |
| B cells | NA | 871 | 477 | 1681 | NA | |
| AML16 | Myeloblasts | 1550 | 865 | 2981 | 2876 | NA |
| B cells | 522 | 1921 | 925 | 2166 | NA | |
| AML17 | Myeloblasts | 1472 | 864 | 1374 | 3181 | NA |
| B cells | 617 | 504 | 2822 | 854 | 5632 | |
| Total | 78,672 | 31,354 | 50,651 | 34,400 | 9099 |
| Patient ID | Sorted Cells | Igγ | Igμ | Igα | Igδ | Igε |
|---|---|---|---|---|---|---|
| AML2 | Myeloblasts | 1752 | NA | 1775 | NA | NA |
| AML4 | Myeloblasts | 3690 | 1640 | 1478 | NA | N/A |
| Monocytes | 4130 | NA | 317 | NA | NA | |
| AML5 | Myeloblasts | 4643 | NA | NA | NA | NA |
| AML8 | Myeloblasts | 4611 | NA | 2775 | NA | NA |
| Monocytes | NA | NA | 6112 | NA | NA | |
| AML9 | Myeloblasts | NA | NA | 6493 | 6908 | NA |
| Monocytes | 9215 | NA | NA | NA | NA | |
| AML10 | Myeloblasts | 3486 | NA | 4114 | 61 | NA |
| Monocytes | 5912 | NA | NA | NA | NA | |
| AML12 | Myeloblasts | 7804 | NA | NA | NA | NA |
| Monocytes | 17,073 | NA | NA | NA | NA | |
| AML13 | Myeloblasts | NA | 468 | 3198 | NA | NA |
| Monocytes | NA | 266 | 1757 | 3266 | NA | |
| AML15 | Myeloblasts | NA | NA | 6865 | NA | NA |
| Monocytes | NA | 990 | 4010 | 65 | NA | |
| AML16 | Monocytes | 4771 | NA | NA | NA | NA |
| AML17 | Myeloblasts | 4170 | NA | NA | NA | 4886 |
| Total | 71,257 | 3364 | 38,894 | 10,300 | 4886 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, H.; Wu, L.; Wang, P.; Xia, M.; Shi, Z.; Huang, X.; Yin, S.; Jiang, Q.; Yin, C.C.; Zhao, X.; et al. A Comparative Analysis of the Immunoglobulin Repertoire in Leukemia Cells and B Cells in Chinese Acute Myeloid Leukemia by High-Throughput Sequencing. Biology 2024, 13, 613. https://doi.org/10.3390/biology13080613
Yan H, Wu L, Wang P, Xia M, Shi Z, Huang X, Yin S, Jiang Q, Yin CC, Zhao X, et al. A Comparative Analysis of the Immunoglobulin Repertoire in Leukemia Cells and B Cells in Chinese Acute Myeloid Leukemia by High-Throughput Sequencing. Biology. 2024; 13(8):613. https://doi.org/10.3390/biology13080613
Chicago/Turabian StyleYan, Huige, Lina Wu, Pingzhang Wang, Miaoran Xia, Zhan Shi, Xinmei Huang, Sha Yin, Qian Jiang, C. Cameron Yin, Xiangyu Zhao, and et al. 2024. "A Comparative Analysis of the Immunoglobulin Repertoire in Leukemia Cells and B Cells in Chinese Acute Myeloid Leukemia by High-Throughput Sequencing" Biology 13, no. 8: 613. https://doi.org/10.3390/biology13080613
APA StyleYan, H., Wu, L., Wang, P., Xia, M., Shi, Z., Huang, X., Yin, S., Jiang, Q., Yin, C. C., Zhao, X., & Qiu, X. (2024). A Comparative Analysis of the Immunoglobulin Repertoire in Leukemia Cells and B Cells in Chinese Acute Myeloid Leukemia by High-Throughput Sequencing. Biology, 13(8), 613. https://doi.org/10.3390/biology13080613






