Cell-Free Extracts of the Ginseng Soil Bacterium Pseudomonas plecoglossicida Promote Suppression of Resistance of American Ginseng (Panax quinquefolius) to Root Rot Caused by Ilyonectria mors-panacis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Biological Materials
2.2. Bacterial Isolation from Soil and Identification of Pseudomonas plecoglossicida
2.3. Genome Sequencing and Assembly of Pseudomonas plecoglossicida
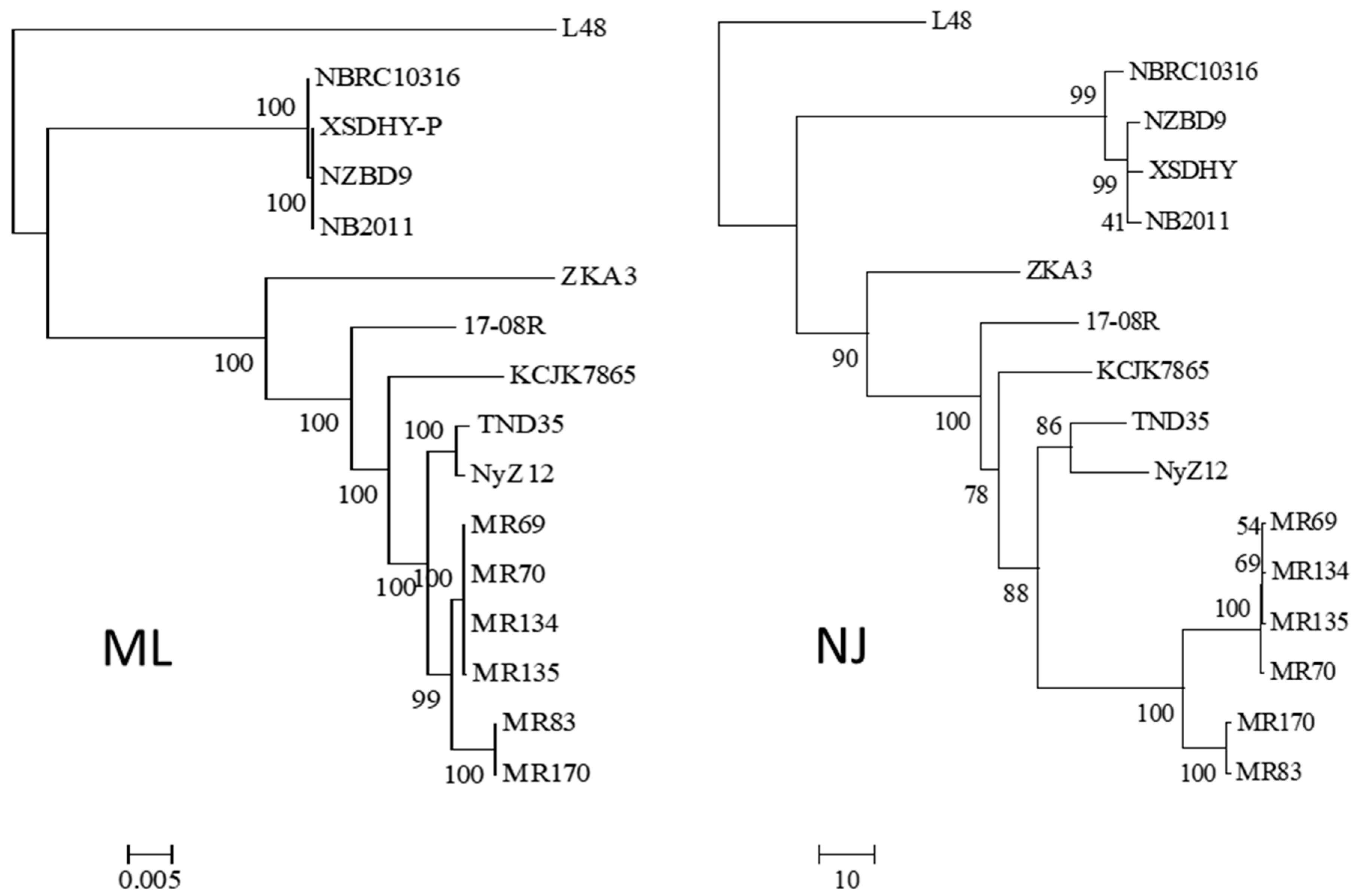
2.4. Phylogeny of Pseudomonas plecoglossicida
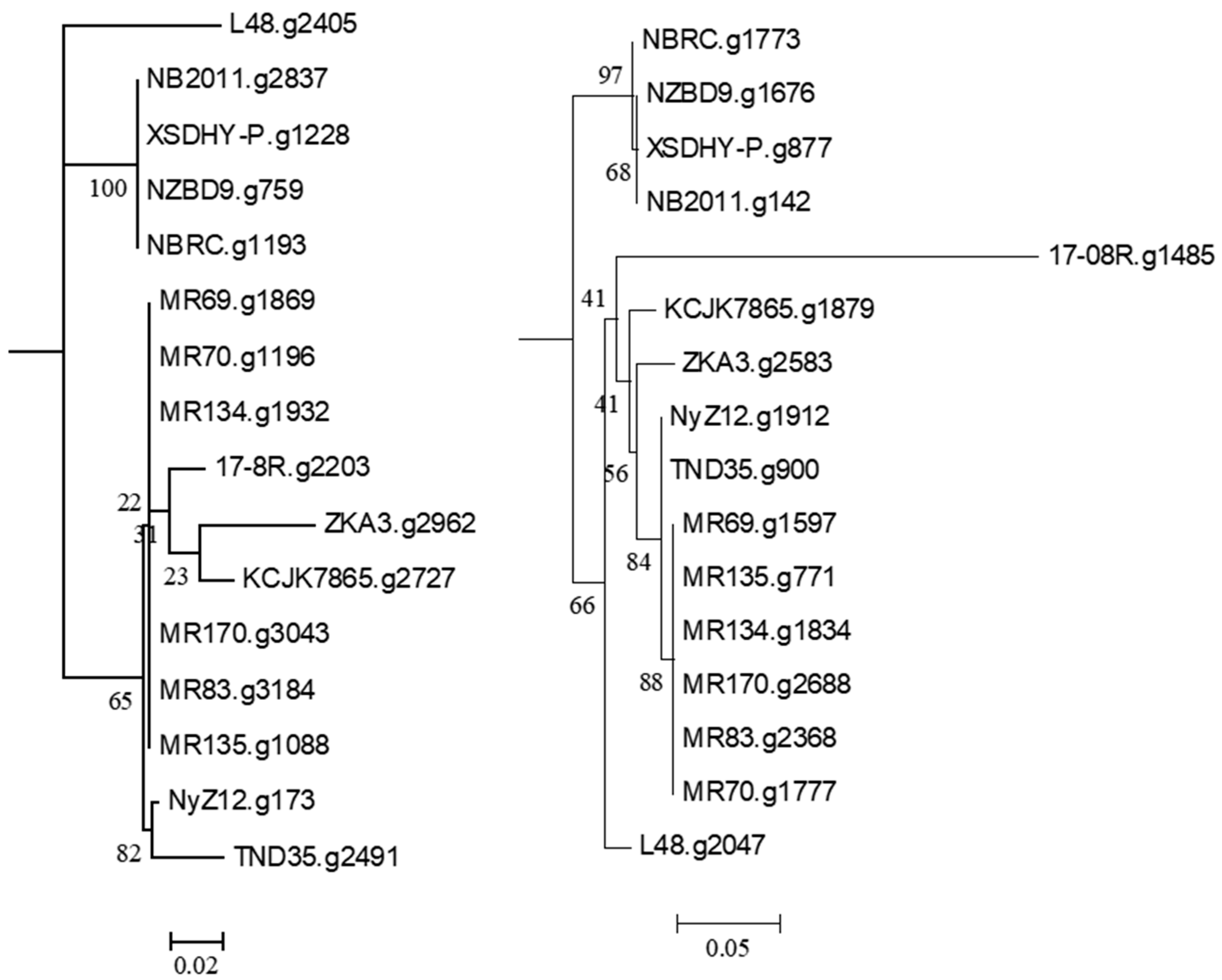
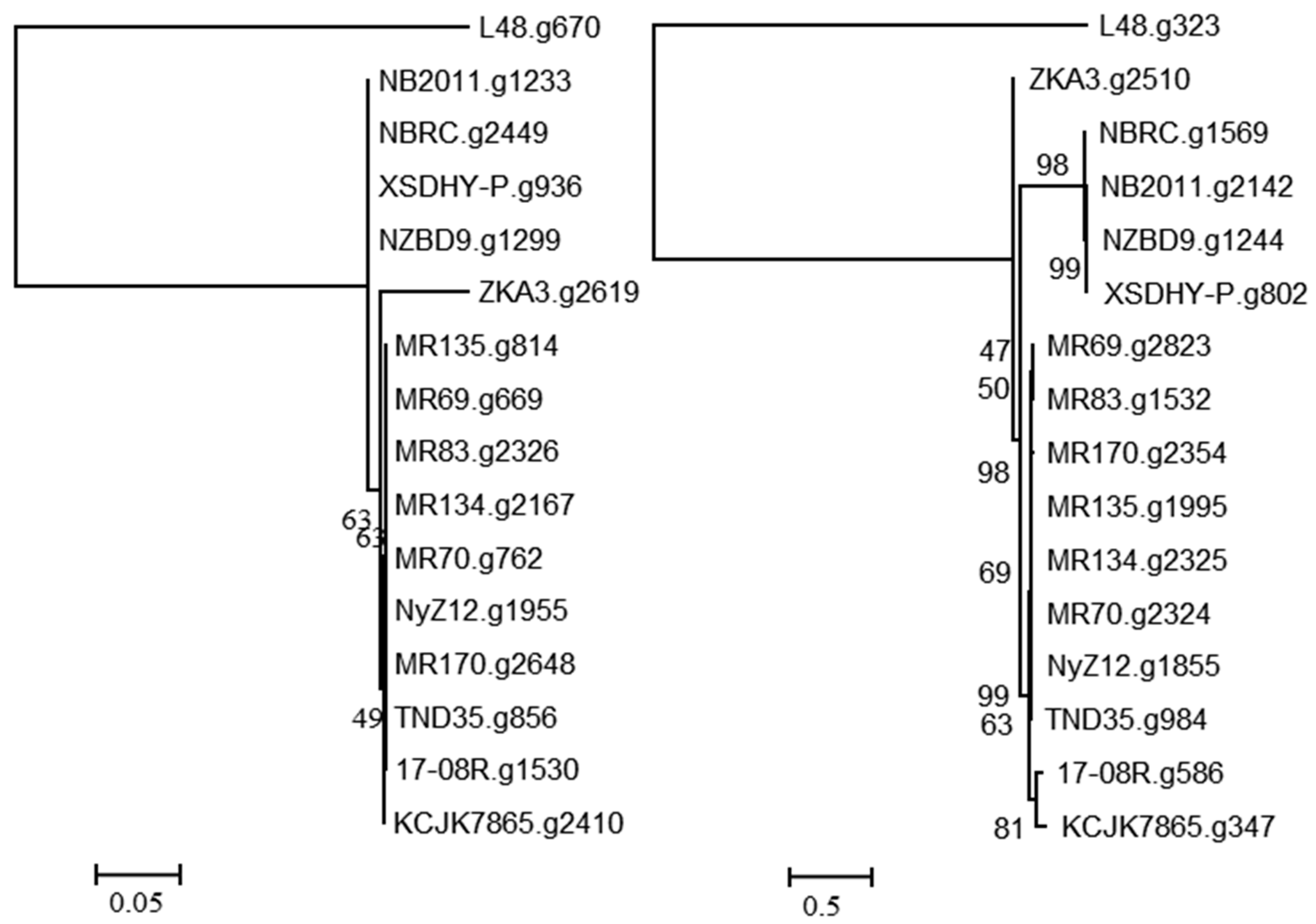
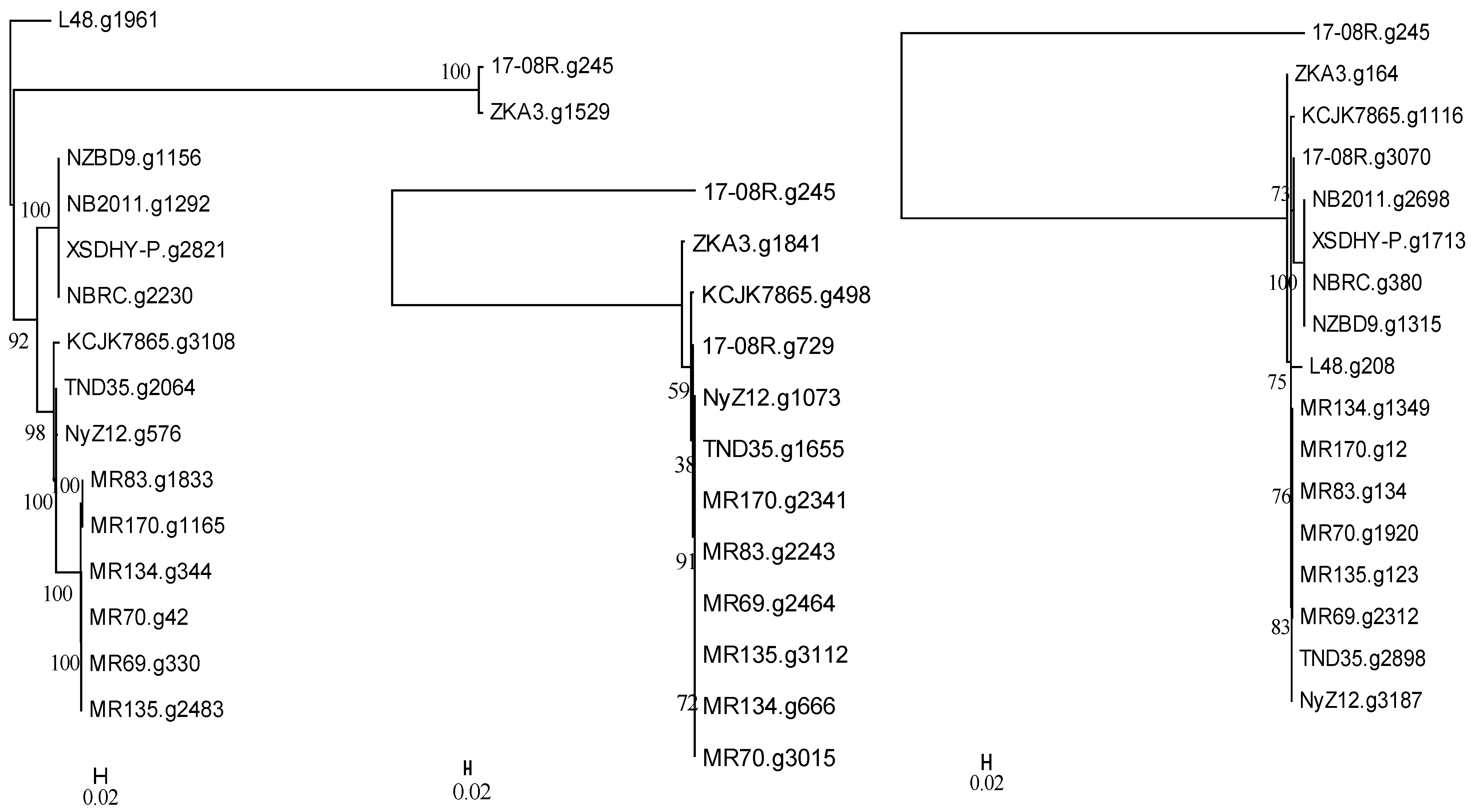
2.5. Selected Gene Predictions from the Genomes of Pseudomonas plecoglossicida
2.6. Ginseng Root Extract
2.7. Bacterial Growth and Ginsenoside Transformation
2.8. Detached Root Assay
2.9. Statistical Analysis
3. Results
3.1. Identification and Phylogenetic Relationships of Pseudomonas plecoglossicida Isolates
3.2. Genomic Analysis of P. plecoglossicida Isolates
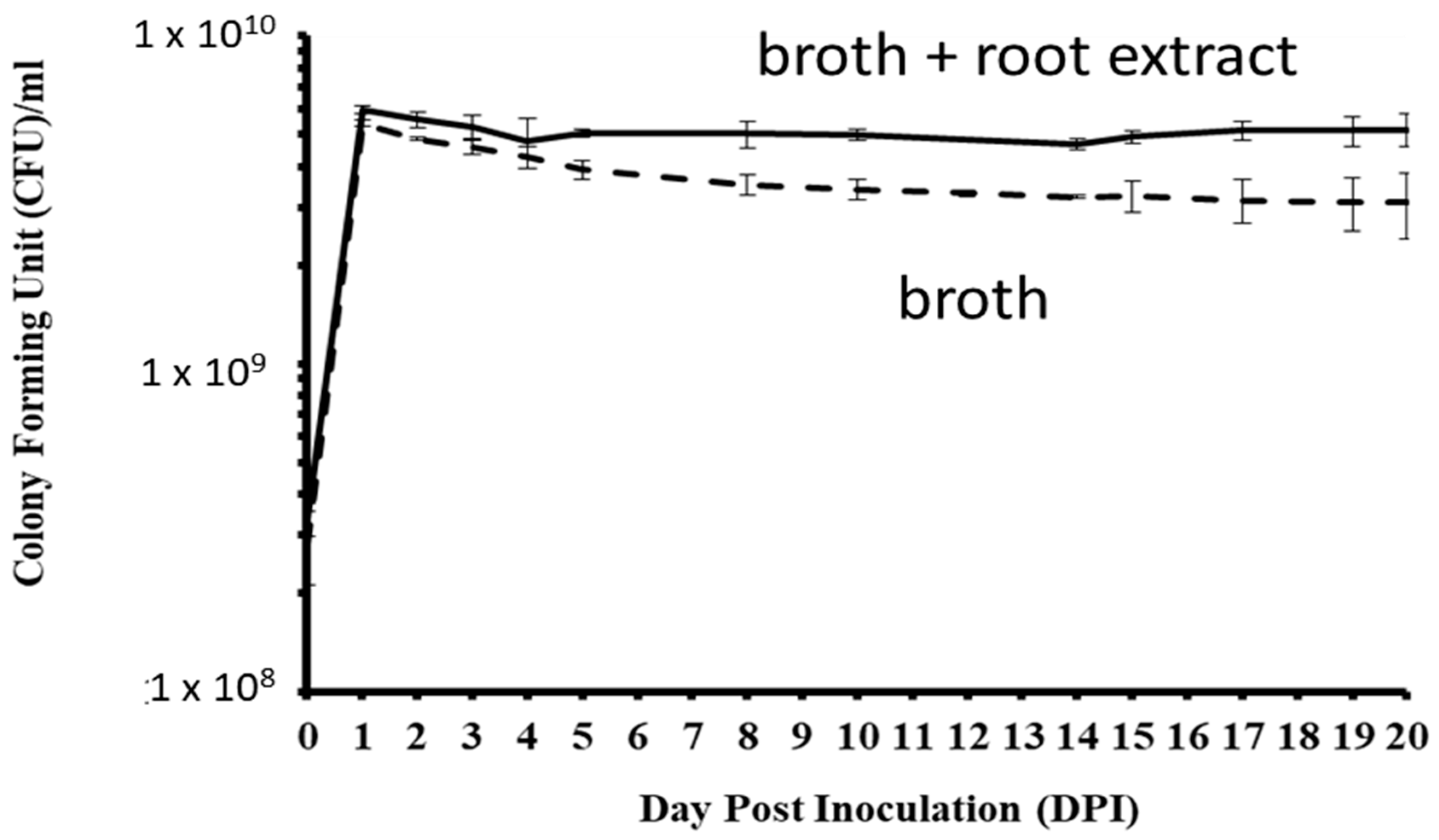
3.3. Pseudomonas Plecoglossicida Growth with Ginseng Root Extract
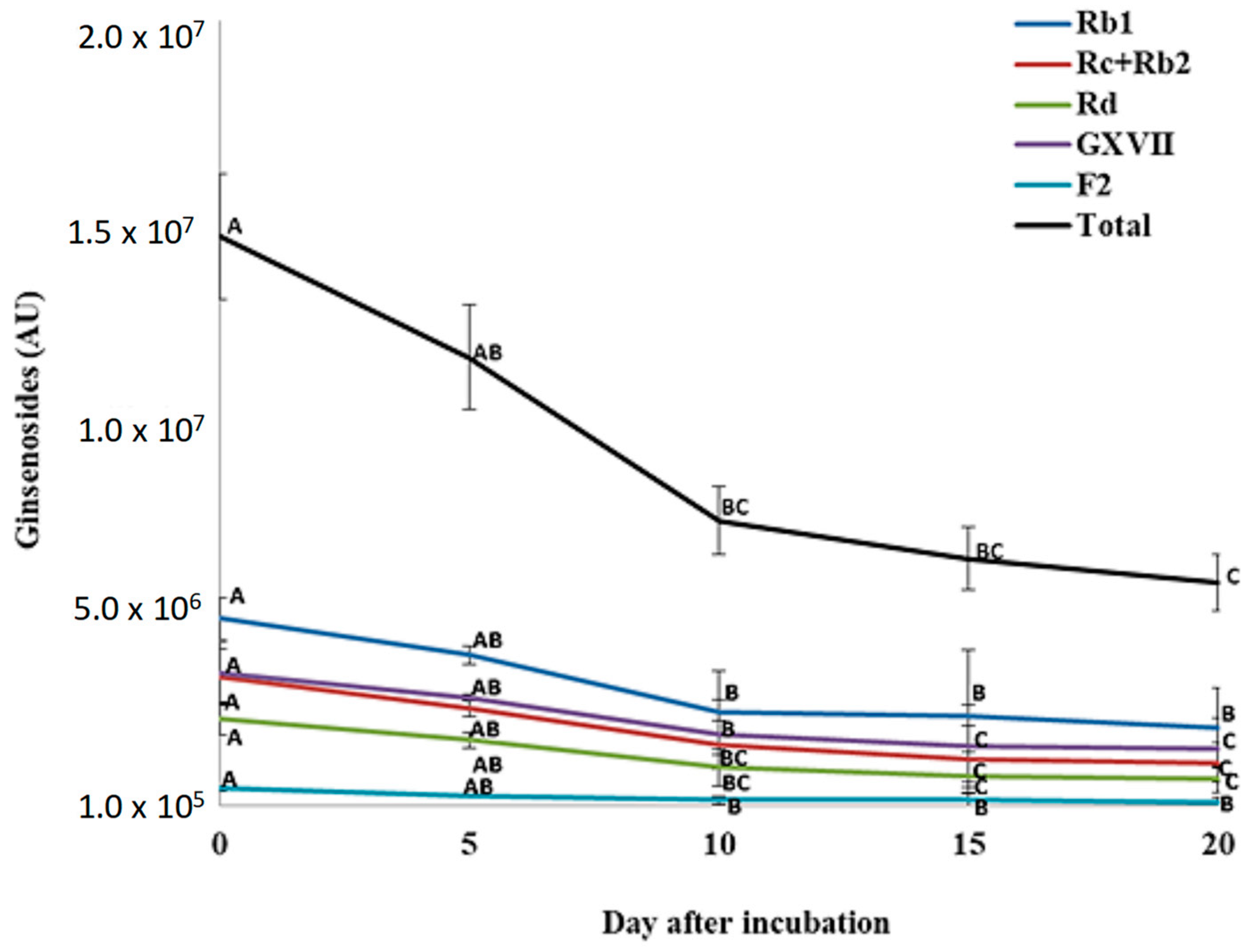
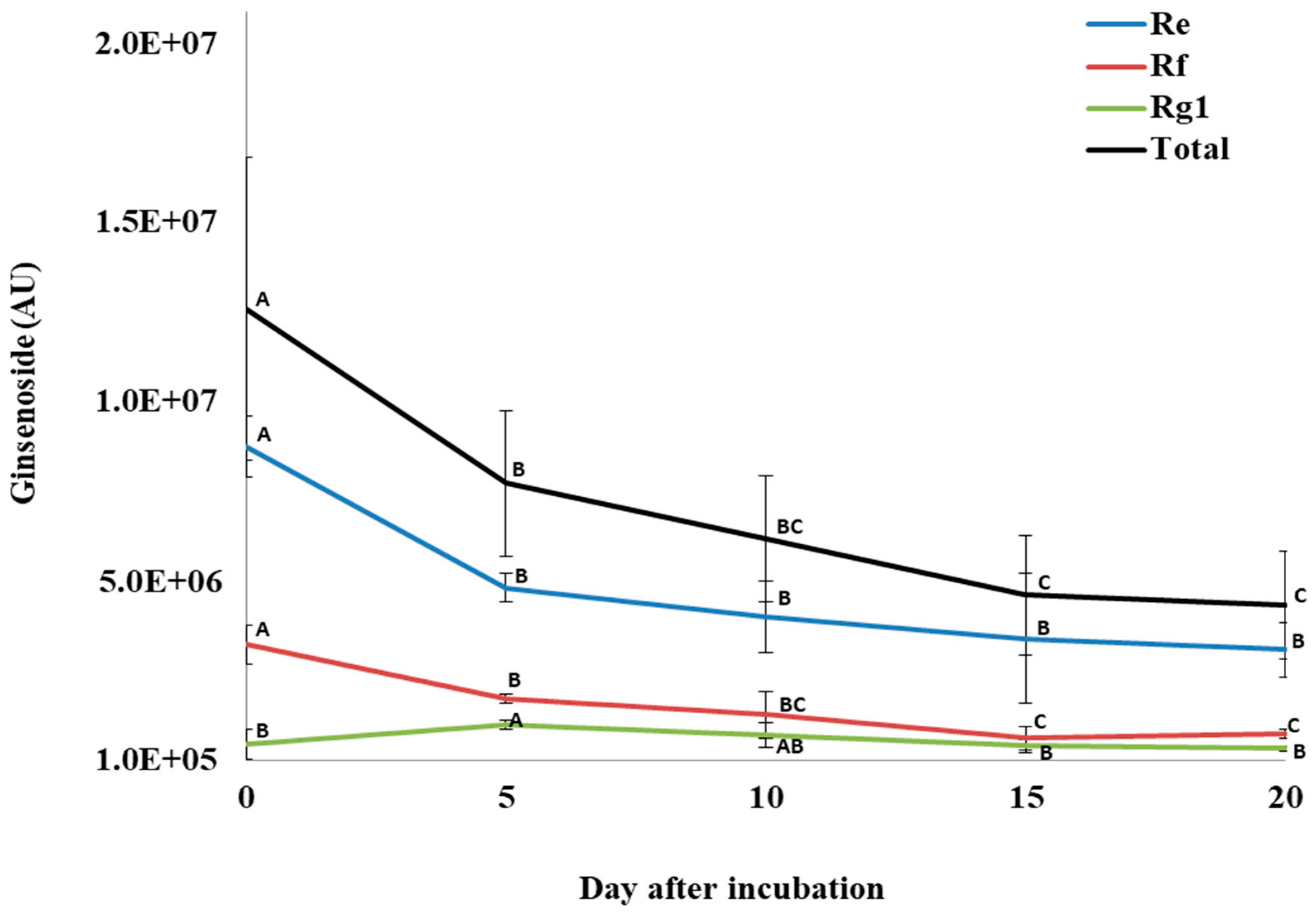
3.4. Changes in Ginsenosides during P. plecoglossicida Growth with Ginseng Root Extract
3.5. Effect of Ginseng Root Extract Incubated with P. plecoglossicida on Root Rot Disease
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Westerveld, S.M.; Shi, F. The history, etiology, and management of ginseng replant disease: A Canadian perspective in review. Can. J. Plant Sci. 2021, 101, 886–901. [Google Scholar] [CrossRef]
- Goodwin, P.H. The rhizosphere microbiome of ginseng. Microorganisms 2022, 10, 1152. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.; Kim, Y.; Hoang, V.; Subramaniyam, S.; Kang, J.; Kang, C.; Yang, D. Bacterial diversity and community structure in Korean ginseng field soil are shifted by cultivation time. PLoS ONE 2016, 11, e0155055. [Google Scholar] [CrossRef]
- Dong, L.; Xu, J.; Li, Y.; Fang, H.; Niu, W.; Li, X.; Zhang, Y.; Ding, W.; Chen, S. Manipulation of microbial community in the rhizosphere alleviates the replanting issues in Panax ginseng. Soil Biol. Biochem. 2018, 125, 64–74. [Google Scholar] [CrossRef]
- Zhang, J.; Fan, S.; Qin, J.; Dai, J.; Zhao, F.; Gao, L.; Lian, X.; Shang, W.; Xu, X.; Hu, X. Changes in the microbiome in the soil of an American ginseng continuous plantation. Front. Plant Sci. 2020, 11, 572199. [Google Scholar] [CrossRef] [PubMed]
- Brimecombe, M.J.; De Leij, F.A.; Lynch, J.M. The effect of root exudates on rhizosphere microbial populations. In The Rhizosphere; CRC Press: London, UK, 2000; pp. 111–156. [Google Scholar]
- Nicol, R.W.; Yousef, L.; Traquair, J.A.; Bernards, M.A. Ginsenosides stimulate the growth of soilborne pathogens of American ginseng. Phytochemistry 2003, 64, 257–264. [Google Scholar] [CrossRef]
- Taira, S.; Ikeda, R.; Yokota, N.; Osaka, I.; Sakamoto, M.; Mitsuro, K.; Sahashi, Y. Mass spectrometric imaging of ginsenosides localization in Panax ginseng root. Am. J. Chin. Med. 2010, 38, 485–493. [Google Scholar] [CrossRef]
- Miao, X.; Wang, E.; Zhou, Y.; Zhan, Y.; Yan, N.; Chen, C.; Li, Q. Effect of ginsenosides on microbial community and enzyme activity in continuous cropping soil of ginseng. Front. Microbiol. 2023, 14, 1060282. [Google Scholar] [CrossRef]
- Luo, L.F.; Yang, L.; Yan, Z.X.; Jiang, B.B.; Li, S.; Huang, H.C.; Liu, Y.X.; Zhu, S.S.; Yang, M. Ginsenosides in root exudates of Panax notoginseng drive the change of soil microbiota through carbon source different utilization. Plant Soil. 2020, 455, 139–153. [Google Scholar] [CrossRef]
- Eom, S.J.; Kim, K.T.; Paik, H.K. Microbial bioconversion of ginsenosides in Panax ginseng and their improved bioactivities. Food Rev. Int. 2018, 34, 698–712. [Google Scholar] [CrossRef]
- Wang, L.; An, D.S.; Kim, S.G.; Jin, F.X.; Lee, S.T.; Im, W.T. Rhodanobacter panaciterrae sp. nov., a bacterium with ginsenoside-converting activity isolated from soil of a ginseng field. Int. J. Syst. Evol. Microbiol. 2011, 61, 3028–3032. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.M.; Kim, J.; Seo, J.H.; Park, J.S.; Kim, D.H.; Kim, B.G. Identification and characterization of the Rhizobium sp. strain GIN611 glycoside oxidoreductase resulting in the deglycosylation of ginsenosides. Appl. Environ. Microbiol. 2012, 78, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Behdarvandi, B.; Goodwin, P.H. Effect of soil and root extracts on the innate immune response of American ginseng (Panax quinquefolius) to root rot caused by Ilyonectria mors-panacis. Plants 2023, 12, 2540. [Google Scholar] [CrossRef]
- Behdarvandi, B.; Hsiang, T.; Valliani, M.; Goodwin, P.H. Differences in saprophytic growth, virulence, genomes, and secretomes of Ilyonectria robusta and I. mors-panacis isolates from roots of American ginseng (Panax quinquefolius). Horticulturae 2023, 9, 713. [Google Scholar] [CrossRef]
- Barba, J.L.R.; Maldonado, A.; Siaz, R.J. Small-scale total DNA extraction from bacteria and yeast for PCR applications. Anal. Biochem. 2005, 347, 333–335. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; Wiley & Sons: Chichester, UK, 1991; pp. 115–175. [Google Scholar]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Clarke, L.; Millar, B.C.; Moore, J.E. Extraction of genomic DNA from Pseudomonas aeruginosa: A comparison of three methods. Br. J. Biomed. Sci. 2003, 60, 34–35. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K. BLAST plus: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Wu, X.; Monchy, S.; Taghavi, S.; Zhu, W.; Ramos, J.; van der Lelie, D. Comparative genomics and functional analysis of niche-specific adaptation in Pseudomonas putida. FEMS Microbiol. Rev. 2011, 35, 299–323. [Google Scholar] [CrossRef]
- Mulet, M.; Gomila, M.; Lemaitra, B.; Lalucat, J.; Valdes, E.G. Taxonomic characterisation of Pseudomonas strain L48 and formal proposal of Pseudomonas entomophila sp. nov. Syst. Appl. Microbiol. 2012, 35, 145–149. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, 636–641. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Paradis, E.; Schliep, K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2018, 35, 526–528. [Google Scholar] [CrossRef]
- Dai, J.; Orsat, V. Extraction of ginsenosides from American ginseng (Panax quinquefolium L.) root. Int. J. Food Eng. 2010, 6, 3. [Google Scholar] [CrossRef]
- Cui, L.; Wu, S.Q.; Zhao, C.A.; Yin, C.R. Microbial conversion of major ginsenosides in ginseng total saponins by Platycodon grandiflorum endophytes. J. Ginseng Res. 2016, 40, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Suárez, I.D.S.; Valliani, M.; Hsiang, T.; Goodwin, P.H. Decay of root debris after harvesting American ginseng (Panax quinquefolius) and changes in soil chemistry and microbiology. Soil Syst. 2023, 7, 108. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, J.; Wang, T.; Li, Z.; Liang, H.; Jiang, C.; Tang, H.; Gao, J.; Jiang, Y.; Chen, C. The novel Pseudomonas thivervalensis strain JI6 promotes growth and controls rusty root rot disease in Panax ginseng. Biol. Control. 2024, 193, 105514. [Google Scholar] [CrossRef]
- Shen, L.; Zhu, G.; Guo, S.; Li, X.; Xiao, S.; Xu, J.; Chen, S. Isolation of a Pseudomonas putida strain that degrades p-hydroxybenzoic acid from the soil of a Panax ginseng field. Res. Sq. 2020, preprint. [Google Scholar] [CrossRef]
- Park, K.H.; Lee, C.Y.; Son, H.J. Mechanism of insoluble phosphate solubilization by Pseudomonas fluorescens RAF15 isolated from ginseng rhizosphere and its plant growth-promoting activities. Lett. Appl. Microbiol. 2009, 49, 222–228. [Google Scholar] [CrossRef]
- Nishimori, E.; Kita-Tsukamoto, K.; Wakabayashi, H. Pseudomonas plecoglossicida sp. nov., the causative agent of bacterial haemorrhagic ascites of ayu, Plecoglossus altivelis. Int. J. Syst. Evol. Microbiol. 2000, 50, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Hussein, K.A.; Joo, J.H. Isolation and detection of genes responsible for pyoverdines biosynthesis in Pseudomonas putida KNUK9. Korean J. Soil Sci. Fert. 2015, 48, 119–124. [Google Scholar] [CrossRef]
- Mao, Z.; Li, M.; Chen, J. Draft genome sequence of Pseudomonas plecoglossicida strain NB2011, the causative agent of white nodules in large yellow croaker (Larimichthys crocea). Genome Announc. 2013, 1, e00586-13. [Google Scholar] [CrossRef]
- Tao, Z.; Wang, G.; Zhou, S. Complete genome sequence of Pseudomonas plecoglossicida XSDHY-P, a strain that is pathogenic for the marine fish Larimichthys crocea. Microbiol. Resour. Announc. 2018, 7, e01228-18. [Google Scholar] [CrossRef]
- Kyrpides, N.; Huntemann, M.; Han, J.; Chen, A.; Mavromatis, K.; Markowitz, V.; Palaniappan, K.; Ivanova, N.; Schaumberg, A.; Pati, A.; et al. Pseudomonas plecoglossicida NBRC 103162; Unpublished Direct Submission; National Center for Biotechnology Information: Bethesda, MD, USA, 2014. [Google Scholar]
- Li, X.; Li, C.Z.; Mao, L.Q.; Yan, D.Z.; Zhou, N.Y. Complete genome sequence of the cyclohexylamine-degrading Pseudomonas plecoglossicida NyZ12. Biotechnology 2015, 199, 29–30. [Google Scholar] [CrossRef] [PubMed]
- Raman, G.; Sakthivel, N.; Park, S. Draft genome sequence of a novel nicotine-degrading bacterium, Pseudomonas plecoglossicida TND35. Genome Announc. 2015, 3, 10–1128. [Google Scholar] [CrossRef]
- Hatzinikolaou, D. Genome Sequencing of Biomass-Degrading Isolates from Greek Habitats; Unpublished Direct Submission; National Center for Biotechnology Information: Bethesda, MD, USA, 2017. [Google Scholar]
- Adelowo, O.; Vollmers, J.; Mäusezahl, I.; Kaster, A.; Müller, J. Detection of the carbapenemase gene blaVIM-5 in members of the Pseudomonas putida group isolated from polluted Nigerian wetlands. Sci. Rep. 2018, 8, 15116. [Google Scholar] [CrossRef]
- Jeong, K.C. Genome of Pseudomonas plecoglossicida KCJK7865; Unpublished Direct Submission; National Center of Biotechnology Information: Bethesda, MD, USA, 2018. [Google Scholar]
- Beacham, I.R. Periplasmic enzymes in gram-negative bacteria. Int. J. Biochem. 1979, 10, 877–883. [Google Scholar] [CrossRef]
- Ichikawa, T.; Sugita, T.; Wang, L.; Yokoyama, K.; Nishimura, K.; Akemi Nishikawa, A. Phenotypic switching and -N-acetylhexosaminidase activity of the pathogenic yeast Trichosporon asahii. Microbiol. Immunol. 2004, 48, 237–242. [Google Scholar] [CrossRef]
- Kim, E.M.; Seo, J.H.; Kim, J.; Park, J.S.; Kim, D.H.; Kim, B.G. Production of ginsenoside aglycons and Rb1 deglycosylation pathway profiling by HPLC and ESI-MS/MS using Sphingobacterium multivorum GIN723. Appl. Microbiol. Biotechnol. 2013, 97, 8031–8039. [Google Scholar] [CrossRef]
- Sarris, P.F.; Zoumadakis, C.; Panopolous, N.J.; Scoulica, E.V. Distribution of the putative type VI secretion system core genes in Klebsiella spp. Infect. Genet. Evol. 2011, 11, 157–166. [Google Scholar] [CrossRef]
- Martens, E.C.; Koropatkin, N.M.; Smith, T.J.; Gordon, J.I. Complex glycan catabolism by the human gut microbiota: The bacteroidetes SUS-like paradigm. J. Biol. Chem. 2009, 284, 24673–24677. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Liu, F.; Sun, L.; Liu, J.; Wang, M.; Chen, X.; Xu, X.; Ma, R.; Feng, K.; Jiang, R. Proteomic analysis of amino acid metabolism differences between wild and cultivated Panax ginseng. J. Ginseng Res. 2016, 40, 113–120. [Google Scholar] [CrossRef]
- Court, W.A.; Reynolds, L.B.; Hendel, J.G. Influence of root age on the concentration of ginsenosides of American ginseng (Panax quinquefolium). Can. J. Plant Sci. 1996, 76, 853–855. [Google Scholar] [CrossRef]
- Shi, Z.Y.; Zeng, J.Z.; Wong, A.S.T. Chemical structures and pharmacological profiles of ginseng saponins. Molecules 2019, 24, 2443. [Google Scholar] [CrossRef] [PubMed]
- An, D.S.; Cui, C.H.; Lee, H.G.; Wang, L.; Kim, S.C.; Lee, S.T.; Jin, F.; Yu, H.; Chin, Y.W.; Lee, H.K.; et al. Identification and characterization of a novel Terrabacter ginsenosidimutans sp. nov. beta-glucosidase that transforms ginsenoside Rb1 into the rare gypenosides XVII and LXXV. Appl. Environ. Microbiol. 2010, 76, 5827–5836. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Lee, W.J.; Gebru, Y.A.; Upadhyaya, J.; Ko, S.R.; Kim, Y.H.; Kim, M.K. Production of minor ginsenosides CK and CY from naturally occurring major ginsenosides using crude β-glucosidase preparation from submerged culture of Fomitella fraxinea. Molecules 2021, 26, 4820. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, X.; Xu, Y.; Mei, X.; Jiang, B.; Liao, J.; Yin, Z.; Zheng, J.; Zhao, Z.; Fan, L.; et al. Autotoxic ginsenosides in the rhizosphere contribute to the replant failure of Panax notoginseng. PLoS ONE 2015, 10, e0118555. [Google Scholar] [CrossRef] [PubMed]
- Jiao, X.L.; Bi, X.B.; Zhang, X.S.; Gao, W.W. Autotoxic effect of ginsenoside extracts on growth of American ginseng in different medium. China J. Mat. Med. 2015, 40, 1433–1438. [Google Scholar]
- Cipollini, D.; Rigsby, C.M.; Barto, E.K. Microbes as targets and mediators of allelopathy in plants. J. Chem. Ecol. 2012, 38, 714–727. [Google Scholar] [CrossRef]
- Pélissier, R.; Violle, C.; Morel, J.B. Plant immunity: Good fences make good neighbors? Curr. Opin. Plant Biol. 2021, 62, 102045. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Kong, C.H.; Hu, F.; Xu, X.H. Allantoin involved in species interactions with rice and other organisms in paddy soil. Plant Soil. 2007, 296, 43–51. [Google Scholar] [CrossRef]
- Takagi, H.; Ishiga, Y.; Watanabe, S.; Konishi, T.; Egusa, M.; Akiyoshi, N.; Matsuura, T.; Mori, I.C.; Hirayama, T.; Kaminaka, H.; et al. Allantoin, a stress-related purine metabolite, can activate jasmonate signaling in a MYC2-regulated and abscisic acid-dependent manner. J. Exp. Bot. 2016, 67, 2519–2532. [Google Scholar] [CrossRef]
| Organism | Isolate | Assembly Accession | Location | Source | Source Description |
|---|---|---|---|---|---|
| P. plecoglossicida | NZBD9 | GCA_002814195.1 | Ningde/China | fish | Larimichthys crocea |
| P. plecoglossicida | XSDHY-P | GCA_003391255.1 | Ningbo/China | fish | Larimichthys polyactis |
| P. plecoglossicida | NB2011 | GCA_000412715.1 | East coast/China | fish | Larimichthys polyactis |
| P. plecoglossicida | NBRC10316 | GCA_000730665.1 | Tokushima Pref./Japan | fish | Plecoglossus altivelis |
| P. plecoglossicida | NyZ12 | GCA_000831585.1 | Wuhan/China | soil | Cyclohexylamine-degrading bacterium |
| P. plecoglossicida | KCJK7865 | GCA_003062165.1 | Florida/USA | soil | Not determined |
| P. plecoglossicida | ZKA3 | GCA_003633555.1 | Zakynthos/Greece | soil | Biomass-degrading bacterium |
| P. plecoglossicida | TND35 | GCA_000764405.1 | Tamil Nadu/India | soil | Nicotine-degrading bacterium |
| P. plecoglossicida | MR69 | GCA_002864775.1 | Ibadan/Nigeria | soil | Polluted wetlands |
| P. plecoglossicida | MR135 | GCA_002864795.1 | Ibadan/Nigeria | soil | Polluted wetlands |
| P. plecoglossicida | MR170 | GCA_002864845.1 | Ibadan/Nigeria | soil | Polluted wetlands |
| P. plecoglossicida | MR83 | GCA_002864865.1 | Ibadan/Nigeria | soil | Polluted wetlands |
| P. plecoglossicida | MR134 | GCA_002864885.1 | Ibadan/Nigeria | soil | Polluted wetlands |
| P. plecoglossicida | MR70 | GCA_002864905.1 | Ibadan/Nigeria | soil | Polluted wetlands |
| P. plecoglossicida | 17-08R | PRJNA1153425 | Simcoe/Canada | soil | 3-year post-harvest ginseng soil |
| P. entomophila | L48 | GCA_000026105.1 | Western Guadeloupe | fruit fly | Drosophila melanogaster |
| Isolate | Soil Year after Harvest | Colony Size a | Colony Color b | Lesion Size (cm2) |
|---|---|---|---|---|
| 17-06R | 3 | very small | creamy to colorless | 0.15 E |
| 17-07R | 3 | small | creamy | 0.19 CDE |
| 17-08R | 3 | small | creamy to yellow | 0.27 A |
| 17-09R | 3 | small | orange | 0.16 CDE |
| 17-10R | 25 | large | light brown to gray | 0.21 BCD |
| 17-11R | 25 | very large | whitish | 0.22 ABC |
| 17-12R | 5 | large | creamy | 0.19 CDE |
| 17-13R | 1 | small | white | 0.18 CDE |
| 17-14R | 0.08 | very small | white | 0.16 DE |
| 17-15R | 0.08 | large | white | 0.16 E |
| 17-16R | 1 | large | creamy | 0.25 AB |
| 17-17R | 10 | large | white–creamy centre | 0.19 CDE |
| 17-18R | 10 | large | orange | 0.19 CDE |
| Control | - | - | - | 0.18 CDE |
| Media | P. plecoglossicida 17-08R | Lesion Size (cm2) |
|---|---|---|
| LL broth | − | 0.16 B |
| LLG broth | − | 0.18 B |
| LL broth | + | 0.17 B |
| LLG broth | + | 0.23 A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goodwin, P.H.; Hsiang, T. Cell-Free Extracts of the Ginseng Soil Bacterium Pseudomonas plecoglossicida Promote Suppression of Resistance of American Ginseng (Panax quinquefolius) to Root Rot Caused by Ilyonectria mors-panacis. Biology 2024, 13, 671. https://doi.org/10.3390/biology13090671
Goodwin PH, Hsiang T. Cell-Free Extracts of the Ginseng Soil Bacterium Pseudomonas plecoglossicida Promote Suppression of Resistance of American Ginseng (Panax quinquefolius) to Root Rot Caused by Ilyonectria mors-panacis. Biology. 2024; 13(9):671. https://doi.org/10.3390/biology13090671
Chicago/Turabian StyleGoodwin, Paul H., and Tom Hsiang. 2024. "Cell-Free Extracts of the Ginseng Soil Bacterium Pseudomonas plecoglossicida Promote Suppression of Resistance of American Ginseng (Panax quinquefolius) to Root Rot Caused by Ilyonectria mors-panacis" Biology 13, no. 9: 671. https://doi.org/10.3390/biology13090671
APA StyleGoodwin, P. H., & Hsiang, T. (2024). Cell-Free Extracts of the Ginseng Soil Bacterium Pseudomonas plecoglossicida Promote Suppression of Resistance of American Ginseng (Panax quinquefolius) to Root Rot Caused by Ilyonectria mors-panacis. Biology, 13(9), 671. https://doi.org/10.3390/biology13090671






