Prospects and Current Challenges of Extracellular Vesicle-Based Biomarkers in Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Prospects
EVs as a Promising Approach to Cancer Detection
3. Challenges
3.1. EV Heterogeneity
| Challenges | Indications | Potential Solutions |
|---|---|---|
| Extensive EV physical heterogeneity (size, density, and composition) makes tdEV quantification difficult. | - Within glioma, proneural stem cells release EVs largely devoid of markers, while those derived from mesenchymal stem cells uniquely express CD9, CD63, and CD81, indicating intra-disease heterogeneity [60]. - Up to 5000 distinct protein signals have been detected in the EV-associated proteome of a typical cancer cell population [61,62,63,64,65,66,67]. | - Identify and collate appropriate reference genes for the analysis of EV populations derived from multiple tissues and cell types. - Utilise highly sensitive gene amplification technologies (such as RT-qPCR) for accurate EV nucleic acid quantification, targeting known gene variants. - Identification of tdEV-specific proteins with subsequent proteomic profiling, using highly specific antibody-based approaches. |
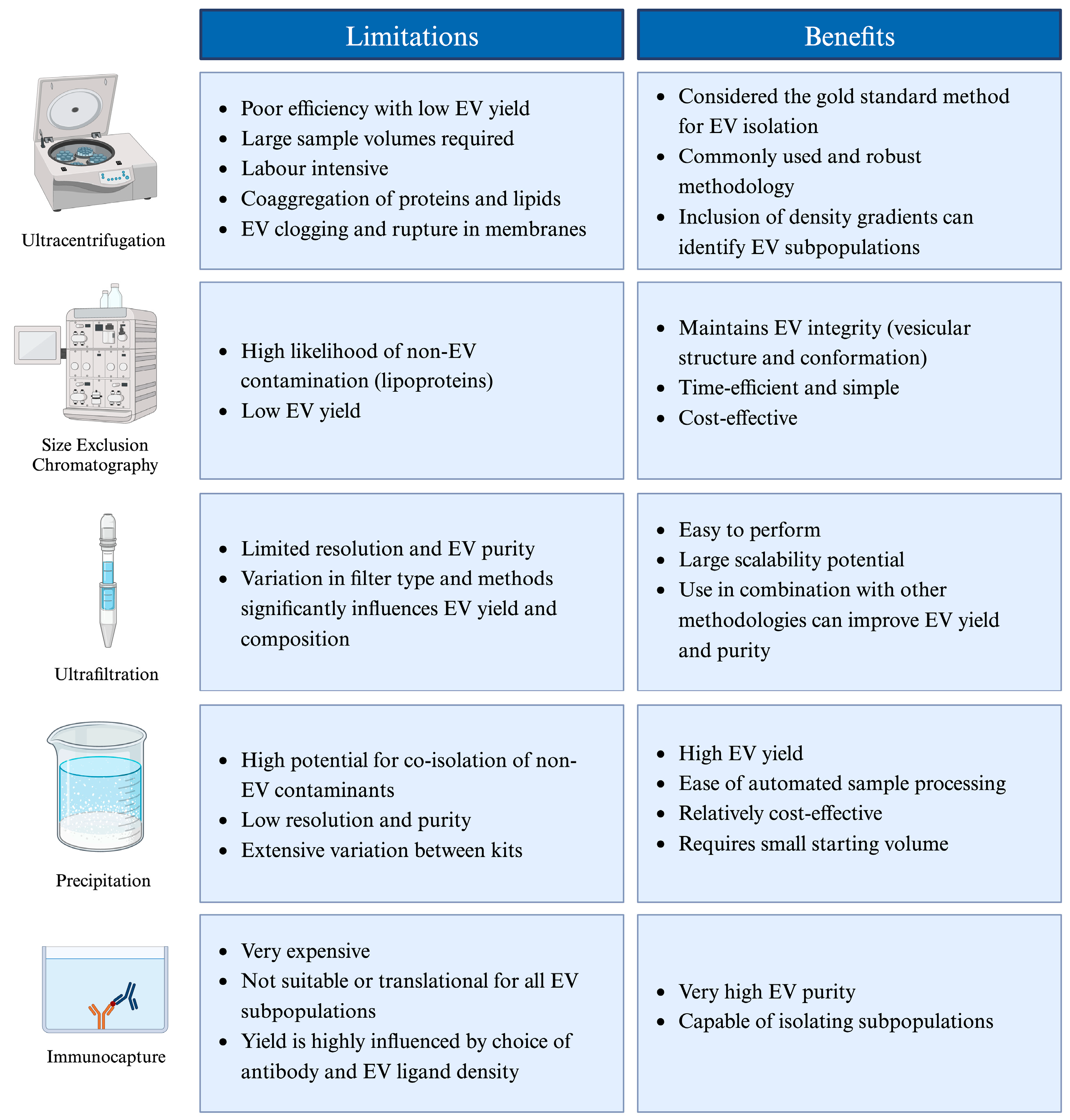
| Enriching EVs often results in the co-isolation of contaminating proteins, which interfere with downstream analysis. | - Maintaining cellular integrity during EV enrichment to prevent intracellular debris release is a technical challenge. - The commonly used method of size exclusion chromatography (SEC) frequently results in the co-isolation of contaminating lipoproteins [68]. | - Employ additional isolation methods such as density gradient separation. - Optimise immunocapture techniques combined with light scattering flow cytometry for high-purity EV isolation. |
| The lack of standardised enrichment methodologies leads to excessive variation and inconsistencies in tdEV detection. | - Variations in pore sizes for SEC, or in relative centrifugal force for density gradient separation, will enrich different sub-fractions of tdEVs [69]. | - Develop a universal approach for enriching different EV types, shifting to standardised, scalable, and accessible technologies and facilitating cost-effective scale-up opportunities. |
| EV Type | Marker | References |
|---|---|---|
| Exosome | CD9, CD63, CD81, TSG101, ALIX, HSP70, HSP90, HSP60, HSP27, HSPA8, Rab27a, Rab27b, syntenin-1, flotillins, ceramides, cholesterol, sphingomyelin, GPC1, CD147 | [76,77] |
| Microvesicle | CD9, CD63, CD81, TSG101, ALIX, HSP70, HSP90, HSP60, HSP27, HSPA8, actin, myosin, ADP-ribosylation factor 6, annexin A1 | [59,76,77,78] |
| Apoptotic body | CD9, CD63, CD81, TSG101, ALIX, HSP70, HSP90, HSP60, HSP27, HSPA8, histone H3, Caspase 3, Phosphatidylserine, annexin V | [59,76,77] |
| Cell Type Specific Markers | CD41 (platelets), CD235a (erythrocytes), EpCAM (epithelial cells), EGFR (cancer cells) | [79,80,81] |
3.2. Contamination during Isolation
| Marker | Sample Type | Cancer Type | Value | Detection Methodology | Relevance |
|---|---|---|---|---|---|
| Phosphatidylinositol-4,5-biphosphate 3-kinase catalytic subunit alpha (PIK3CA) | Freshly frozen tissue biopsy | Breast | Sens. 100% Spec. 100% | Digital droplet PCR | Activating PIK3CA mutations occurs in 20–30% of all breast cancer cases. Specific mutations act as prognostic factors for relapse-free survival [93]. |
| Circulating tumour cells | Blood plasma | Breast, prostate, colorectal | Sens. 85% Spec. 94.45% | Antibody | CTC enumeration can help assess therapeutic response and prognosis in metastatic cancers [94]. |
| Circulating cfDNA | Blood plasma | Colorectal | Sens. 83.1% Spec. 90% | NGS | Colorectal cancer screening in individuals at average risk for the disease [95]. |
| Exosomal RNA (SPDEF, PCA3, ERG) | Urine | Prostate | Sens. 92% Spec. 34% NPV: 91% PPV: 36% | RT-qPCR | Able to discriminate between high-grade, low-grade, and benign disease [90]. |
| Faecal haemoglobin | Stool | Colorectal | Sens. 92.1% Spec. 85.8% | Antibody | Detect the degradation products of blood in faeces and can help identify patients requiring investigation with the highest priority [96]. |
3.3. Method Standardisation and Validation
4. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Deo, S.V.S.; Sharma, J.; Kumar, S. GLOBOCAN 2020 Report on Global Cancer Burden: Challenges and Opportunities for Surgical Oncologists. Ann. Surg. Oncol. 2022, 29, 6497–6500. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J. Tumor markers in clinical practice: A review focusing on common solid cancers. Med. Princ. Pract. 2013, 22, 4–11. [Google Scholar] [CrossRef] [PubMed]
- NHS. The NHS Long Term Plan. Available online: https://www.longtermplan.nhs.uk/publication/nhs-long-term-plan/ (accessed on 23 May 2023).
- Ilic, D.; Djulbegovic, M.; Jung, J.H.; Hwang, E.C.; Zhou, Q.; Cleves, A.; Agoritsas, T.; Dahm, P. Prostate cancer screening with prostate-specific antigen (PSA) test: A systematic review and meta-analysis. BMJ 2018, 362, k3519. [Google Scholar] [CrossRef]
- Saini, S. PSA and beyond: Alternative prostate cancer biomarkers. Cell. Oncol. 2016, 39, 97–106. [Google Scholar] [CrossRef] [PubMed]
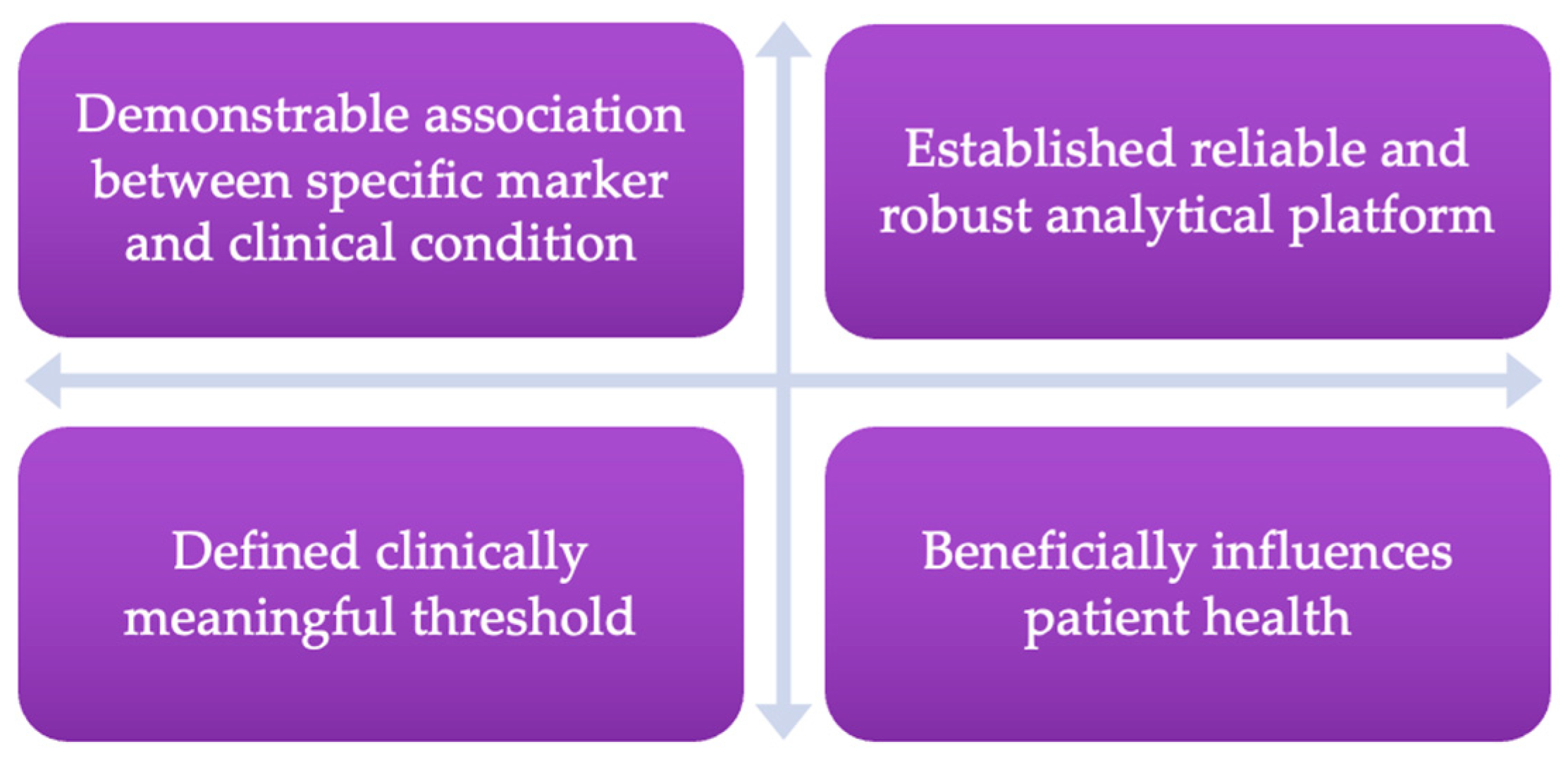
- Holland, R. What makes a good biomarker? Adv. Precis. Med. 2016, 1, 4–11. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Tian, L.; Lu, J.; Ng, I.O. Extracellular vesicles and cancer stemness in hepatocellular carcinoma—Is there a link? Front. Immunol. 2024, 15, 1368898. [Google Scholar] [CrossRef]
- Zhang, W.; Ou, M.; Yang, P.; Ning, M. The role of extracellular vesicle immune checkpoints in cancer. Clin. Exp. Immunol. 2024, 216, 230–239. [Google Scholar] [CrossRef]
- Zhang, C.; Qin, C.; Dewanjee, S.; Bhattacharya, H.; Chakraborty, P.; Jha, N.K.; Gangopadhyay, M.; Jha, S.K.; Liu, Q. Tumor-derived small extracellular vesicles in cancer invasion and metastasis: Molecular mechanisms, and clinical significance. Mol. Cancer 2024, 23, 18. [Google Scholar] [CrossRef]
- Marar, C.; Starich, B.; Wirtz, D. Extracellular vesicles in immunomodulation and tumor progression. Nat. Immunol. 2021, 22, 560–570. [Google Scholar] [CrossRef]
- Crescitelli, R.; Filges, S.; Karimi, N.; Urzi, O.; Alonso-Agudo, T.; Stahlberg, A.; Lotvall, J.; Lasser, C.; Olofsson Bagge, R. Extracellular vesicle DNA from human melanoma tissues contains cancer-specific mutations. Front. Cell Dev. Biol. 2022, 10, 1028854. [Google Scholar] [CrossRef]
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V.; Consortium, C. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef]
- Tatischeff, I. Extracellular Vesicle-DNA: The Next Liquid Biopsy Biomarker for Early Cancer Diagnosis? Cancers 2023, 15, 1456. [Google Scholar] [CrossRef]
- Qian, Z.; Shen, Q.; Yang, X.; Qiu, Y.; Zhang, W. The Role of Extracellular Vesicles: An Epigenetic View of the Cancer Microenvironment. BioMed Res. Int. 2015, 2015, 649161. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Watanabe, Y.; Oikawa, R.; Morita, R.; Yoshida, Y.; Maehata, T.; Yasuda, H.; Itoh, F. BARHL2 Methylation Using Gastric Wash DNA or Gastric Juice Exosomal DNA is a Useful Marker For Early Detection of Gastric Cancer in an H. pylori-Independent Manner. Clin. Transl. Gastroenterol. 2016, 7, e184. [Google Scholar] [CrossRef] [PubMed]
- Maire, C.L.; Fuh, M.M.; Kaulich, K.; Fita, K.D.; Stevic, I.; Heiland, D.H.; Welsh, J.A.; Jones, J.C.; Gorgens, A.; Ricklefs, T.; et al. Genome-wide methylation profiling of glioblastoma cell-derived extracellular vesicle DNA allows tumor classification. Neuro Oncol. 2021, 23, 1087–1099. [Google Scholar] [CrossRef] [PubMed]
- Minic, Z.; Li, Y.; Huttmann, N.; Uppal, G.K.; D’Mello, R.; Berezovski, M.V. Lysine Acetylome of Breast Cancer-Derived Small Extracellular Vesicles Reveals Specific Acetylation Patterns for Metabolic Enzymes. Biomedicines 2023, 11, 1076. [Google Scholar] [CrossRef]
- Al-Nedawi, K.; Meehan, B.; Micallef, J.; Lhotak, V.; May, L.; Guha, A.; Rak, J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008, 10, 619–624. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, W.; Zhang, H.; Sun, B.; Chen, T.; Hu, M.; Zhou, H.; Cao, Y.; Han, B.; Wu, L. Extracellular vesicle long RNA markers of early-stage lung adenocarcinoma. Int. J. Cancer 2023, 152, 1490–1500. [Google Scholar] [CrossRef]
- Reduced Lung-Cancer Mortality with Low-Dose Computed Tomographic Screening. N. Engl. J. Med. 2011, 365, 395–409. [CrossRef]
- The Mercy Halo™ Test. Available online: https://mercybio.com/the-mercy-halo-test/ (accessed on 2 June 2023).
- Lone, S.N.; Nisar, S.; Masoodi, T.; Singh, M.; Rizwan, A.; Hashem, S.; El-Rifai, W.; Bedognetti, D.; Batra, S.K.; Haris, M.; et al. Liquid biopsy: A step closer to transform diagnosis, prognosis and future of cancer treatments. Mol. Cancer 2022, 21, 79. [Google Scholar] [CrossRef]
- Connal, S.; Cameron, J.M.; Sala, A.; Brennan, P.M.; Palmer, D.S.; Palmer, J.D.; Perlow, H.; Baker, M.J. Liquid biopsies: The future of cancer early detection. J. Transl. Med. 2023, 21, 118. [Google Scholar] [CrossRef]
- Ilié, M.; Hofman, P. Pros: Can tissue biopsy be replaced by liquid biopsy? Transl. Lung Cancer Res. 2016, 5, 420–423. [Google Scholar] [CrossRef]
- Sheridan, R.; Brennan, K.; Bazou, D.; O’Gorman, P.; Matallanas, D.; Mc Gee, M.M. Multiple Myeloma Derived Extracellular Vesicle Uptake by Monocyte Cells Stimulates IL-6 and MMP-9 Secretion and Promotes Cancer Cell Migration and Proliferation. Cancers 2024, 16, 1011. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Duo, Y.; Zheng, L.; Zhao, N.; Wang, J.; Liu, Z.; Qiu, L.; Bi, F. CAFs-derived exosomes promote the development of cervical cancer by regulating miR-18a-5p-TMEM170B signaling axis. Biochem. Biophys. Res. Commun. 2024, 694, 149403. [Google Scholar] [CrossRef]
- Deng, C.; Huo, M.; Chu, H.; Zhuang, X.; Deng, G.; Li, W.; Wei, H.; Zeng, L.; He, Y.; Liu, H.; et al. Exosome circATP8A1 induces macrophage M2 polarization by regulating the miR-1-3p/STAT6 axis to promote gastric cancer progression. Mol. Cancer 2024, 23, 49. [Google Scholar] [CrossRef]
- Woodhouse, R.; Li, M.; Hughes, J.; Delfosse, D.; Skoletsky, J.; Ma, P.; Meng, W.; Dewal, N.; Milbury, C.; Clark, T.; et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS ONE 2020, 15, e0237802. [Google Scholar] [CrossRef]
- Kogej, K.; Božič, D.; Kobal, B.; Herzog, M.; Černe, K. Application of Dynamic and Static Light Scattering for Size and Shape Characterization of Small Extracellular Nanoparticles in Plasma and Ascites of Ovarian Cancer Patients. Int. J. Mol. Sci. 2021, 22, 12946. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.P.; Pan, Y.H.; Cai, M.Y.; Cen, J.M.; Chen, C.; Zheng, L.; Liu, X.; Xiong, X.D. Plasma-Derived Exosomal Circular RNA hsa_circ_0005540 as a Novel Diagnostic Biomarker for Coronary Artery Disease. Dis. Markers 2020, 2020, 3178642. [Google Scholar] [CrossRef]
- Nanou, A.; Miller, M.C.; Zeune, L.L.; de Wit, S.; Punt, C.J.A.; Groen, H.J.M.; Hayes, D.F.; de Bono, J.S.; Terstappen, L.W.M.M. Tumour-derived extracellular vesicles in blood of metastatic cancer patients associate with overall survival. Br. J. Cancer 2020, 122, 801–811. [Google Scholar] [CrossRef]
- Isebia, K.T.; Dathathri, E.; Verschoor, N.; Nanou, A.; De Jong, A.C.; Coumans, F.A.W.; Terstappen, L.; Kraan, J.; Martens, J.W.M.; Bansal, R.; et al. Characterizing Circulating Tumor Cells and Tumor-Derived Extracellular Vesicles in Metastatic Castration-Naive and Castration-Resistant Prostate Cancer Patients. Cancers 2022, 14, 4404. [Google Scholar] [CrossRef] [PubMed]
- Kotrbová, A.; Štěpka, K.; Maška, M.; Pálenik, J.J.; Ilkovics, L.; Klemová, D.; Kravec, M.; Hubatka, F.; Dave, Z.; Hampl, A.; et al. TEM ExosomeAnalyzer: A computer-assisted software tool for quantitative evaluation of extracellular vesicles in transmission electron microscopy images. J. Extracell. Vesicles 2019, 8, 1560808. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Martínez, A.; de Miguel-Pérez, D.; Ortega, F.G.; García-Puche, J.L.; Robles-Fernández, I.; Exposito, J.; Martorell-Marugan, J.; Carmona-Sáez, P.; Garrido-Navas, M.D.C.; Rolfo, C.; et al. Exosomal miRNA profile as complementary tool in the diagnostic and prediction of treatment response in localized breast cancer under neoadjuvant chemotherapy. Breast Cancer Res. 2019, 21, 21. [Google Scholar] [CrossRef]
- Szatanek, R.; Baj-Krzyworzeka, M.; Zimoch, J.; Lekka, M.; Siedlar, M.; Baran, J. The Methods of Choice for Extracellular Vesicles (EVs) Characterization. Int. J. Mol. Sci. 2017, 18, 1153. [Google Scholar] [CrossRef]
- Khan, M.A.; Anand, S.; Deshmukh, S.K.; Singh, S.; Singh, A.P. Determining the Size Distribution and Integrity of Extracellular Vesicles by Dynamic Light Scattering. Methods Mol. Biol. 2022, 2413, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Chaikov, L.L.; Kirichenko, M.N.; Krivokhizha, S.V.; Zaritskiy, A.R. Dynamics of statistically confident particle sizes and concentrations in blood plasma obtained by the dynamic light scattering method. J. Biomed. Opt. 2015, 20, 57003. [Google Scholar] [CrossRef]
- Hartjes, T.A.; Mytnyk, S.; Jenster, G.W.; van Steijn, V.; van Royen, M.E. Extracellular Vesicle Quantification and Characterization: Common Methods and Emerging Approaches. Bioengineering 2019, 6, 7. [Google Scholar] [CrossRef]
- Rikkert, L.G.; Beekman, P.; Caro, J.; Coumans, F.A.W.; Enciso-Martinez, A.; Jenster, G.; Le Gac, S.; Lee, W.; van Leeuwen, T.G.; Loozen, G.B.; et al. Cancer-ID: Toward Identification of Cancer by Tumor-Derived Extracellular Vesicles in Blood. Front. Oncol. 2020, 10, 608. [Google Scholar] [CrossRef]
- Dufrene, Y.F.; Ando, T.; Garcia, R.; Alsteens, D.; Martinez-Martin, D.; Engel, A.; Gerber, C.; Muller, D.J. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat. Nanotechnol. 2017, 12, 295–307. [Google Scholar] [CrossRef]
- Ye, S.; Li, W.; Wang, H.; Zhu, L.; Wang, C.; Yang, Y. Quantitative Nanomechanical Analysis of Small Extracellular Vesicles for Tumor Malignancy Indication. Adv. Sci. 2021, 8, e2100825. [Google Scholar] [CrossRef]
- Ridolfi, A.; Brucale, M.; Montis, C.; Caselli, L.; Paolini, L.; Borup, A.; Boysen, A.T.; Loria, F.; van Herwijnen, M.J.C.; Kleinjan, M.; et al. AFM-Based High-Throughput Nanomechanical Screening of Single Extracellular Vesicles. Anal. Chem. 2020, 92, 10274–10282. [Google Scholar] [CrossRef]
- Sokolova, V.; Ludwig, A.K.; Hornung, S.; Rotan, O.; Horn, P.A.; Epple, M.; Giebel, B. Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids Surf. B Biointerfaces 2011, 87, 146–150. [Google Scholar] [CrossRef]
- Nieuwland, R.; Siljander, P.R.M.; Falcón-Pérez, J.M.; Witwer, K.W. Reproducibility of extracellular vesicle research. Eur. J. Cell Biol. 2022, 101, 151226. [Google Scholar] [CrossRef]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrugger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef] [PubMed]
- Ge, Q.; Zhou, Y.; Lu, J.; Bai, Y.; Xie, X.; Lu, Z. miRNA in plasma exosome is stable under different storage conditions. Molecules 2014, 19, 1568–1575. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Ban, J.-J.; Im, W.; Kim, M. Influence of storage condition on exosome recovery. Biotechnol. Bioprocess. Eng. 2016, 21, 299–304. [Google Scholar] [CrossRef]
- Lőrincz Á, M.; Timár, C.I.; Marosvári, K.A.; Veres, D.S.; Otrokocsi, L.; Kittel, Á.; Ligeti, E. Effect of storage on physical and functional properties of extracellular vesicles derived from neutrophilic granulocytes. J. Extracell. Vesicles 2014, 3, 25465. [Google Scholar] [CrossRef] [PubMed]
- Jayachandran, M.; Miller, V.M.; Heit, J.A.; Owen, W.G. Methodology for isolation, identification and characterization of microvesicles in peripheral blood. J. Immunol. Methods 2012, 375, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Jeon, H.; Yoo, S.-M.; Lee, M.-S. The effect of storage temperature on the biological activity of extracellular vesicles for the complement system. Vitr. Cell. Dev. Biol. Anim. 2018, 54, 423–429. [Google Scholar] [CrossRef]
- Cheng, Y.; Zeng, Q.; Han, Q.; Xia, W. Effect of pH, temperature and freezing-thawing on quantity changes and cellular uptake of exosomes. Protein Cell 2019, 10, 295–299. [Google Scholar] [CrossRef]
- Ghadami, S.; Dellinger, K. The lipid composition of extracellular vesicles: Applications in diagnostics and therapeutic delivery. Front. Mol. Biosci. 2023, 10, 1198044. [Google Scholar] [CrossRef] [PubMed]
- IVD Clinical Evidence. Available online: https://mdic.org/project/ivd-clinical-evidence/ (accessed on 3 June 2023).
- Füzéry, A.K.; Levin, J.; Chan, M.M.; Chan, D.W. Translation of proteomic biomarkers into FDA approved cancer diagnostics: Issues and challenges. Clin. Proteom. 2013, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Shin, K.J.; Chae, Y.C. Regulation of cargo selection in exosome biogenesis and its biomedical applications in cancer. Exp. Mol. Med. 2024, 56, 877–889. [Google Scholar] [CrossRef]
- Brennan, K.; Martin, K.; FitzGerald, S.P.; O’Sullivan, J.; Wu, Y.; Blanco, A.; Richardson, C.; Mc Gee, M.M. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci. Rep. 2020, 10, 1039. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; Carter, D.R.F.; Clayton, A.; Lambert, D.W.; Raposo, G.; Vader, P. Challenges and directions in studying cell–cell communication by extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2022, 23, 369–382. [Google Scholar] [CrossRef]
- Sheta, M.; Taha, E.A.; Lu, Y.; Eguchi, T. Extracellular Vesicles: New Classification and Tumor Immunosuppression. Biology 2023, 12, 110. [Google Scholar] [CrossRef]
- Spinelli, C.; Montermini, L.; Meehan, B.; Brisson, A.R.; Tan, S.; Choi, D.; Nakano, I.; Rak, J. Molecular subtypes and differentiation programmes of glioma stem cells as determinants of extracellular vesicle profiles and endothelial cell-stimulating activities. J. Extracell. Vesicles 2018, 7, 1490144. [Google Scholar] [CrossRef]
- Choi, D.; Spinelli, C.; Montermini, L.; Rak, J. Oncogenic Regulation of Extracellular Vesicle Proteome and Heterogeneity. Proteomics 2019, 19, e1800169. [Google Scholar] [CrossRef]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Théry, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [PubMed]
- Poliakov, A.; Spilman, M.; Dokland, T.; Amling, C.L.; Mobley, J.A. Structural heterogeneity and protein composition of exosome-like vesicles (prostasomes) in human semen. Prostate 2009, 69, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Rak, J.; Strzadala, L. Heterogeneity of Extracellular Vesicles and Particles: Molecular Voxels in the Blood Borne “Hologram” of Organ Function, Disfunction and Cancer. Arch. Immunol. Et Ther. Exp. 2023, 71, 5. [Google Scholar] [CrossRef]
- van der Pol, E.; Coumans, F.; Varga, Z.; Krumrey, M.; Nieuwland, R. Innovation in detection of microparticles and exosomes. J. Thromb. Haemost. 2013, 11, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Karimi, N.; Cvjetkovic, A.; Jang, S.C.; Crescitelli, R.; Hosseinpour Feizi, M.A.; Nieuwland, R.; Lötvall, J.; Lässer, C. Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cell. Mol. Life Sci. 2018, 75, 2873–2886. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; He, J. Characterization of Extracellular Vesicles by Size-Exclusion High-Performance Liquid Chromatography (HPLC). Methods Mol. Biol. 2017, 1660, 191–199. [Google Scholar] [CrossRef]
- Bernard, P.S.; Wittwer, C.T. Real-time PCR technology for cancer diagnostics. Clin. Chem. 2002, 48, 1178–1185. [Google Scholar] [CrossRef]
- Benz, F.; Roderburg, C.; Vargas Cardenas, D.; Vucur, M.; Gautheron, J.; Koch, A.; Zimmermann, H.; Janssen, J.; Nieuwenhuijsen, L.; Luedde, M.; et al. U6 is unsuitable for normalization of serum miRNA levels in patients with sepsis or liver fibrosis. Exp. Mol. Med. 2013, 45, e42. [Google Scholar] [CrossRef]
- Chiba, M.; Kimura, M.; Asari, S. Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol. Rep. 2012, 28, 1551–1558. [Google Scholar] [CrossRef]
- Gouin, K.; Peck, K.; Antes, T.; Johnson, J.L.; Li, C.; Vaturi, S.D.; Middleton, R.; de Couto, G.; Walravens, A.S.; Rodriguez-Borlado, L.; et al. A comprehensive method for identification of suitable reference genes in extracellular vesicles. J. Extracell. Vesicles 2017, 6, 1347019. [Google Scholar] [CrossRef]
- Hoshino, A.; Kim, H.S.; Bojmar, L.; Gyan, K.E.; Cioffi, M.; Hernandez, J.; Zambirinis, C.P.; Rodrigues, G.; Molina, H.; Heissel, S.; et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell 2020, 182, 1044–1061. [Google Scholar] [CrossRef] [PubMed]
- Kwizera, E.A.; O’Connor, R.; Vinduska, V.; Williams, M.; Butch, E.R.; Snyder, S.E.; Chen, X.; Huang, X. Molecular Detection and Analysis of Exosomes Using Surface-Enhanced Raman Scattering Gold Nanorods and a Miniaturized Device. Theranostics 2018, 8, 2722–2738. [Google Scholar] [CrossRef]
- Andras, I.E.; Toborek, M. Extracellular vesicles of the blood-brain barrier. Tissue Barriers 2016, 4, e1131804. [Google Scholar] [CrossRef]
- Malvicini, R.; Santa-Cruz, D.; Tolomeo, A.M.; Muraca, M.; Yannarelli, G.; Pacienza, N. Ion exchange chromatography as a simple and scalable method to isolate biologically active small extracellular vesicles from conditioned media. PLoS ONE 2023, 18, e0291589. [Google Scholar] [CrossRef] [PubMed]
- Ukrainskaya, V.M.; Rubtsov, Y.P.; Knorre, V.D.; Maschan, M.A.; Gabibov, A.G.; Stepanov, A.V. The Role of Tumor-Derived Vesicles in the Regulation of Antitumor Immunity. Acta Nat. 2019, 11, 33–41. [Google Scholar] [CrossRef]
- Ye, C.; Li, H.; Bao, M.; Zhuo, R.; Jiang, G.; Wang, W. Alveolar macrophage - derived exosomes modulate severity and outcome of acute lung injury. Aging 2020, 12, 6120–6128. [Google Scholar] [CrossRef]
- Ferlizza, E.; Romaniello, D.; Borrelli, F.; Pagano, F.; Girone, C.; Gelfo, V.; Kuhre, R.S.; Morselli, A.; Mazzeschi, M.; Sgarzi, M.; et al. Extracellular Vesicles and Epidermal Growth Factor Receptor Activation: Interplay of Drivers in Cancer Progression. Cancers 2023, 15, 2970. [Google Scholar] [CrossRef]
- Ekstrom, K.; Crescitelli, R.; Petursson, H.I.; Johansson, J.; Lasser, C.; Olofsson Bagge, R. Characterization of surface markers on extracellular vesicles isolated from lymphatic exudate from patients with breast cancer. BMC Cancer 2022, 22, 50. [Google Scholar] [CrossRef] [PubMed]
- Coumans, F.A.W.; Brisson, A.R.; Buzas, E.I.; Dignat-George, F.; Drees, E.E.E.; El-Andaloussi, S.; Emanueli, C.; Gasecka, A.; Hendrix, A.; Hill, A.F.; et al. Methodological Guidelines to Study Extracellular Vesicles. Circ. Res. 2017, 120, 1632–1648. [Google Scholar] [CrossRef]
- Crescitelli, R.; Lässer, C.; Lötvall, J. Isolation and characterization of extracellular vesicle subpopulations from tissues. Nat. Protoc. 2021, 16, 1548–1580. [Google Scholar] [CrossRef]
- Merij, L.B.; Andrade, F.B.; Silva, A.R.; Hottz, E.D. Isolation of Microvesicles from Plasma Samples Avoiding Lipoprotein Contamination. Methods Mol. Biol. 2022, 2409, 245–255. [Google Scholar] [CrossRef]
- Chen, M.; Huang, J.; Yang, X.; Liu, B.; Zhang, W.; Huang, L.; Deng, F.; Ma, J.; Bai, Y.; Lu, R.; et al. Serum starvation induced cell cycle synchronization facilitates human somatic cells reprogramming. PLoS ONE 2012, 7, e28203. [Google Scholar] [CrossRef] [PubMed]
- Mollereau, B.; Ma, D. The p53 control of apoptosis and proliferation: Lessons from Drosophila. Apoptosis 2014, 19, 1421–1429. [Google Scholar] [CrossRef]
- Rashid, M.-u.; Coombs, K.M. Serum-reduced media impacts on cell viability and protein expression in human lung epithelial cells. J. Cell. Physiol. 2019, 234, 7718–7724. [Google Scholar] [CrossRef] [PubMed]
- Brandi, J.; Manfredi, M.; Speziali, G.; Gosetti, F.; Marengo, E.; Cecconi, D. Proteomic approaches to decipher cancer cell secretome. Semin. Cell Dev. Biol. 2018, 78, 93–101. [Google Scholar] [CrossRef]
- Mørk, M.; Handberg, A.; Pedersen, S.; Jørgensen, M.M.; Bæk, R.; Nielsen, M.K.; Kristensen, S.R. Prospects and limitations of antibody-mediated clearing of lipoproteins from blood plasma prior to nanoparticle tracking analysis of extracellular vesicles. J. Extracell. Vesicles 2017, 6, 1308779. [Google Scholar] [CrossRef]
- McKiernan, J.; Donovan, M.J.; O’Neill, V.; Bentink, S.; Noerholm, M.; Belzer, S.; Skog, J.; Kattan, M.W.; Partin, A.; Andriole, G.; et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol. 2016, 2, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Pal, M.; Muinao, T.; Boruah, H.P.D.; Mahindroo, N. Current advances in prognostic and diagnostic biomarkers for solid cancers: Detection techniques and future challenges. Biomed. Pharmacother. 2022, 146, 112488. [Google Scholar] [CrossRef]
- Kosanovic, M.; Jankovic, M. Isolation of urinary extracellular vesicles from Tamm-Horsfall protein-depleted urine and their application in the development of a lectin-exosome-binding assay. Biotechniques 2014, 57, 143–149. [Google Scholar] [CrossRef]
- Shimoi, T.; Hamada, A.; Yamagishi, M.; Hirai, M.; Yoshida, M.; Nishikawa, T.; Sudo, K.; Shimomura, A.; Noguchi, E.; Yunokawa, M.; et al. PIK3CA mutation profiling in patients with breast cancer, using a highly sensitive detection system. Cancer Sci. 2018, 109, 2558–2566. [Google Scholar] [CrossRef]
- Satelli, A.; Brownlee, Z.; Mitra, A.; Meng, Q.H.; Li, S. Circulating tumor cell enumeration with a combination of epithelial cell adhesion molecule- and cell-surface vimentin-based methods for monitoring breast cancer therapeutic response. Clin. Chem. 2015, 61, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.C.; Gray, D.M., 2nd; Singh, H.; Issaka, R.B.; Raymond, V.M.; Eagle, C.; Hu, S.; Chudova, D.I.; Talasaz, A.; Greenson, J.K.; et al. A Cell-free DNA Blood-Based Test for Colorectal Cancer Screening. N. Engl. J. Med. 2024, 390, 973–983. [Google Scholar] [CrossRef]
- Monahan, K.J.; Davies, M.M.; Abulafi, M.; Banerjea, A.; Nicholson, B.D.; Arasaradnam, R.; Barker, N.; Benton, S.; Booth, R.; Burling, D.; et al. Faecal immunochemical testing (FIT) in patients with signs or symptoms of suspected colorectal cancer (CRC): A joint guideline from the Association of Coloproctology of Great Britain and Ireland (ACPGBI) and the British Society of Gastroenterology (BSG). Gut 2022, 71, 1939–1962. [Google Scholar] [CrossRef]
- Dhondt, B.; Geeurickx, E.; Tulkens, J.; Van Deun, J.; Vergauwen, G.; Lippens, L.; Miinalainen, I.; Rappu, P.; Heino, J.; Ost, P.; et al. Unravelling the proteomic landscape of extracellular vesicles in prostate cancer by density-based fractionation of urine. J. Extracell. Vesicles 2020, 9, 1736935. [Google Scholar] [CrossRef]
- Ramirez-Garrastacho, M.; Bajo-Santos, C.; Line, A.; Martens-Uzunova, E.S.; de la Fuente, J.M.; Moros, M.; Soekmadji, C.; Tasken, K.A.; Llorente, A. Extracellular vesicles as a source of prostate cancer biomarkers in liquid biopsies: A decade of research. Br. J. Cancer 2022, 126, 331–350. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, C.; Di Vizio, D.; Sahoo, S.; Théry, C.; Witwer, K.W.; Wauben, M.; Hill, A.F. Techniques used for the isolation and characterization of extracellular vesicles: Results of a worldwide survey. J. Extracell. Vesicles 2016, 5, 32945. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabières, C.; Pantel, K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021, 11, 858–873. [Google Scholar] [CrossRef] [PubMed]
- Stam, J.; Bartel, S.; Bischoff, R.; Wolters, J.C. Isolation of extracellular vesicles with combined enrichment methods. J. Chromatogr. B 2021, 1169, 122604. [Google Scholar] [CrossRef]
- Monguió-Tortajada, M.; Gálvez-Montón, C.; Bayes-Genis, A.; Roura, S.; Borràs, F.E. Extracellular vesicle isolation methods: Rising impact of size-exclusion chromatography. Cell. Mol. Life Sci. 2019, 76, 2369–2382. [Google Scholar] [CrossRef]
- Chames, P.; Van Regenmortel, M.; Weiss, E.; Baty, D. Therapeutic antibodies: Successes, limitations and hopes for the future. Br. J. Pharmacol. 2009, 157, 220–233. [Google Scholar] [CrossRef]
- Liangsupree, T.; Multia, E.; Riekkola, M.-L. Modern isolation and separation techniques for extracellular vesicles. J. Chromatogr. A 2021, 1636, 461773. [Google Scholar] [CrossRef] [PubMed]
- Cantin, R.; Diou, J.; Bélanger, D.; Tremblay, A.M.; Gilbert, C. Discrimination between exosomes and HIV-1: Purification of both vesicles from cell-free supernatants. J. Immunol. Methods 2008, 338, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Skog, J.; Hsu, C.H.; Lessard, R.T.; Balaj, L.; Wurdinger, T.; Carter, B.S.; Breakefield, X.O.; Toner, M.; Irimia, D. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab. Chip 2010, 10, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Chhoy, P.; Brown, C.W.; Amante, J.J.; Mercurio, A.M. Protocol for the separation of extracellular vesicles by ultracentrifugation from in vitro cell culture models. STAR Protoc. 2021, 2, 100303. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.W.; Simpson, R.J. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018, 15, 617–638. [Google Scholar] [CrossRef]
- Chen, J.; Tan, Q.; Yang, Z.; Jin, Y. Engineered extracellular vesicles: Potentials in cancer combination therapy. J. Nanobiotechnol. 2022, 20, 132. [Google Scholar] [CrossRef]
- Klein, C.; Brinkmann, U.; Reichert, J.M.; Kontermann, R.E. The present and future of bispecific antibodies for cancer therapy. Nat. Rev. Drug Discov. 2024, 23, 301–319. [Google Scholar] [CrossRef]
- Gu, Y.; Wang, Z.; Wang, Y. Bispecific antibody drug conjugates: Making 1+1>2. Acta Pharm. Sin. B 2024, 14, 1965–1986. [Google Scholar] [CrossRef]

| Contaminant | Subtype |
|---|---|
| Lipoproteins | High-density |
| Low-density | |
| Very low-density | |
| Chylomicrons | |
| Proteins | Aggregates |
| Soluble proteins | |
| Ribonucleoproteins | |
| Nucleic acids | Circulating free RNA/DNA |
| Other | Cellular debris |
| Apoptotic bodies | |
| Viruses |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lawrence, S.R.; Shah, K.M. Prospects and Current Challenges of Extracellular Vesicle-Based Biomarkers in Cancer. Biology 2024, 13, 694. https://doi.org/10.3390/biology13090694
Lawrence SR, Shah KM. Prospects and Current Challenges of Extracellular Vesicle-Based Biomarkers in Cancer. Biology. 2024; 13(9):694. https://doi.org/10.3390/biology13090694
Chicago/Turabian StyleLawrence, Samuel R., and Karan M. Shah. 2024. "Prospects and Current Challenges of Extracellular Vesicle-Based Biomarkers in Cancer" Biology 13, no. 9: 694. https://doi.org/10.3390/biology13090694
APA StyleLawrence, S. R., & Shah, K. M. (2024). Prospects and Current Challenges of Extracellular Vesicle-Based Biomarkers in Cancer. Biology, 13(9), 694. https://doi.org/10.3390/biology13090694






