Optimization of Conditions for Production of Soluble E. coli Poly(A)-Polymerase for Biotechnological Applications
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cloning of E. coli Poly(A) Polymerase
2.2. Expression of E. coli Poly(A) Polymerase
2.3. Solubility Test
2.4. Polyadenylation Assay
2.5. Quantitative PCR
2.6. Data Analysis
3. Results
3.1. Expression of E. coli PAP 1
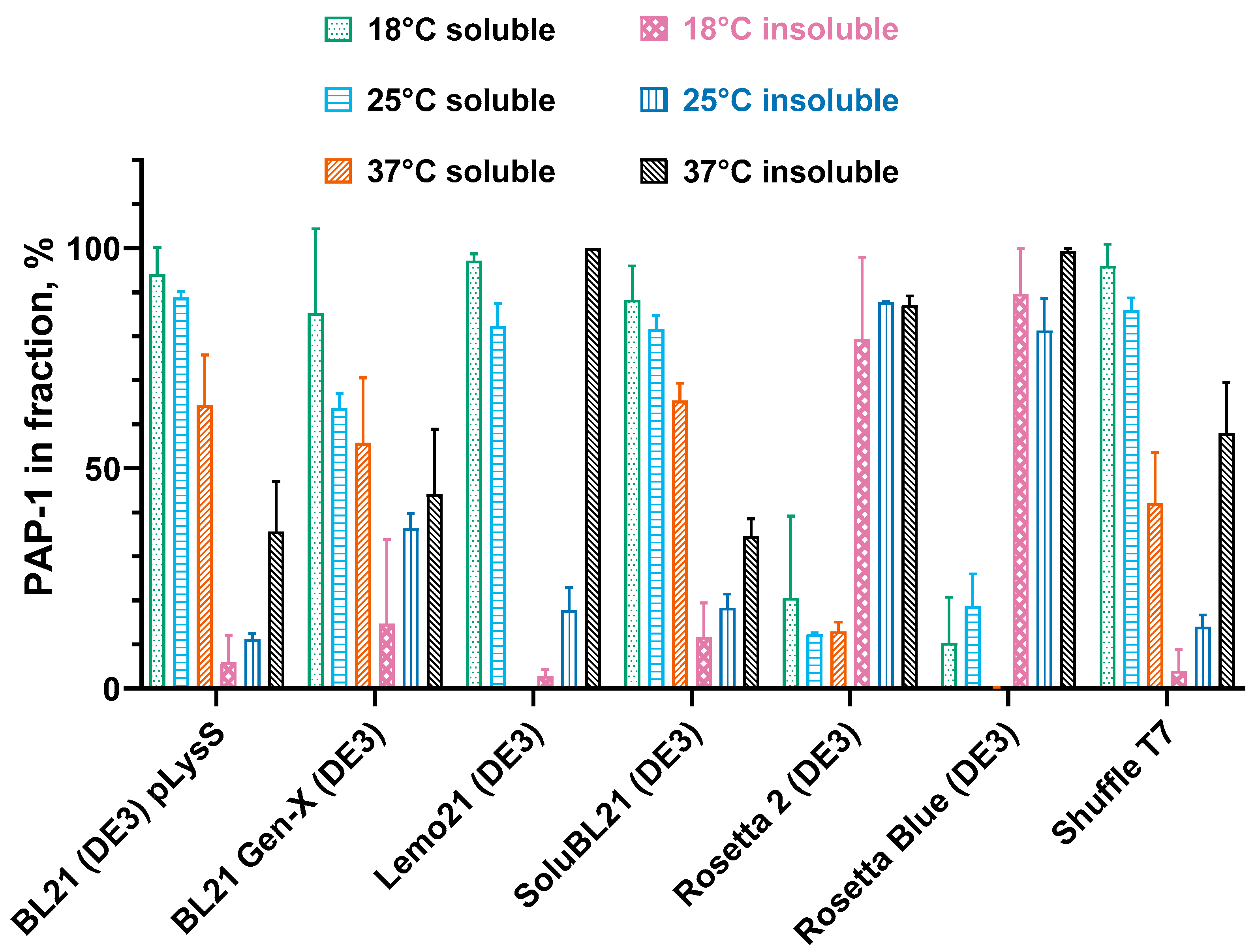
3.2. Solubility of E. coli PAP 1
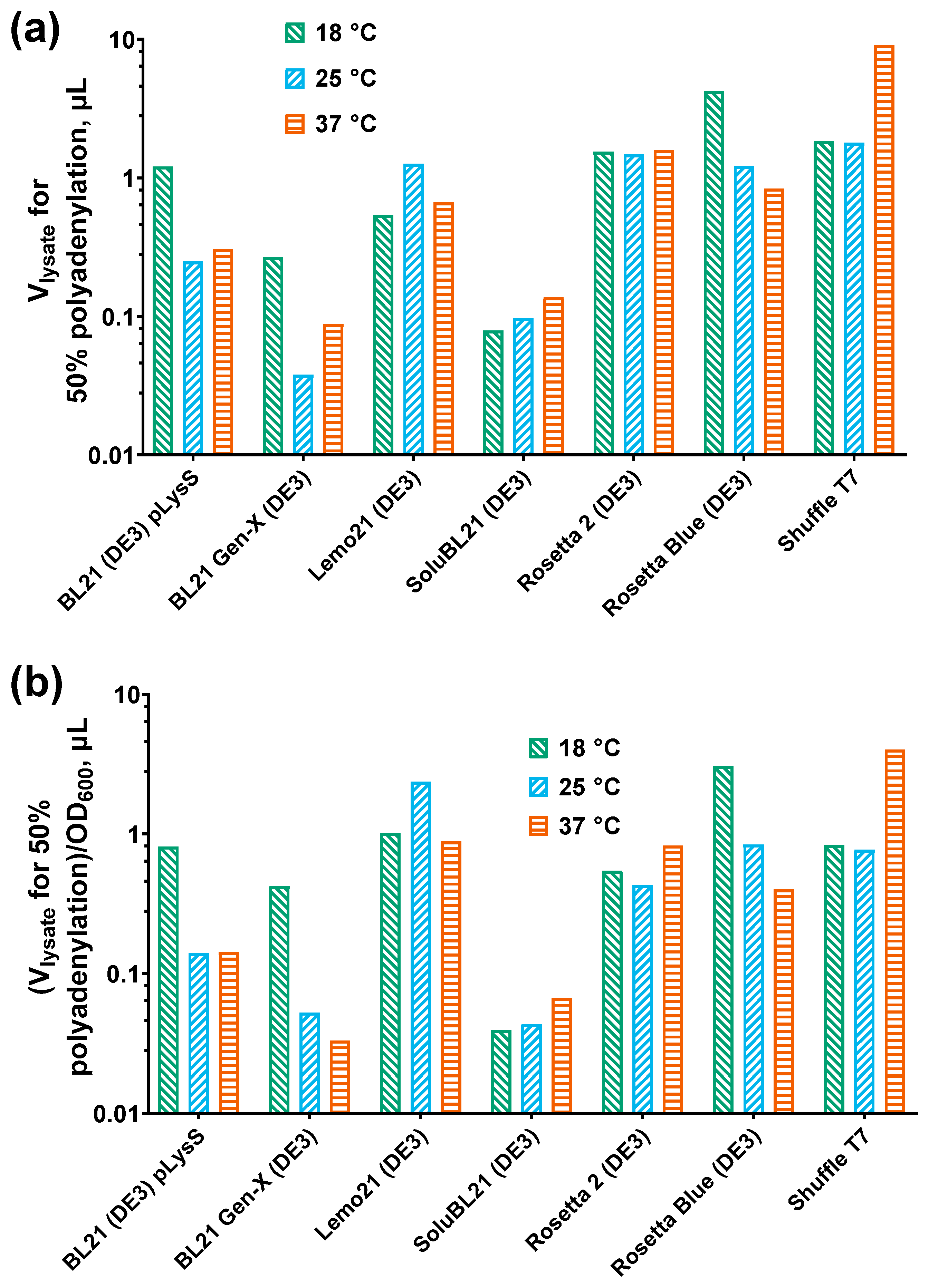
3.3. Specific Activity of E. coli PAP 1
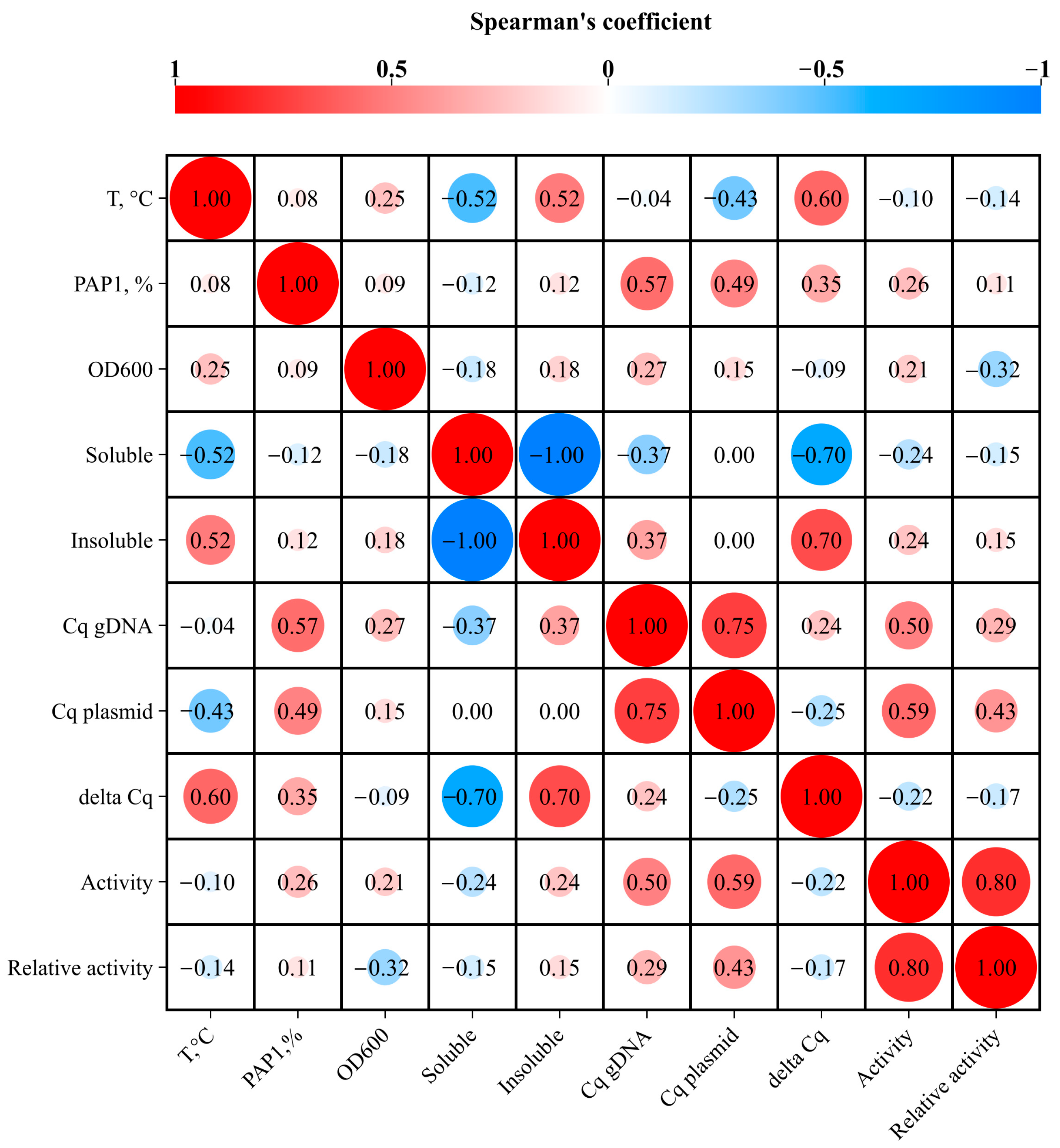
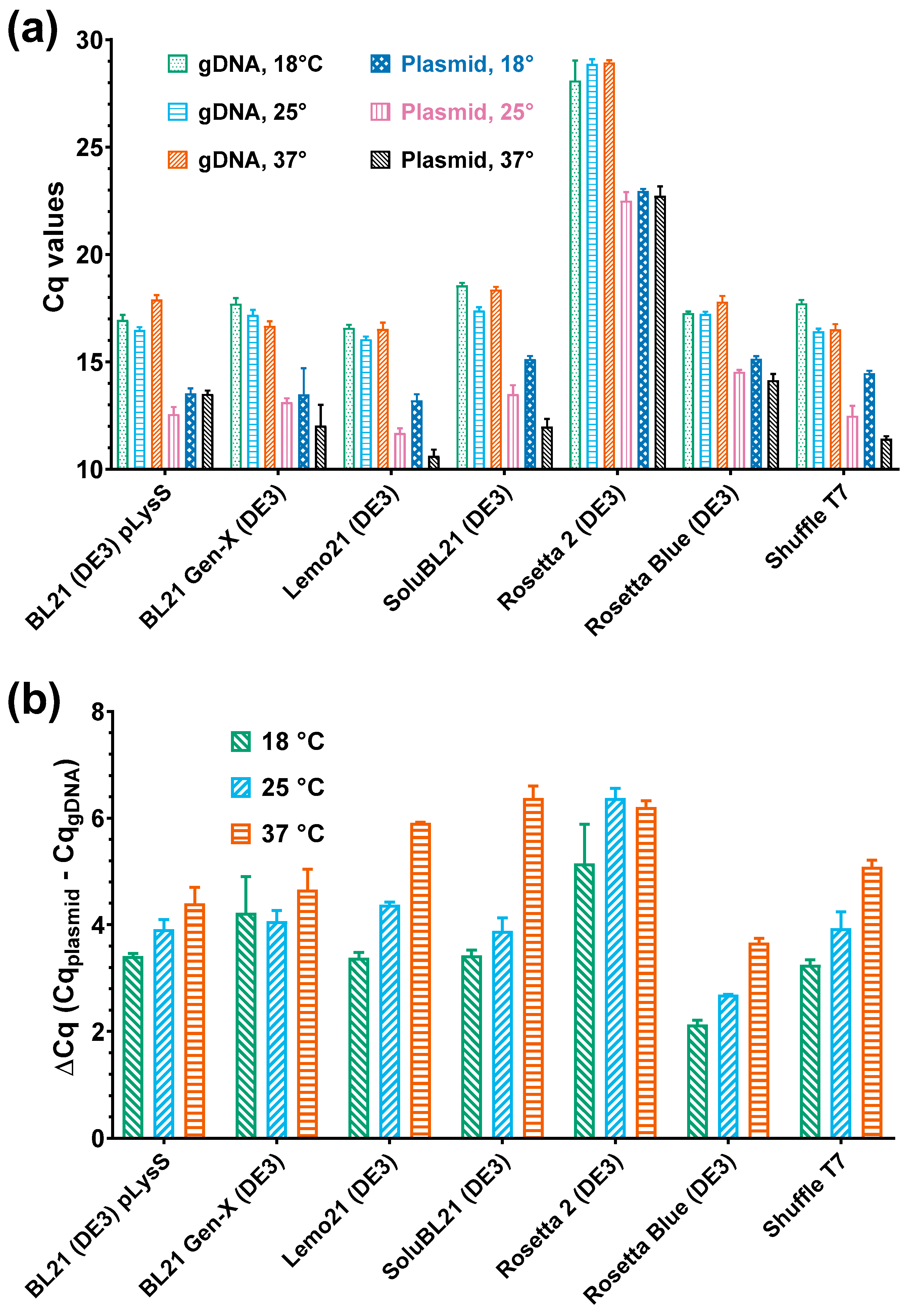
3.4. Relation Between Plasmid Copy Number and Level of Recombinant PAP 1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hajnsdorf, E.; Kaberdin, V.R. RNA Polyadenylation and Its Consequences in Prokaryotes. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20180166. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, B.K.; Kushner, S.R. Polynucleotide Phosphorylase Functions Both as a 3′ → 5′ Exonuclease and a Poly(A) Polymerase in Escherichia coli. Proc. Natl. Acad. Sci. USA 2000, 97, 11966–11971. [Google Scholar] [CrossRef] [PubMed]
- Bralley, P.; Cozad, M.; Jones, G.H. Geobacter Sulfurreducens Contains Separate C- and A-Adding TRNA Nucleotidyltransferases and a Poly(A) Polymerase. J. Bacteriol. 2009, 191, 109–114. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lopilato, J.; Bortner, S.; Beckwith, J. Mutations in a New Chromosomal Gene of Escherichia coli K-12, PcnB, Reduce Plasmid Copy Number of PBR322 and Its Derivatives. Mol. Gen. Genet. MGG 1986, 205, 285–290. [Google Scholar] [CrossRef]
- Cao, G.J.; Sarkar, N. Identification of the Gene for an Escherichia coli Poly(A) Polymerase. Proc. Natl. Acad. Sci. USA 1992, 89, 10380–10384. [Google Scholar] [CrossRef]
- Mohanty, B.K.; Kushner, S.R. Analysis of the Function of Escherichia coli Poly(A) Polymerase I in RNA Metabolism. Mol. Microbiol. 1999, 34, 1094–1108. [Google Scholar] [CrossRef]
- Maes, A.; Gracia, C.; Innocenti, N.; Zhang, K.; Aurell, E.; Hajnsdorf, E. Landscape of RNA Polyadenylation in E. coli. Nucleic Acids Res. 2017, 45, 2746–2756. [Google Scholar] [CrossRef]
- Jones, G.H. Phylogeny and Evolution of RNA 3′-Nucleotidyltransferases in Bacteria. J. Mol. Evol. 2019, 87, 254–270. [Google Scholar] [CrossRef]
- August, J.T.; Ortiz, P.J.; Hurwitz, J. Ribonucleic Acid-Dependent Ribonucleotide Incorporation. I. Purification and Properties of the Enzyme. J. Biol. Chem. 1962, 237, 3786–3793. [Google Scholar] [CrossRef]
- Kalapos, M.P.; Cao, G.J.; Kushner, S.R.; Sarkar, N. Identification of a Second Poly(A) Polymerase in Escherichia coli. Biochem. Biophys. Res. Commun. 1994, 198, 459–465. [Google Scholar] [CrossRef]
- Payne, K.J.; Boezi, J.A. The Purification and Characterization of Adenosine Triphosphate Ribonucleic Acid Adenyltransferase from Pseudomonas putida. J. Biol. Chem. 1970, 245, 1378–1387. [Google Scholar] [CrossRef] [PubMed]
- Sippel, A.E. Purification and Characterization of Adenosine Triphosphate: Ribonucleic Acid Adenyltransferase from Escherichia coli. Eur. J. Biochem. 1973, 37, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Yehudai-Resheff, S. Characterization of the E. coli Poly(A) Polymerase: Nucleotide Specificity, RNA-Binding Affinities and RNA Structure Dependence. Nucleic Acids Res. 2000, 28, 1139–1144. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.D.; Parkinson, J.S. Genetics and Sequence Analysis of the PcnB Locus, an Escherichia coli Gene Involved in Plasmid Copy Number Control. J. Bacteriol. 1989, 171, 1254–1261. [Google Scholar] [CrossRef]
- Jones, G.H. Acquisition of PcnB [Poly(A) Polymerase I] Genes via Horizontal Transfer from the β, γ-Proteobacteria. Microb. Genomics 2021, 7, 000508. [Google Scholar] [CrossRef]
- Chaudhary, N.; Weissman, D.; Whitehead, K.A. MRNA Vaccines for Infectious Diseases: Principles, Delivery and Clinical Translation. Nat. Rev. Drug Discov. 2021, 20, 817–838. [Google Scholar] [CrossRef]
- Sayour, E.J.; Boczkowski, D.; Mitchell, D.A.; Nair, S.K. Cancer MRNA Vaccines: Clinical Advances and Future Opportunities. Nat. Rev. Clin. Oncol. 2024, 21, 489–500. [Google Scholar] [CrossRef]
- Rosano, G.L.; Morales, E.S.; Ceccarelli, E.A. New Tools for Recombinant Protein Production in Escherichia coli: A 5-year Update. Protein Sci. 2019, 28, 1412–1422. [Google Scholar] [CrossRef]
- İncir, İ.; Kaplan, Ö. Escherichia coli as a Versatile Cell Factory: Advances and Challenges in Recombinant Protein Production. Protein Expr. Purif. 2024, 219, 106463. [Google Scholar] [CrossRef]
- Tegel, H.; Tourle, S.; Ottosson, J.; Persson, A. Increased Levels of Recombinant Human Proteins with the Escherichia coli Strain Rosetta(DE3). Protein Expr. Purif. 2010, 69, 159–167. [Google Scholar] [CrossRef]
- Redda, Y.T.; Venkatesh, G.; Kalaiyarasu, S.; Bhatia, S.; Kumar, D.S.; Nagarajan, S.; Pillai, A.; Tripathi, S.; Kulkarni, D.D.; Dubey, S.C. Expression and Purification of Recombinant H5HA1 Protein of H5N1 Avian Influenza Virus in E. coli and Its Application in Indirect ELISA. J. Immunoass. Immunochem. 2016, 37, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Ossysek, K.; Uchański, T.; Kulesza, M.; Bzowska, M.; Klaus, T.; Woś, K.; Madej, M.; Bereta, J. A New Expression Vector Facilitating Production and Functional Analysis of ScFv Antibody Fragments Selected from Tomlinson I+J Phagemid Libraries. Immunol. Lett. 2015, 167, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Huynh Thi Yen, L.; Park, S.-Y.; Kim, J.-S. Cloning, Crystallization and Preliminary X-Ray Diffraction Analysis of an Intact DNA Methyltransferase of a Type I Restriction-Modification Enzyme from Vibrio Vulnificus. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2014, 70, 489–492. [Google Scholar] [CrossRef] [PubMed]
- Hata, S.; Kitamura, F.; Sorimachi, H. Efficient Expression and Purification of Recombinant Human μ-Calpain Using an Escherichia coli Expression System. Genes Cells 2013, 18, 753–763. [Google Scholar] [CrossRef]
- Yamaguchi, R.; Akter, S.; Kanehama, A.; Iwamoto, T.; Hasegawa, M.; Ito, A.; Nishimukai, M.; Yamada, M.; Kashiwagi, A. Improvement of Solubility of Phospholipase D from Streptomyces antibioticus in Recombinant Escherichia coli and Its Application for the Enzymatic Synthesis of a Non-Natural Plasmalogen. Lett. Appl. Microbiol. 2023, 76, ovad049. [Google Scholar] [CrossRef]
- Diankristanti, P.A.; Effendi, S.S.W.; Hsiang, C.-C.; Ng, I.-S. High-Level Itaconic Acid (IA) Production Using Engineered Escherichia coli Lemo21(DE3) toward Sustainable Biorefinery. Enzyme Microb. Technol. 2023, 167, 110231. [Google Scholar] [CrossRef]
- Schlegel, S.; Löfblom, J.; Lee, C.; Hjelm, A.; Klepsch, M.; Strous, M.; Drew, D.; Slotboom, D.J.; de Gier, J.-W. Optimizing Membrane Protein Overexpression in the Escherichia coli Strain Lemo21(DE3). J. Mol. Biol. 2012, 423, 648–659. [Google Scholar] [CrossRef]
- Nasiri, M.; Babaie, J.; Amiri, S.; Azimi, E.; Shamshiri, S.; Khalaj, V.; Golkar, M.; Fard-Esfahani, P. SHuffleTM T7 Strain Is Capable of Producing High Amount of Recombinant Human Fibroblast Growth Factor-1 (RhFGF-1) with Proper Physicochemical and Biological Properties. J. Biotechnol. 2017, 259, 30–38. [Google Scholar] [CrossRef]
- Lobstein, J.; Emrich, C.A.; Jeans, C.; Faulkner, M.; Riggs, P.; Berkmen, M. SHuffle, a Novel Escherichia coli Protein Expression Strain Capable of Correctly Folding Disulfide Bonded Proteins in Its Cytoplasm. Microb. Cell Fact. 2012, 11, 56. [Google Scholar] [CrossRef]
- Evans, G.A. Molecular cloning: A laboratory manual. Second edition. Volumes 1, 2, and 3. Current protocols in molecular biology. Volumes 1 and 2: By J. Sambrook, E. F. Fritsch, and T. Maniatis. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press. (1989). 1626 pp. $115.00. Edited by F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. New York: Greene Publishing Associates and John Wiley & Sons. (1989). 1120 pp. $255.00. Cell 1990, 61, 17–18. [Google Scholar]
- Boël, G.; Letso, R.; Neely, H.; Price, W.N.; Wong, K.-H.; Su, M.; Luff, J.D.; Valecha, M.; Everett, J.K.; Acton, T.B.; et al. Codon Influence on Protein Expression in E. coli Correlates with MRNA Levels. Nature 2016, 529, 358–363. [Google Scholar] [CrossRef]
- Rosano, G.L.; Ceccarelli, E.A. Rare Codon Content Affects the Solubility of Recombinant Proteins in a Codon Bias-Adjusted Escherichia coli Strain. Microb. Cell Fact. 2009, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Price, W.N.; Handelman, S.K.; Everett, J.K.; Tong, S.N.; Bracic, A.; Luff, J.D.; Naumov, V.; Acton, T.; Manor, P.; Xiao, R.; et al. Large-Scale Experimental Studies Show Unexpected Amino Acid Effects on Protein Expression and Solubility in Vivo in E. coli. Microb. Inform. Exp. 2011, 1, 6. [Google Scholar] [CrossRef] [PubMed]
- Chhetri, G.; Kalita, P.; Tripathi, T. An Efficient Protocol to Enhance Recombinant Protein Expression Using Ethanol in Escherichia coli. MethodsX 2015, 2, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Oganesyan, N.; Ankoudinova, I.; Kim, S.-H.; Kim, R. Effect of Osmotic Stress and Heat Shock in Recombinant Protein Overexpression and Crystallization. Protein Expr. Purif. 2007, 52, 280–285. [Google Scholar] [CrossRef]
- Prasad, S.; Khadatare, P.B.; Roy, I. Effect of Chemical Chaperones in Improving the Solubility of Recombinant Proteins in Escherichia coli. Appl. Environ. Microbiol. 2011, 77, 4603–4609. [Google Scholar] [CrossRef]
- Leandro, P.; Lechner, M.C.; Tavares de Almeida, I.; Konecki, D. Glycerol Increases the Yield and Activity of Human Phenylalanine Hydroxylase Mutant Enzymes Produced in a Prokaryotic Expression System. Mol. Genet. Metab. 2001, 73, 173–178. [Google Scholar] [CrossRef]

| Primer | 5′-Sequence-3′ | Restriction Endonuclease |
|---|---|---|
| PcnB-Eco-F1 | CACACCATATGTACAGGCTCAATAAAGCGGG | NdeI |
| PcnB-Eco-R1 | CACACCTCGAGCGACGTGGTGCGCG | XhoI |
| pET-F | CCTATAGTGAGTCGTATTAATTTC | |
| pET-R | CAACTCAGCTTCCTTTCGG | |
| Eco20-F | GTTCGAGAAGAAACTCGAAGC | |
| Eco20-R | CCAGATATACCTTAACAATCTTCAG | |
| Eco20-P | FAM-CGGTGCCAGATCGTCGCCC-BHQ1 | |
| CANP-F | ATTCTTCTAATACCTGGAATGCTGT | |
| CANP-R | TTTATCCGTACTCCTGATGATGC | |
| CANP-P | ROX-CGGGGATCGCAGTGGTGAGT-BHQ2 |
| Strain | Lineage | Genotype |
|---|---|---|
| BL21 (DE3) pLysS | BL21 | F− ompT gal dcm lon hsdSB(rB− mB−) λ(DE3 [lacI lacUV5-T7p07 ind1 sam7 nin5]) [malB+]K-12(λS) pLysS[T7p20 orip15A](CmR) |
| BL21 Gen-X (DE3) | BL21 | F− ompT hsdSB (rB− mB−) gal dcm (DE3) |
| Lemo21 (DE3) | BL21 | fhuA2 [lon] ompT gal (λ DE3) [dcm] ∆hsdS/pLemo(CamR) λ DE3 = λ sBamHIo ∆EcoRI-B int::(lacI::PlacUV5::T7 gene1) i21 ∆nin5 pLemo = pACYC184-PrhaBAD-lysY |
| SoluBL21 (DE3) | BL21 | F− ompT hsdSB (rB− mB−) gal dcm (DE3) |
| Shuffle T7 | K-12 | F′ lac, pro, lacIq/Δ(ara-leu)7697 araD139 fhuA2 lacZ::T7 gene1 Δ(phoA)PvuII phoR ahpC galE (or U) galK λatt::pNEB3-r1-cDsbC (SpecR, lacIq) ΔtrxB rpsL150(StrR) Δgor Δ(malF)3 |
| Rosetta 2 (DE3) | BL21 | F− ompT hsdSB(rB− mB−) gal dcm (DE3) pRARE2 (CamR) |
| Rosetta Blue (DE3) | K-12 | endA1 hsdR17 (rK12− mK12+) supE44 thi-1 recA1 gyrA96 relA1 lac (DE3) F′[proA+B+ lacIqZΔM15::Tn10] pRARE (CamR, TetR) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oscorbin, I.P.; Kunova, M.S.; Filipenko, M.L. Optimization of Conditions for Production of Soluble E. coli Poly(A)-Polymerase for Biotechnological Applications. Biology 2025, 14, 48. https://doi.org/10.3390/biology14010048
Oscorbin IP, Kunova MS, Filipenko ML. Optimization of Conditions for Production of Soluble E. coli Poly(A)-Polymerase for Biotechnological Applications. Biology. 2025; 14(1):48. https://doi.org/10.3390/biology14010048
Chicago/Turabian StyleOscorbin, Igor P., Maria S. Kunova, and Maxim L. Filipenko. 2025. "Optimization of Conditions for Production of Soluble E. coli Poly(A)-Polymerase for Biotechnological Applications" Biology 14, no. 1: 48. https://doi.org/10.3390/biology14010048
APA StyleOscorbin, I. P., Kunova, M. S., & Filipenko, M. L. (2025). Optimization of Conditions for Production of Soluble E. coli Poly(A)-Polymerase for Biotechnological Applications. Biology, 14(1), 48. https://doi.org/10.3390/biology14010048







