DICER1: The Argonaute Endonuclease Family Member and Its Role in Pediatric and Youth Pathology
Simple Summary
Abstract
1. Introduction
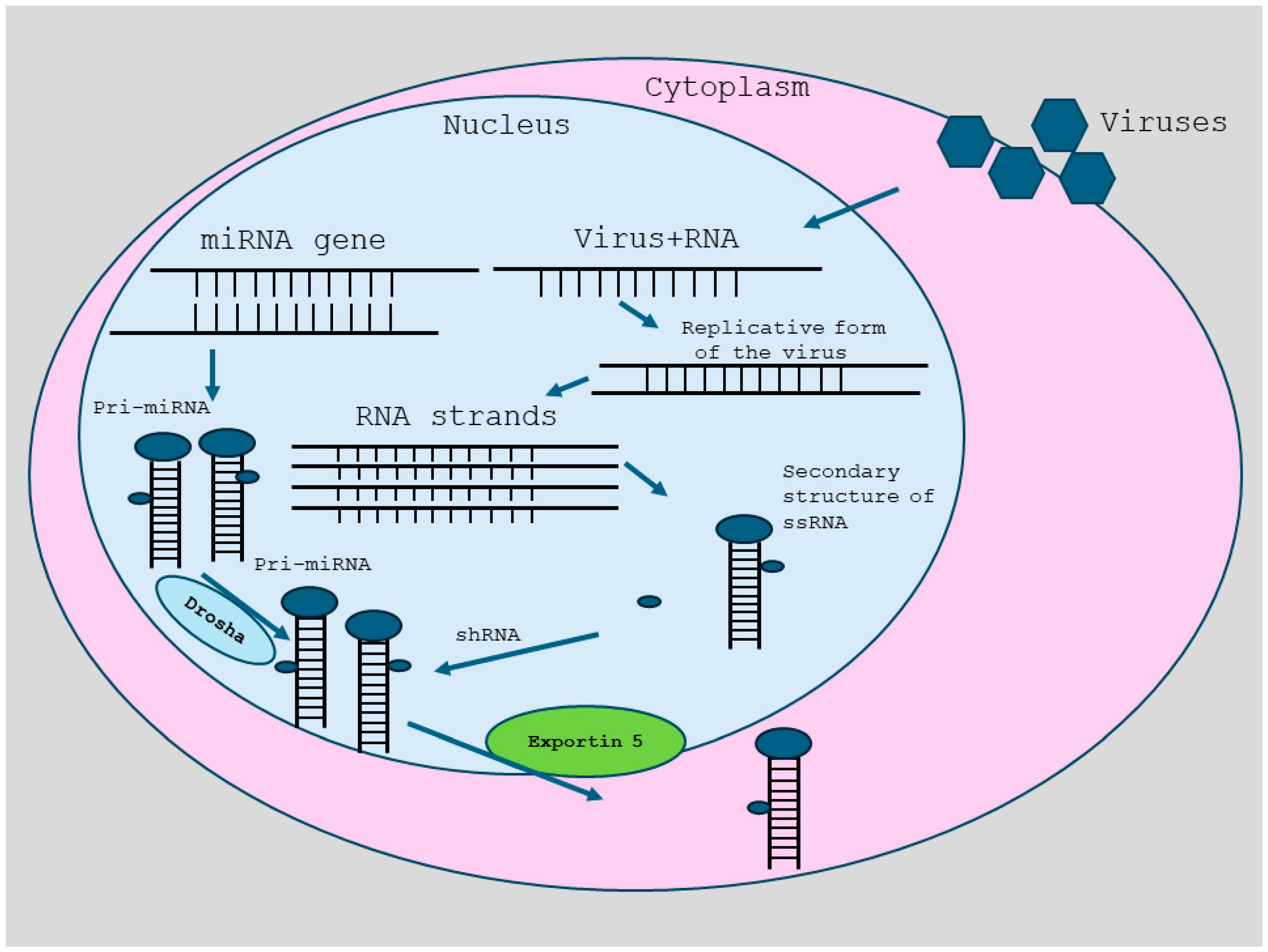
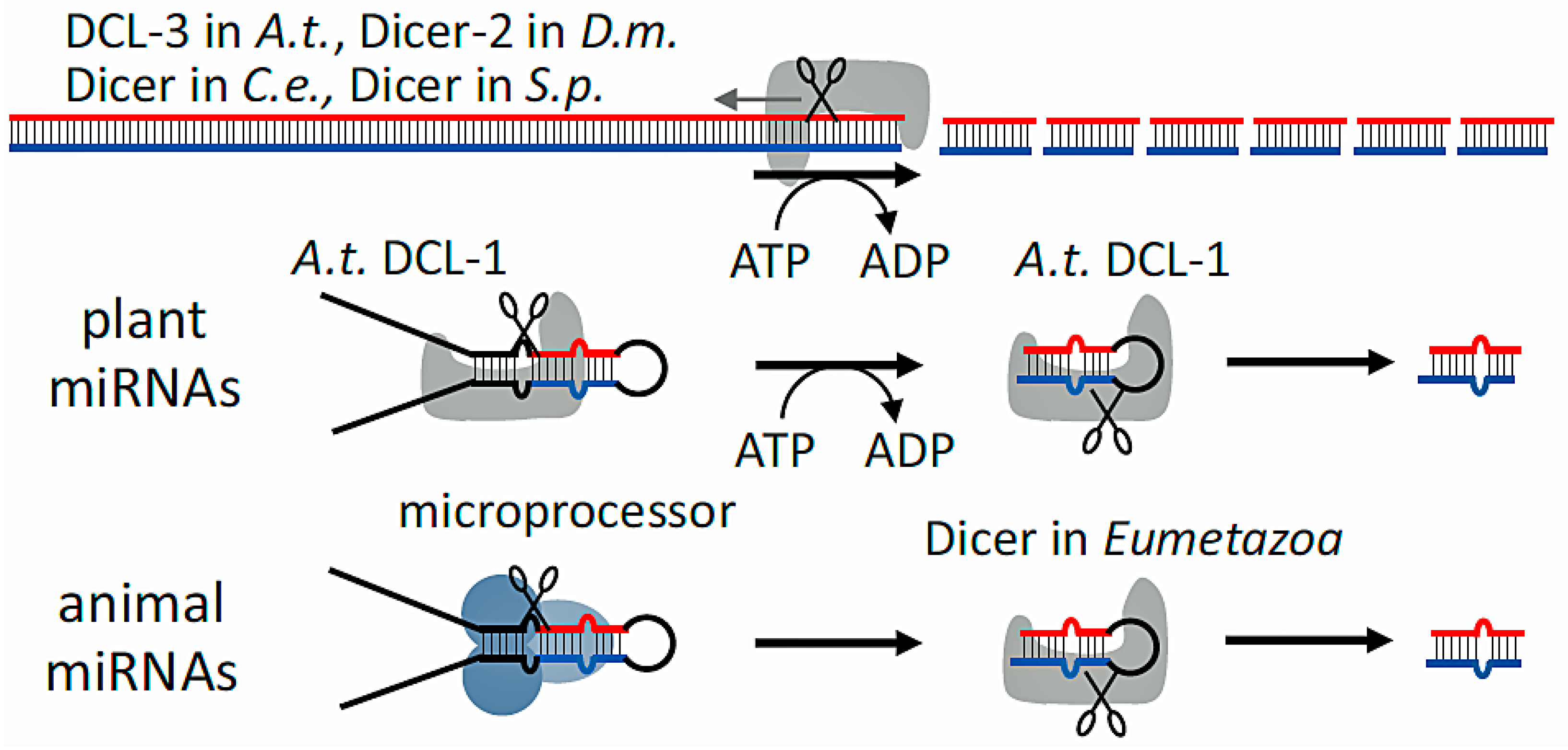
2. RNA Interference
3. Antiviral Properties
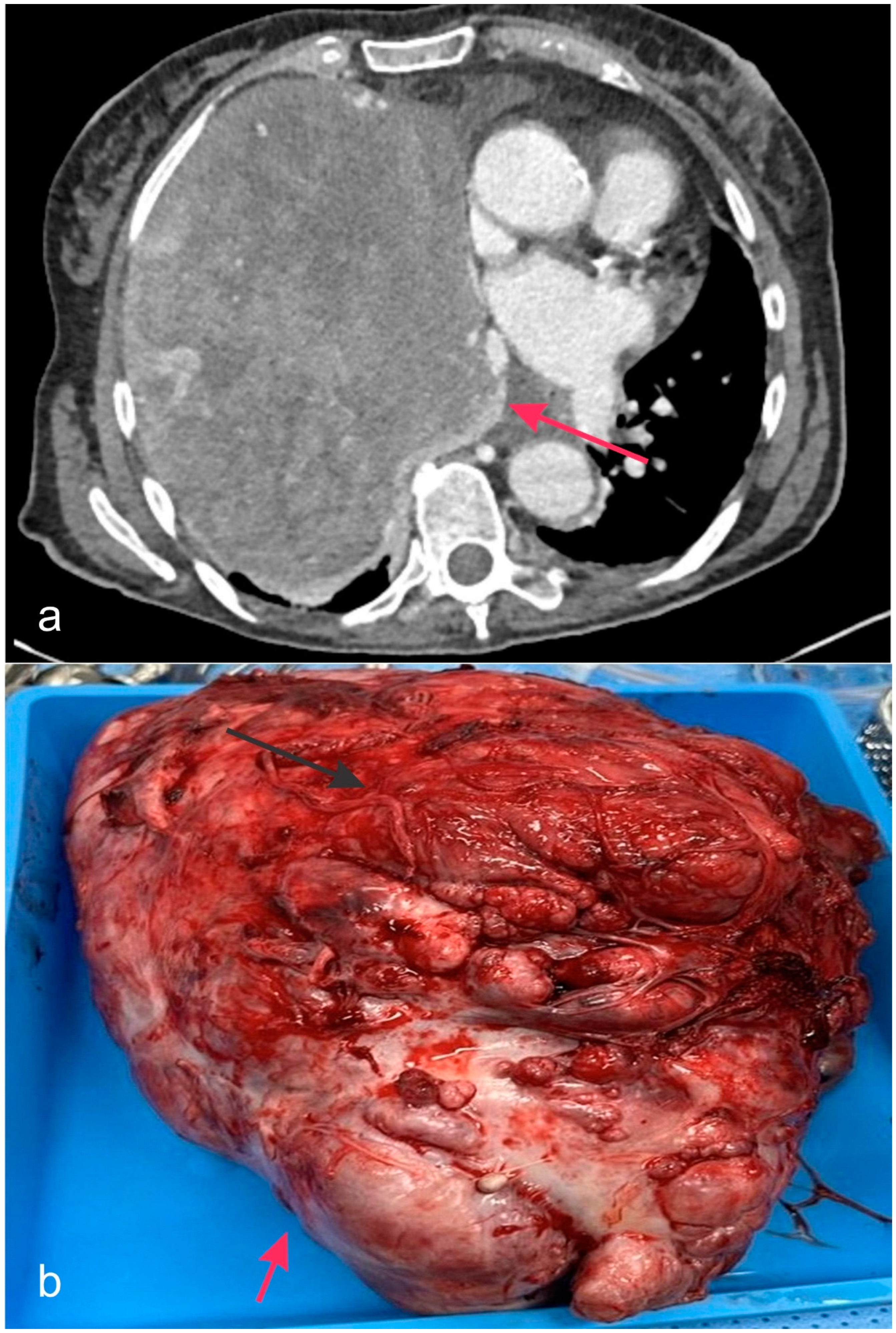
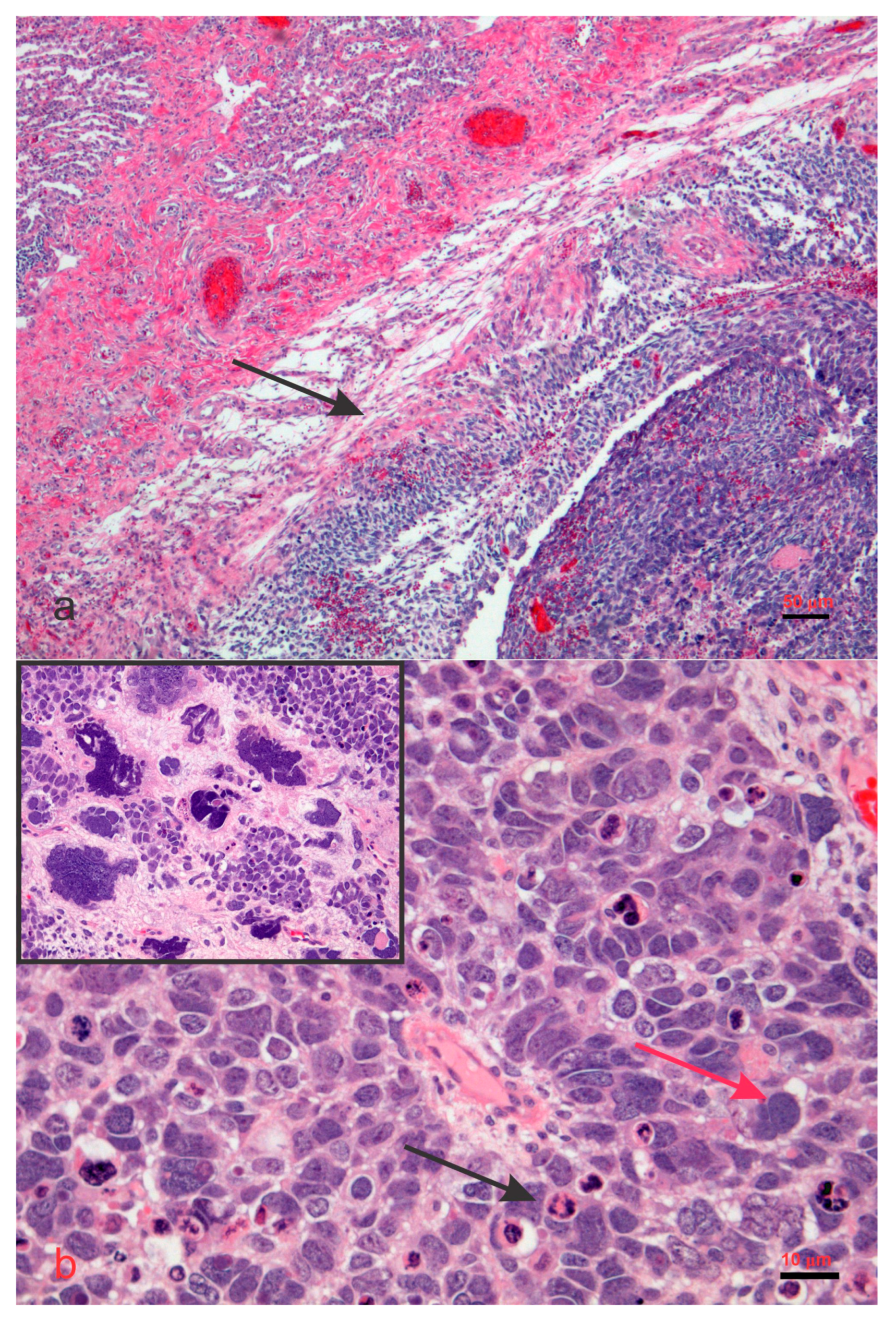
4. DICER1 in Human Disease and Its Pathobiology
5. Disorders Associated with DICER1 Gene Mutations
6. DICER1 Syndrome
7. GLOW Syndrome
8. Assessing for DICER1 Gene Mutations: Timing and Methodology
9. DICER and Therapies
9.1. The Two-Hit Hypothesis: Insights into Molecular Mechanisms of DICER1 Mutations
9.2. Limitations
9.3. Future Directions
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, D.; Rivera, B.; Fabian, M.R.; Foulkes, W.D. miRNA biogenesis and inherited disorders: Clinico-molecular insights. Trends Genet. 2023, 39, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Zapletal, D.; Kubicek, K.; Svoboda, P.; Stefl, R. Dicer structure and function: Conserved and evolving features. EMBO Rep. 2023, 24, e57215. [Google Scholar] [CrossRef] [PubMed]
- Doench, J.G.; Fusi, N.; Sullender, M.; Hegde, M.; Vaimberg, E.W.; Donovan, K.F.; Smith, I.; Tothova, Z.; Wilen, C.; Orchard, R.; et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Silvestri, A.; Bansal, C.; Rubio-Somoza, I. After silencing suppression: miRNA targets strike back. Trends Plant Sci. 2024, 29, 1266–1276. [Google Scholar] [CrossRef] [PubMed]
- Nadhan, R.; Isidoro, C.; Song, Y.S.; Dhanasekaran, D.N. LncRNAs and the cancer epigenome: Mechanisms and therapeutic potential. Cancer Lett. 2024, 605, 217297. [Google Scholar] [CrossRef]
- Quilez-Molina, A.I.; Niño Sanchez, J.; Merino, D. The role of polymers in enabling RNAi-based technology for sustainable pest management. Nat. Commun. 2024, 15, 9158. [Google Scholar] [CrossRef] [PubMed]
- Anwar, A.A.; Jalan-Sakrikar, N.; Huebert, R.C. LncRNAs, RNA Therapeutics, and Emerging Technologies in Liver Pathobiology. Semin. Liver Dis. 2024. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.B.; Harrison, P.T.; Scholefield, J. Prime editing: Therapeutic advances and mechanistic insights. Gene Ther. 2024. [Google Scholar] [CrossRef] [PubMed]
- Koralewska, N.; Szczepanska, A.; Ciechanowska, K.; Wojnicka, M.; Pokornowska, M.; Milewski, M.C.; Gudanis, D.; Baranowski, D.; Nithin, C.; Bujnicki, J.M.; et al. RNA and DNA G-quadruplexes bind to human dicer and inhibit its activity. Cell Mol. Life Sci. 2021, 78, 3709–3724. [Google Scholar] [CrossRef] [PubMed]
- Pokornowska, M.; Milewski, M.C.; Ciechanowska, K.; Szczepanska, A.; Wojnicka, M.; Radogostowicz, Z.; Figlerowicz, M.; Kurzynska-Kokorniak, A. The RNA-RNA base pairing potential of human Dicer and Ago2 proteins. Cell Mol. Life Sci. 2020, 77, 3231–3244. [Google Scholar] [CrossRef] [PubMed]
- Szczepanska, A.; Wojnicka, M.; Kurzynska-Kokorniak, A. The Significance of the DUF283 Domain for the Activity of Human Ribonuclease Dicer. Int. J. Mol. Sci. 2021, 22, 8690. [Google Scholar] [CrossRef] [PubMed]
- Lau, P.W.; Guiley, K.Z.; De, N.; Potter, C.S.; Carragher, B.; MacRae, I.J. The molecular architecture of human Dicer. Nat. Struct. Mol. Biol. 2012, 19, 436–440. [Google Scholar] [CrossRef] [PubMed]
- Lau, P.W.; Potter, C.S.; Carragher, B.; MacRae, I.J. Structure of the human Dicer-TRBP complex by electron microscopy. Structure 2009, 17, 1326–1332. [Google Scholar] [CrossRef] [PubMed]
- Aryal, N.K.; Pant, V.; Wasylishen, A.R.; Rimel, B.J.; Baseler, L.; El-Naggar, A.K.; Mutch, D.G.; Goodfellow, P.J.; Arur, S.; Lozano, G. Dicer1 Phosphomimetic Promotes Tumor Progression and Dissemination. Cancer Res. 2019, 79, 2662–2668. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.F.; Ren, H. The role of dicer in DNA damage repair. Int. J. Mol. Sci. 2012, 13, 16769–16778. [Google Scholar] [CrossRef] [PubMed]
- Theotoki, E.I.; Pantazopoulou, V.I.; Georgiou, S.; Kakoulidis, P.; Filippa, V.; Stravopodis, D.J.; Anastasiadou, E. Dicing the Disease with Dicer: The Implications of Dicer Ribonuclease in Human Pathologies. Int. J. Mol. Sci. 2020, 21, 7223. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Matskevich, A.A.; Moelling, K. Stimuli-dependent cleavage of Dicer during apoptosis. Biochem. J. 2008, 412, 527–534. [Google Scholar] [CrossRef]
- Bogerd, H.P.; Whisnant, A.W.; Kennedy, E.M.; Flores, O.; Cullen, B.R. Derivation and characterization of Dicer- and microRNA-deficient human cells. RNA 2014, 20, 923–937. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lu, R.; Yang, Q.; He, J.; Huang, C.; Cao, Y.; Zhou, Z.; Huang, J.; Li, L.; Chen, R.; et al. USP7 reduces the level of nuclear DICER, impairing DNA damage response and promoting cancer progression. Mol. Oncol. 2024, 18, 170–189. [Google Scholar] [CrossRef] [PubMed]
- Francia, S.; Cabrini, M.; Matti, V.; Oldani, A.; d’Adda di Fagagna, F. DICER, DROSHA and DNA damage response RNAs are necessary for the secondary recruitment of DNA damage response factors. J. Cell Sci. 2016, 129, 1468–1476. [Google Scholar] [CrossRef]
- Garnier, N.; Sane, F.; Massara, L.; Soncin, F.; Gosset, P.; Hober, D.; Szunerits, S.; Engelmann, I. Genes Involved in miRNA Biogenesis Are Not Downregulated in SARS-CoV-2 Infection. Viruses 2023, 15, 1177. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.; Huang, M.; Li, T. Screening the Potential Biomarkers of COVID-19-Related Thrombosis Through Bioinformatics Analysis. Front. Genet. 2022, 13, 889348. [Google Scholar] [CrossRef] [PubMed]
- Keikha, R.; Hashemi-Shahri, S.M.; Jebali, A. The relative expression of miR-31, miR-29, miR-126, and miR-17 and their mRNA targets in the serum of COVID-19 patients with different grades during hospitalization. Eur. J. Med. Res. 2021, 26, 75. [Google Scholar] [CrossRef] [PubMed]
- Song, M.S.; Rossi, J.J. Molecular mechanisms of Dicer: Endonuclease and enzymatic activity. Biochem. J. 2017, 474, 1603–1618. [Google Scholar] [CrossRef]
- Borowczyk, M.; Sypniewski, M.; Szyda, J.; Braszka, M.; Ziemnicka, K.; Ruchala, M.; Oszywa, M.; Krol, Z.J.; Dobosz, P. Genetic predisposition to differentiated thyroid cancer among Polish population. Pol. Arch. Intern. Med. 2024, 134, 16654. [Google Scholar] [CrossRef] [PubMed]
- Riascos, M.C.; Huynh, A.; Faquin, W.C.; Nose, V. Expanding Our Knowledge of DICER1 Gene Alterations and Their Role in Thyroid Diseases. Cancers 2024, 16, 347. [Google Scholar] [CrossRef]
- Yang, B.; Chour, W.; Salazar, C.G.; Zamiara, P.; Schmidt, R.J.; Raca, G.; Shillingford, N.; Zhou, S.; Warren, M.; Parham, D.M.; et al. Pediatric Sertoli-Leydig Cell Tumors of the Ovary: An Integrated Study of Clinicopathological Features, Pan-cancer-Targeted Next-generation Sequencing and Chromosomal Microarray Analysis From a Single Institution. Am. J. Surg. Pathol. 2024, 48, 194–203. [Google Scholar] [CrossRef]
- Bug, D.S.; Moiseev, I.S.; Porozov, Y.B.; Petukhova, N.V. Shedding light on the DICER1 mutational spectrum of uncertain significance in malignant neoplasms. Front. Mol. Biosci. 2024, 11, 1441180. [Google Scholar] [CrossRef]
- Del Baldo, G.; Mastronuzzi, A.; Cipri, S.; Agolini, E.; Matraxia, M.; Novelli, A.; Cacchione, A.; Serra, A.; Carai, A.; Boccuto, L.; et al. The coexistence of a BRCA2 germline and a DICER1 somatic variant in two first-degree cousins suggests their potential synergic effect. Sci. Rep. 2024, 14, 21435. [Google Scholar] [CrossRef] [PubMed]
- Llorens, F.; Banez-Coronel, M.; Pantano, L.; del Rio, J.A.; Ferrer, I.; Estivill, X.; Marti, E. A highly expressed miR-101 isomiR is a functional silencing small RNA. BMC Genom. 2013, 14, 104. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, H.; Dridi, S.; Tarallo, V.; Gelfand, B.D.; Fowler, B.J.; Cho, W.G.; Kleinman, M.E.; Ponicsan, S.L.; Hauswirth, W.W.; Chiodo, V.A.; et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 2011, 471, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Saeki, M.; Watanabe, M.; Inoue, N.; Tokiyoshi, E.; Takuse, Y.; Arakawa, Y.; Hidaka, Y.; Iwatani, Y. DICER and DROSHA gene expression and polymorphisms in autoimmune thyroid diseases. Autoimmunity 2016, 49, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Ojcius, D.M.; Jafari, A.; Yeruva, L.; Schindler, C.W.; Abdul-Sater, A.A. Dicer regulates activation of the NLRP3 inflammasome. PLoS ONE 2019, 14, e0215689. [Google Scholar] [CrossRef]
- De Cauwer, A.; Mariotte, A.; Sibilia, J.; Bahram, S.; Georgel, P. DICER1: A Key Player in Rheumatoid Arthritis, at the Crossroads of Cellular Stress, Innate Immunity, and Chronic Inflammation in Aging. Front. Immunol. 2018, 9, 1647. [Google Scholar] [CrossRef]
- Sergi, C.M. MASLD and aspartame: Are new studies in the horizon? Front. Med. 2023, 10, 1266918. [Google Scholar] [CrossRef] [PubMed]
- Sergi, C.M. NLRP-3 Inflammasome: A Key Target, but Mostly Overlooked following SARS-CoV-2 Infection. Vaccines 2022, 10, 1307. [Google Scholar] [CrossRef] [PubMed]
- Sergi, C.M.; Chiu, B. Targeting NLRP3 inflammasome in an animal model for Coronavirus Disease 2019 (COVID-19) caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J. Med. Virol. 2021, 93, 669–670. [Google Scholar] [CrossRef]
- Chiu, B.; Jantuan, E.; Shen, F.; Chiu, B.; Sergi, C. Autophagy-Inflammasome Interplay in Heart Failure: A Systematic Review on Basics, Pathways, and Therapeutic Perspectives. Ann. Clin. Lab. Sci. 2017, 47, 243–252. [Google Scholar] [PubMed]
- Kumar, M.S.; Pester, R.E.; Chen, C.Y.; Lane, K.; Chin, C.; Lu, J.; Kirsch, D.G.; Golub, T.R.; Jacks, T. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009, 23, 2700–2704. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.; Lee, H.; Ghahremani, S.; Kempert, P.; Ischander, M.; Teitell, M.A.; Nelson, S.F.; Martinez-Agosto, J.A. Expanding the phenotype of mutations in DICER1: Mosaic missense mutations in the RNase IIIb domain of DICER1 cause GLOW syndrome. J. Med. Genet. 2014, 51, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Canberk, S.; Correia, M.; Lima, A.R.; Bongiovanni, M.; Sobrinho-Simões, M.; Soares, P.; Máximo, V. The Multifaceted Profile of Thyroid Disease in the Background of DICER1 Germline and Somatic Mutations: Then, Now and Future Perspectives. J. Mol. Pathol. 2022, 3, 1–14. [Google Scholar] [CrossRef]
- Yuan, X.; Mu, N.; Wang, N.; Straat, K.; Sofiadis, A.; Guo, Y.; Stenman, A.; Li, K.; Cheng, G.; Zhang, L.; et al. GABPA inhibits invasion/metastasis in papillary thyroid carcinoma by regulating DICER1 expression. Oncogene 2019, 38, 965–979. [Google Scholar] [CrossRef] [PubMed]
- Dobrijevic, Z.; Stevanovic, J.; Sunderic, M.; Penezic, A.; Miljus, G.; Danilovic Lukovic, J.; Janjic, F.; Matijasevic Jokovic, S.; Brkusanin, M.; Savic-Pavicevic, D.; et al. Diagnostic properties of miR-146a-5p from liquid biopsies in prostate cancer: A meta-analysis. Pathol. Res. Pract. 2024, 262, 155522. [Google Scholar] [CrossRef]
- Condello, V.; Poma, A.M.; Macerola, E.; Vignali, P.; Paulsson, J.O.; Zedenius, J.; Basolo, F.; Juhlin, C.C. Prevalence, Molecular Landscape and Clinical Impact of DICER1 and DGCR8 Mutated Follicular-Patterned Thyroid Nodules. J. Clin. Endocrinol. Metab. 2024, 109, 1733–1744. [Google Scholar] [CrossRef]
- de Sousa, M.S.A.; Nunes, I.N.; Christiano, Y.P.; Sisdelli, L.; Cerutti, J.M. Genetic alterations landscape in paediatric thyroid tumours and/or differentiated thyroid cancer: Systematic review. Rev. Endocr. Metab. Disord. 2024, 25, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.K.; Liu, Z.; Hirokawa, M.; Bychkov, A. Histological clues of DICER1 mutations in thyroid nodules. Virchows Arch. 2024, 485, 755–757. [Google Scholar] [CrossRef] [PubMed]
- Thunders, M.; Delahunt, B. Gene of the month: DICER1: Ruler and controller. J. Clin. Pathol. 2021, 74, 69–72. [Google Scholar] [CrossRef]
- de Kock, L.; Wu, M.K.; Foulkes, W.D. Ten years of DICER1 mutations: Provenance, distribution, and associated phenotypes. Hum. Mutat. 2019, 40, 1939–1953. [Google Scholar] [CrossRef] [PubMed]
- Yndestad, S.; Haugland, H.K.; Goplen, D.; Wojcik, D.; Knappskog, S.; Lonning, P.E. Germline variants in patients diagnosed with pediatric soft tissue sarcoma. Acta. Oncol. 2024, 63, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Narin Tongal, S.; Yilmaz, O.; Ozguven, A.A.; Yuksel, H. DICER1 Syndrome: Coocurrence of Pleuropulmonary Blastoma and Cystic Nephroma. J. Bronchol. Interv. Pulmonol. 2024, 31, 94–97. [Google Scholar] [CrossRef] [PubMed]
- Berber Hamamci, M.; Yesil, S.; Bicakcioglu, P.; Gursoy, T.R.; Agackiran, Y.; Kurucu, B.; Kilci, A.C.; Gok, S.U.; Fettah, A.; Sahin, G. DICER1 syndrome with hepatoblastoma and pleuropulmonary blastoma. Pediatr. Blood Cancer 2024, 71, e31285. [Google Scholar] [CrossRef]
- Chernock, R.D.; Rivera, B.; Borrelli, N.; Hill, D.A.; Fahiminiya, S.; Shah, T.; Chong, A.S.; Aqil, B.; Mehrad, M.; Giordano, T.J.; et al. Poorly differentiated thyroid carcinoma of childhood and adolescence: A distinct entity characterized by DICER1 mutations. Mod. Pathol. 2020, 33, 1264–1274. [Google Scholar] [CrossRef] [PubMed]
- Schultz, K.A.P.; Nelson, A.T.; Mallinger, P.H.R.; Harris, A.K.; Kamihara, J.; Baldinger, S.; Chen, K.S.; Pond, D.; Hatton, J.N.; Dybvik, A.; et al. DICER1-Related Tumor Predisposition: Identification of At-risk Individuals and Recommended Surveillance Strategies. Clin. Cancer Res. 2024, 30, 5681–5692. [Google Scholar] [CrossRef] [PubMed]
- Sergi, C. Pathology of Childhood and Adolescence—An Illustrated Guide; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar] [CrossRef]
- Alfawaz, B.; Koujok, K.; Eamer, G.; Sergi, C.M. Mediastinal Teratoma with Nephroblastomatous Elements: Case Report, Literature Review, and Comparison with Maturing Fetal Glomerulogenic Zone/Definitive Zone Ratio and Nephrogenic Rests. Int. J. Mol. Sci. 2024, 25, 12427. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Feulefack, J.; Sergi, C.M. Exposure to pesticides and pediatric Wilms’ tumor. A meta-analysis on pre-conception and pregnancy parental exposure with an IARC/WHO commentary. Hum. Exp. Toxicol. 2022, 41, 9603271221136211. [Google Scholar] [CrossRef]
- Venger, K.; Elbracht, M.; Carlens, J.; Deutz, P.; Zeppernick, F.; Lassay, L.; Kratz, C.; Zenker, M.; Kim, J.; Stewart, D.R.; et al. Unusual phenotypes in patients with a pathogenic germline variant in DICER1. Fam. Cancer 2023, 22, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Ponten, E.; Frisk, S.; Taylan, F.; Vaz, R.; Wessman, S.; de Kock, L.; Pal, N.; Foulkes, W.D.; Lagerstedt-Robinson, K.; Nordgren, A. A complex DICER1 syndrome phenotype associated with a germline pathogenic variant affecting the RNase IIIa domain of DICER1. J. Med. Genet. 2022, 59, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Thiemann, I.; Hoppen, T.; Laur, M.; Ohlert, M.; Lobitz, S.; Schwerk, N.; Dingemann, J.; Langer, F.; Grychtol, R.M.; Nusslein, T. Complete Tracheal Ring Deformity, Recurrent Pneumothoraces and Pleuropulmonary Blastoma in a Child: Coincidence or Common Genetic Cause? Klin. Padiatr. 2022, 234, 311–313. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.D.; Martinez-Agosto, J.A. Hotspot Mutations in DICER1 Causing GLOW Syndrome-Associated Macrocephaly via Modulation of Specific microRNA Populations Result in the Activation of PI3K/ATK/mTOR Signaling. Microrna 2020, 9, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Rojo-Pardillo, M.; Godefroid, L.; Dom, G.; Lefort, A.; Libert, F.; Robaye, B.; Maenhaut, C. Understanding the Dosage-Dependent Role of Dicer1 in Thyroid Tumorigenesis. Int. J. Mol. Sci. 2024, 25, 10701. [Google Scholar] [CrossRef] [PubMed]
- Mastnikova, K.; Bulanova Pekova, B.; Kuklikova, V.; Vaclavikova, E.; Carkova, J.; Katra, R.; Fialova, L.; Vlcek, P.; Kodetova, D.; Chovanec, M.; et al. DICER1 Variants in Pediatric and Young Adult Thyroid Nodules. Thyroid 2024, 34, 1225–1233. [Google Scholar] [CrossRef] [PubMed]
- Chong, A.S.; Nikiforov, Y.E.; Condello, V.; Wald, A.I.; Nikiforova, M.N.; Foulkes, W.D.; Rivera, B. Prevalence and Spectrum of DICER1 Mutations in Adult-onset Thyroid Nodules with Indeterminate Cytology. J. Clin. Endocrinol. Metab. 2021, 106, 968–977. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Moya, J.; Wert-Lamas, L.; Riesco-Eizaguirre, G.; Santisteban, P. Impaired microRNA processing by DICER1 downregulation endows thyroid cancer with increased aggressiveness. Oncogene 2019, 38, 5486–5499. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.; Villanueva, A.; Moutinho, C.; Davalos, V.; Spizzo, R.; Ivan, C.; Rossi, S.; Setien, F.; Casanovas, O.; Simo-Riudalbas, L.; et al. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc. Natl. Acad. Sci. USA 2011, 108, 4394–4399. [Google Scholar] [CrossRef] [PubMed]
- Sergi, C.M. Epigallocatechin gallate for Parkinson’s disease. Clin. Exp. Pharmacol. Physiol. 2022, 49, 1029–1041. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, A.M.; Pyles, H.M.; Diaz-Cruz, E.S.; Barton, C.E. Enoxacin and Epigallocatechin Gallate (EGCG) Act Synergistically to Inhibit the Growth of Cervical Cancer Cells in Culture. Molecules 2019, 24, 1580. [Google Scholar] [CrossRef]
- Nordling, C. A new theory on the cancer-inducing mechanism. Br. J. Cancer 1953, 7, 68. [Google Scholar] [CrossRef]
- Knudson Jr, A.G. Mutation and cancer: Statistical study of retinoblastoma. Proc. Natl. Acad. Sci. USA 1971, 68, 820–823. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.; Eyre, H.; Jacka, F.N.; Dodd, S.; Dean, O.; McEwen, S.; Debnath, M.; McGrath, J.; Maes, M.; Amminger, P. A review of vulnerability and risks for schizophrenia: Beyond the two hit hypothesis. Neurosci. Biobehav. Rev. 2016, 65, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Berthon, A.; Faucz, F.; Bertherat, J.; Stratakis, C.A. Analysis of ARMC5 expression in human tissues. Mol. Cell. Endocrinol. 2017, 441, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Hill, D.A.; Ivanovich, J.; Priest, J.R.; Gurnett, C.A.; Dehner, L.P.; Desruisseau, D.; Jarzembowski, J.A.; Wikenheiser-Brokamp, K.A.; Suarez, B.K.; Whelan, A.J.; et al. DICER1 mutations in familial pleuropulmonary blastoma. Science 2009, 325, 965. [Google Scholar] [CrossRef]
- Canfarotta, M.; Riba-Wolman, R.; Orsey, A.D.; Balarezo, F.; Finck, C. DICER1 syndrome and thyroid disease. J. Pediatr. Surg. Case Rep. 2016, 11, 31–34. [Google Scholar] [CrossRef]
- Wu, M.K.; Sabbaghian, N.; Xu, B.; Addidou-Kalucki, S.; Bernard, C.; Zou, D.; Reeve, A.E.; Eccles, M.R.; Cole, C.; Choong, C.S.; et al. Biallelic DICER1 mutations occur in Wilms tumours. J. Pathol. 2013, 230, 154–164. [Google Scholar] [CrossRef]
- Anglesio, M.S.; Wang, Y.; Yang, W.; Senz, J.; Wan, A.; Heravi-Moussavi, A.; Salamanca, C.; Maines-Bandiera, S.; Huntsman, D.G.; Morin, G.B. Cancer-associated somatic DICER1 hotspot mutations cause defective miRNA processing and reverse-strand expression bias to predominantly mature 3p strands through loss of 5p strand cleavage. J. Pathol. 2013, 229, 400–409. [Google Scholar] [CrossRef] [PubMed]
- de Kock, L.; Bah, I.; Revil, T.; Berube, P.; Wu, M.K.; Sabbaghian, N.; Priest, J.R.; Ragoussis, J.; Foulkes, W.D. Deep Sequencing Reveals Spatially Distributed Distinct Hot Spot Mutations in DICER1-Related Multinodular Goiter. J. Clin. Endocrinol. Metab. 2016, 101, 3637–3645. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Yoshida, K.; Shiraishi, Y.; Shimamura, T.; Sato, Y.; Nishimura, R.; Okuno, Y.; Chiba, K.; Tanaka, H.; Kato, K.; et al. Biallelic DICER1 mutations in sporadic pleuropulmonary blastoma. Cancer Res. 2014, 74, 2742–2749. [Google Scholar] [CrossRef] [PubMed]
- Melendez-Zajgla, J.; Mercado-Celis, G.E.; Gaytan-Cervantes, J.; Torres, A.; Gabino, N.B.; Zapata-Tarres, M.; Juarez-Villegas, L.E.; Lezama, P.; Maldonado, V.; Ruiz-Monroy, K.; et al. Genomics of a pediatric ovarian fibrosarcoma. Association with the DICER1 syndrome. Sci. Rep. 2018, 8, 3252. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Martínez, L.; Villegas, J.A.; Santamaría, Í.; Pitiot, A.S.; Alvarado, M.G.; Fernández, S.; Torres, H.; Paredes, Á.; Blay, P.; Balbín, M. Identification of somatic and germ-line DICER1 mutations in pleuropulmonary blastoma, cystic nephroma and rhabdomyosarcoma tumors within a DICER1 syndrome pedigree. BMC Cancer 2017, 17, 146. [Google Scholar] [CrossRef] [PubMed]
- Brenneman, M.; Field, A.; Yang, J.; Williams, G.; Doros, L.; Rossi, C.; Schultz, K.A.; Rosenberg, A.; Ivanovich, J.; Turner, J.; et al. Temporal order of RNase IIIb and loss-of-function mutations during development determines phenotype in pleuropulmonary blastoma/DICER1 syndrome: A unique variant of the two-hit tumor suppression model. F1000Research 2015, 4, 214. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, Y.; McMonechy, M.K.; Anglesio, M.S.; Yang, W.; Senz, J.; Maines-Bandiera, S.; Rosner, J.; Trigo-Gonzalez, G.; Grace Cheng, S.W.; et al. Recurrent DICER1 hotspot mutations in endometrial tumours and their impact on microRNA biogenesis. J. Pathol. 2015, 237, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Solarski, M.; Rotondo, F.; Foulkes, W.D.; Priest, J.R.; Syro, L.V.; Butz, H.; Cusimano, M.D.; Kovacs, K. DICER1 gene mutations in endocrine tumors. Endocr. Relat. Cancer 2018, 25, R197–R208. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.; Greer, R.; Russ, H.; von Figura, G.; Kim, G.; Busch, A.; Lee, J.; Hertel, K.; Kim, S.; McManus, M. Dicer regulates differentiation and viability during mouse pancreatic cancer initiation. PLoS ONE 2014, 9, e95486. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.J.; McAllister, F.; Bailey, J.M.; Scott, S.G.; Hendley, A.M.; Leach, S.D.; Ghosh, B. Dicer is required for maintenance of adult pancreatic acinar cell identity and plays a role in Kras-driven pancreatic neoplasia. PLoS ONE 2014, 9, e113127. [Google Scholar] [CrossRef] [PubMed]
- Eser, S.; Schnieke, A.; Schneider, G.; Saur, D. Oncogenic KRAS signalling in pancreatic cancer. Br. J. Cancer 2014, 111, 817–822. [Google Scholar] [CrossRef] [PubMed]
- Criscimanna, A.; Speicher, J.; Houshmand, G.; Shiota, C.; Prasadan, K.; Ji, B.; Logsdon, C.; Gittes, G.; Esni, F. Duct cells contribute to regeneration of endocrine and acinar cells following pancreatic damage in adult mice. Gastroenterology 2011, 141, 1451–1462. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, J.; Huang, J.; Tang, H.; Yu, S.; Chen, Y. The regulatory roles of miRNA and methylation on oncogene and tumor suppressor gene expression in pancreatic cancer cells. Biochem. Biophys. Res. Commun. 2012, 425, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Blandino, G.; Valerio, M.; Cioce, M.; Mori, F.; Casadei, L.; Pulito, C.; Sacconi, A.; Biagioni, F.; Cortese, G.; Galanti, S. Metformin elicits anticancer effects through the sequential modulation of DICER and c-MYC. Nat. Commun. 2012, 3, 865. [Google Scholar] [CrossRef]
- Noren Hooten, N.; Martin-Montalvo, A.; Dluzen, D.F.; Zhang, Y.; Bernier, M.; Zonderman, A.B.; Becker, K.G.; Gorospe, M.; de Cabo, R.; Evans, M.K. Metformin-mediated increase in DICER1 regulates microRNA expression and cellular senescence. Aging Cell 2016, 15, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Yang, Y.; Huang, Y.; Chen, Y.; Wang, T.; Wu, S.; Tong, L.; Wang, Y.; Lin, L.; Hao, M.; et al. RNA-binding protein AUF1 suppresses miR-122 biogenesis by down-regulating Dicer1 in hepatocellular carcinoma. Oncotarget 2018, 9, 14815–14827. [Google Scholar] [CrossRef]
- Oxford, A.E.; Jorcyk, C.L.; Oxford, J.T. Neuropathies of Stüve-Wiedemann Syndrome due to mutations in leukemia inhibitory factor receptor (LIFR) gene. J. Neurol. Neuromed. 2016, 1, 37. [Google Scholar] [CrossRef]
- Welch, E.M.; Barton, E.R.; Zhuo, J.; Tomizawa, Y.; Friesen, W.J.; Trifillis, P.; Paushkin, S.; Patel, M.; Trotta, C.R.; Hwang, S. PTC124 targets genetic disorders caused by nonsense mutations. Nature 2007, 447, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Pibiri, I.; Lentini, L.; Melfi, R.; Gallucci, G.; Pace, A.; Spinello, A.; Barone, G.; Di Leonardo, A. Enhancement of premature stop codon readthrough in the CFTR gene by Ataluren (PTC124) derivatives. Eur. J. Med. Chem. 2015, 101, 236–244. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2021, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]


| Time | Main Mechanism(s) |
|---|---|
| Cell cycle kinetics, Caspase 3 activation and apoptosis |
| Cell cycle kinetics |
| Cell cycle kinetics |
| Cell cycle kinetics |
| Caspase 3 activation and apoptosis (programmed cell death)) |
| B lymphocytic cellular function, Stress granule response |
| Cell cycle kinetics |
| Tumor | DICER1 Alteration (Red = Down-Regulation/Green = Up-Regulation) |
|---|---|
| NSCLC | |
| CRC | |
| Prostate Ca | |
| Breast Ca | |
| ALL | |
| Ovary Ca | |
| Ovary Ca | |
| Thyroid Ca | |
| Thyroid Ca |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sergi, C.M.; Minervini, F. DICER1: The Argonaute Endonuclease Family Member and Its Role in Pediatric and Youth Pathology. Biology 2025, 14, 93. https://doi.org/10.3390/biology14010093
Sergi CM, Minervini F. DICER1: The Argonaute Endonuclease Family Member and Its Role in Pediatric and Youth Pathology. Biology. 2025; 14(1):93. https://doi.org/10.3390/biology14010093
Chicago/Turabian StyleSergi, Consolato M., and Fabrizio Minervini. 2025. "DICER1: The Argonaute Endonuclease Family Member and Its Role in Pediatric and Youth Pathology" Biology 14, no. 1: 93. https://doi.org/10.3390/biology14010093
APA StyleSergi, C. M., & Minervini, F. (2025). DICER1: The Argonaute Endonuclease Family Member and Its Role in Pediatric and Youth Pathology. Biology, 14(1), 93. https://doi.org/10.3390/biology14010093






