Bacterial Diversity at Himalayan Pink Salt Extraction Site
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Khewra Salt Mine
2.2. Sample Collection
2.3. Chemicals and Reagents
2.4. Physicochemical Examination of Soil and Brine Samples
2.5. Isolation of Halophilic Microorganisms
2.6. 16S rRNA Amplicon Sequencing
2.7. Sequence Analysis
2.8. Statistical Analysis
3. Results
3.1. Physiochemical Analysis
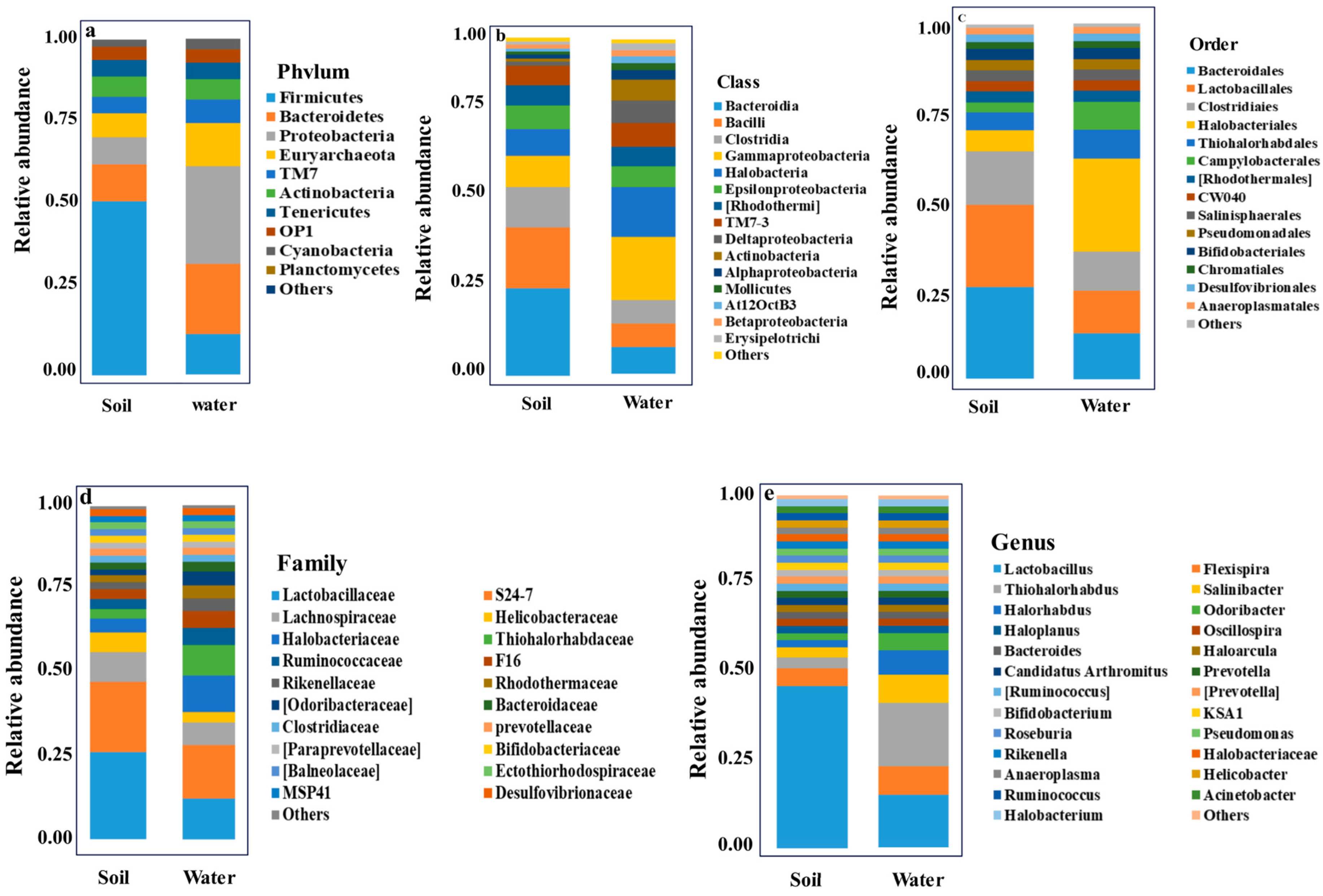
3.2. Taxonomic Analysis of Bacterial Diversity
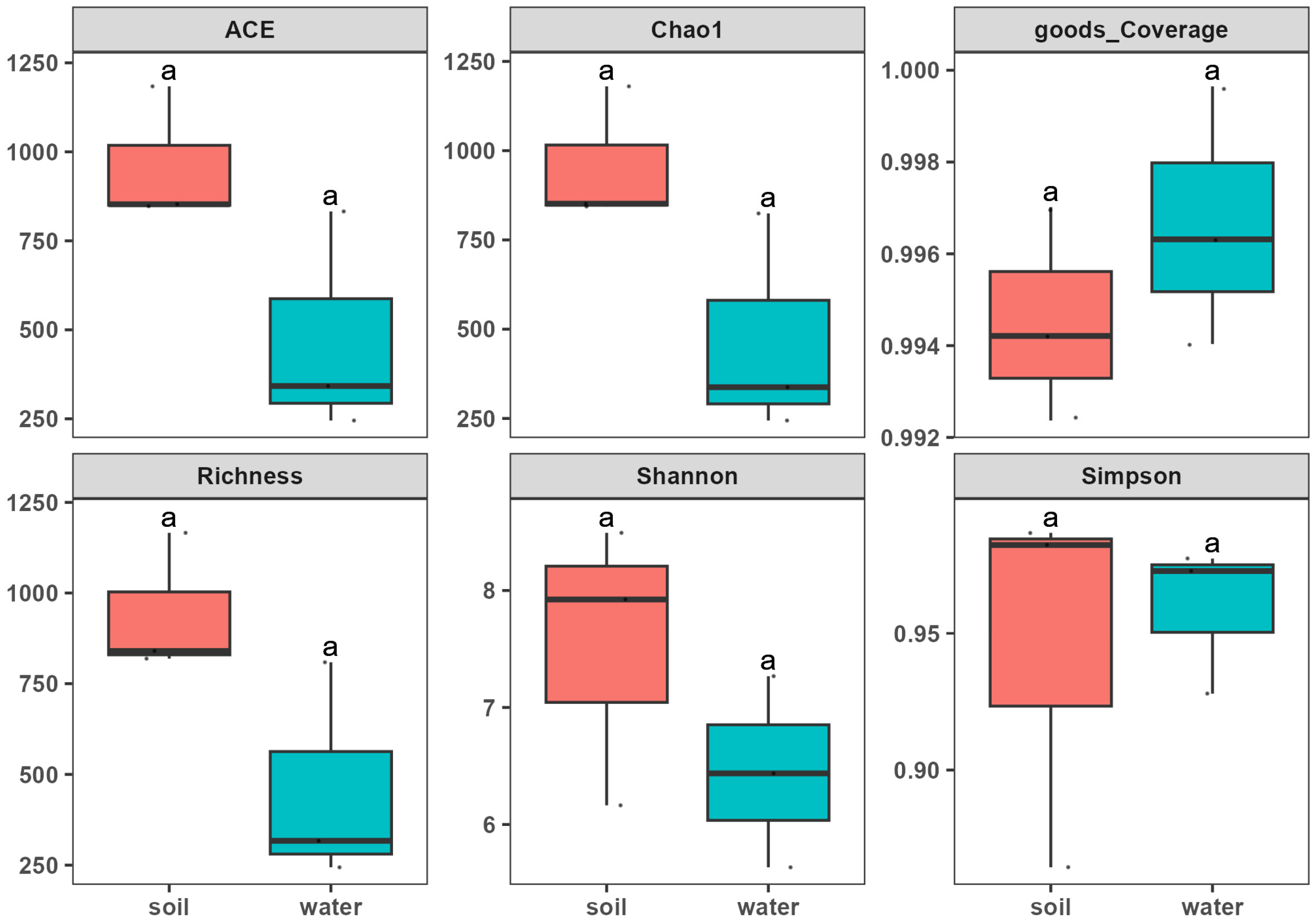
3.3. Alpha and Beta Diversity Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vidal, V.A.; Paglarini, C.S.; Lorenzo, J.M.; Munekata, P.E.; Pollonio, M.A. Salted meat products: Nutritional characteristics, processing and strategies for sodium reduction. Food Rev. Int. 2023, 39, 2183–2202. [Google Scholar] [CrossRef]
- dos Santos, M.; Triviño, A.P.R.; Barros, J.C.; da Cruz, A.G.; Pollonio, M.A.R. Strategies for the reduction of salt in food products. In Food Structure Engineering and Design for Improved Nutrition, Health and Well-Being; Elsevier: Amsterdam, The Netherlands, 2023; pp. 187–218. [Google Scholar]
- Antonites, A. Salt production, use, and trade. In Oxford Research Encyclopedia of Anthropology; Oxford University Press: Oxford, UK, 2020. [Google Scholar]
- Wismanu, R.E.; Prakasa, Y.; Wahyudi, L.E.; Nashihah, D. Stakeholders Collaboration to Stimulate the Economic Empowerment for Salt Farmers in Pamekasan Regency. J. Sos. Ekon. Kelaut. Dan Perikan. 2023, 18, 65–74. [Google Scholar] [CrossRef]
- Thompson, T.P.; Gilmore, B.F. Exploring halophilic environments as a source of new antibiotics. Crit. Rev. Microbiol. 2023, 50, 341–370. [Google Scholar] [CrossRef]
- Ali, N.; Nughman, M.; Shah, S.M. Extremophiles and Limits of Life in a Cosmic Perspective. In Life in Extreme Environments-Diversity, Adaptability and Valuable Resources of Bioactive Molecules; IntechOpen: London, UK, 2023. [Google Scholar]
- Malik, K.; Mehnaz, S.; Shah, H.Z. Characterization of Plant-Growth Promoting Archaea Associated with the Rhizosphere of Salsola stocksii and Atriplex. Authorea Prepr. 2023. [Google Scholar] [CrossRef]
- Favreau, C.; Tribondeau, A.; Marugan, M.; Guyot, F.; Marie, A.; Puppo, R.; Dufour, T.; Huguet, A.; Zirah, S.; Kish, A. Molecular acclimation of Halobacterium salinarum to halite brine inclusions. Front. Microbiol. 2023, 13, 1075274. [Google Scholar]
- Huby, T.J.; Clark, D.R.; McKew, B.A.; McGenity, T.J. Extremely halophilic archaeal communities are resilient to short-term entombment in halite. Environ. Microbiol. 2021, 23, 3370–3383. [Google Scholar] [CrossRef]
- Franchi, F.; Cassaro, A.; Cavalazzi, B.; Lebogang, L.; Tarozzi, A.; Pacelli, C. Microbial abundance across a salinity and mineralogical transect in the Ntwetwe Pan of Botswana: A terrestrial analogue for playa deposits on Mars. Planet. Space Sci. 2025, 255, 106028. [Google Scholar] [CrossRef]
- DasSarma, P.; Laye, V.; Harvey, J.; Reid, C.; Shultz, J.; Yarborough, A.; Lamb, A.; Koske-Phillips, A.; Herbst, A.; Molina, F. Survival of halophilic Archaea in Earth’s cold stratosphere. Int. J. Astrobiol. 2017, 16, 321–327. [Google Scholar] [CrossRef]
- Yoo, Y.; Lee, H.; Lee, J.; Khim, J.S.; Kim, J.-J. Insights into saline adaptation strategies through a novel halophilic bacterium isolated from solar saltern of Yellow sea. Front. Mar. Sci. 2023, 10, 1229444. [Google Scholar] [CrossRef]
- Li, J.; Gao, Y.; Dong, H.; Sheng, G.-P. Haloarchaea, excellent candidates for removing pollutants from hypersaline wastewater. Trends Biotechnol. 2022, 40, 226–239. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Y.; Ma, Y.; Wang, X.; Zhang, B.; Zhang, G.; Bahadur, A.; Chen, T.; Liu, G.; Zhang, W. Research progress regarding the role of halophilic and halotolerant microorganisms in the eco-environmental sustainability and conservation. J. Clean. Prod. 2023, 418, 138054. [Google Scholar] [CrossRef]
- Naik, B.; Kumar, V.; Goyal, S.; Tripati, A.D.; Khan, J.M.; Irfan, M.; Bhatt, S.C.; Gupta, A.K.; Rustagi, S. Production, characterization, and application of novel fungal pullulanase for fruit juice processing. Int. J. Biol. Macromol. 2023, 248, 125936. [Google Scholar]
- Dindhoria, K.; Manyapu, V.; Ali, A.; Kumar, R. Unveiling the role of emerging metagenomics for the examination of hypersaline environments. Biotechnol. Genet. Eng. Rev. 2024, 40, 2090–2128. [Google Scholar] [PubMed]
- Kumari, K.; Sharma, P.K.; Shikha, S.; Singh, R.P. Molecular characterization and in-depth genome analysis of Enterobacter sp. S-16. Funct. Integr. Genom. 2023, 23, 245. [Google Scholar]
- Gostinčar, C.; Sun, X.; Zajc, J.; Fang, C.; Hou, Y.; Luo, Y.; Gunde-Cimerman, N.; Song, Z. Population genomics of an obligately halophilic basidiomycete Wallemia ichthyophaga. Front. Microbiol. 2019, 10, 2019. [Google Scholar]
- Das, O.; Kumar, S.H.; Nayak, B.B. Relative abundance of halophilic archaea and bacteria in diverse salt-fermented fish products. LWT 2020, 117, 108688. [Google Scholar]
- Kothe, C.I.; Bolotin, A.; Kraïem, B.-F.; Dridi, B.; Team, F.M.; Renault, P. Unraveling the world of halophilic and halotolerant bacteria in cheese by combining cultural, genomic and metagenomic approaches. Int. J. Food Microbiol. 2021, 358, 109312. [Google Scholar]
- Ohshima, C.; Takahashi, H.; Insang, S.; Phraephaisarn, C.; Techaruvichit, P.; Khumthong, R.; Haraguchi, H.; Lopetcharat, K.; Keeratipibul, S. Next-generation sequencing reveals predominant bacterial communities during fermentation of Thai fish sauce in large manufacturing plants. LWT 2019, 114, 108375. [Google Scholar]
- Randazzo, C.L.; Todaro, A.; Pino, A.; Pitino, I.; Corona, O.; Caggia, C. Microbiota and metabolome during controlled and spontaneous fermentation of Nocellara Etnea table olives. Food Microbiol. 2017, 65, 136–148. [Google Scholar]
- Stoll, D.A.; Müller, A.; Meinhardt, A.-K.; Dötsch, A.; Greiner, R.; Kulling, S.E.; Huch, M. Influence of salt concentration and iodized table salt on the microbiota of fermented cucumbers. Food Microbiol. 2020, 92, 103552. [Google Scholar]
- Leena, M.; Aamer, A.; Abdul, H.; Fariha, H. Physiological, biochemical and phylogenetic characterization of extremely halophilic bacteria isolated from Khewra mine, Pakistan. Appl. Ecol. Environ. Res. 2018, 16, 1243. [Google Scholar]
- Cycil, L.M.; DasSarma, S.; Pecher, W.; McDonald, R.; AbdulSalam, M.; Hasan, F. Metagenomic insights into the diversity of halophilic microorganisms indigenous to the Karak Salt Mine, Pakistan. Front. Microbiol. 2020, 11, 1567. [Google Scholar]
- Vera-Gargallo, B.; Hernández, M.; Dumont, M.G.; Ventosa, A. Thrive or survive: Prokaryotic life in hypersaline soils. Environ. Microbiome 2023, 18, 17. [Google Scholar]
- Mukhtar, S.; Mehnaz, S.; Mirza, M.S.; Mirza, B.S.; Malik, K.A. Diversity of Bacillus-like bacterial community in the rhizospheric and non-rhizospheric soil of halophytes (Salsola stocksii and Atriplex amnicola), and characterization of osmoregulatory genes in halophilic Bacilli. Can. J. Microbiol. 2018, 64, 567–579. [Google Scholar]
- Leadbeater, D.R.; Bruce, N.C. Functional characterisation of a new halotolerant seawater active glycoside hydrolase family 6 cellobiohydrolase from a salt marsh. Sci. Rep. 2024, 14, 3205. [Google Scholar]
- Roohi, A.; Ahmed, I.; Iqbal, M.; Jamil, M. Preliminary isolation and characterization of halotolerant and halophilic bacteria from salt mines of Karak, Pakistan. Pak. J. Bot. 2012, 44, 365–370. [Google Scholar]
- Javed, A.; Zahoor, S.; Javed, M.M.; Shah, F.S.; Mansoor, F. Characterization of halophilic/halotolerant bacteria isolated from the hypersaline environment of Khewra, district Jhelum. Adv. Life Sci. 2023, 10, 115–121. [Google Scholar]
- Lach, J.; Królikowska, K.; Baranowska, M.; Krupińska, M.; Strapagiel, D.; Matera-Witkiewicz, A.; Stączek, P. A first insight into the Polish Bochnia Salt Mine metagenome. Environ. Sci. Pollut. Res. 2023, 30, 49551–49566. [Google Scholar]
- Zhang, G.; Bai, J.; Zhai, Y.; Jia, J.; Zhao, Q.; Wang, W.; Hu, X. Microbial diversity and functions in saline soils: A review from a biogeochemical perspective. J. Adv. Res. 2024, 59, 129–140. [Google Scholar]
- Dindhoria, K.; Jain, R.; Kumar, R.; Bhargava, B.; Kumar, R.; Kumar, S. Microbial community structure analysis of hypersaline niches and elucidation of their role in the biogeochemical cycling of nitrogen, sulphur and methane. Ecol. Inform. 2023, 75, 102023. [Google Scholar]
- Naseem, S.; Fu, G.L.; Mohsin, M.; Rehman, M.Z.-u.; Baig, S.A. Semi-quantitative environmental impact assessment of khewra salt mine of Pakistan: An application of mathematical approach of environmental sustainability. Min. Metall. Explor. 2020, 37, 1185–1196. [Google Scholar] [CrossRef]
- Hussain, S.A.; Feng-Qing, H.; Yunqi, M.; Khan, H.; Jian, Y.; Hussain, G.; Widory, D. An overview of Pakistan rock salt resources and their chemical characterization. Pak. J. Sci. Ind. Res. Ser. A Phys. Sci. 2021, 64, 137–148. [Google Scholar] [CrossRef]
- Jensen, J.L.; Schjønning, P.; Watts, C.W.; Christensen, B.T.; Peltre, C.; Munkholm, L.J. Relating soil C and organic matter fractions to soil structural stability. Geoderma 2019, 337, 834–843. [Google Scholar] [CrossRef]
- Winegardner, D.L. An Introduction to Soils for Environmental Professionals; Routledge: London, UK, 2019. [Google Scholar]
- Ali, I.; Kanhayuwa, L.; Rachdawong, S.; Rakshit, S.K. Identification, phylogenetic analysis and characterization of obligate halophilic fungi isolated from a man-made solar saltern in Phetchaburi province, Thailand. Ann. Microbiol. 2013, 63, 887–895. [Google Scholar] [CrossRef]
- Delbès, C.; Moletta, R.; Godon, J.J. Monitoring of activity dynamics of an anaerobic digester bacterial community using 16S rRNA polymerase chain reaction–single-strand conformation polymorphism analysis. Environ. Microbiol. 2000, 2, 506–515. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Dillon, M.R.; Zhang, Y.; Rideout, J.R.; Bolyen, E.; Li, H.; Albert, P.S.; Caporaso, J.G. q2-longitudinal: Longitudinal and paired-sample analyses of microbiome data. MSystems 2018, 3, 10–1128. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Faith, D.P.; Baker, A.M. Phylogenetic diversity (PD) and biodiversity conservation: Some bioinformatics challenges. Evol. Bioinform. 2006, 2, 117693430600200007. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [PubMed]
- Sharma, R.; Munns, K.; Alexander, T.; Entz, T.; Mirzaagha, P.; Yanke, L.J.; Mulvey, M.; Topp, E.; McAllister, T. Diversity and distribution of commensal fecal Escherichia coli bacteria in beef cattle administered selected subtherapeutic antimicrobials in a feedlot setting. Appl. Environ. Microbiol. 2008, 74, 6178–6186. [Google Scholar] [PubMed]
- Ali, Z.; Shahzadi, I.; Majeed, A.; Malik, H.M.T.; Waseem, S.; Ahmed, I.; Anis, R.A.; Saeed, S.; Anees, M. Comparative analysis of the serum microbiome of HIV infected individuals. Genomics 2021, 113, 4015–4021. [Google Scholar] [CrossRef] [PubMed]
- Mousaa, I.; Elhady, M.; El-Sayyad, G.S.; Attia, R. Development of a highly hydrophobic and antimicrobial surface for polyester fabrics treated with (vinyl acetate versatic ester/paraffin wax) blend containing sodium chloride using electron beam irradiation. Prog. Org. Coat. 2023, 174, 107230. [Google Scholar]
- Chen, X.; Zhang, W.; Quek, S.Y.; Zhao, L. Flavor–food ingredient interactions in fortified or reformulated novel food: Binding behaviors, manipulation strategies, sensory impacts, and future trends in delicious and healthy food design. Compr. Rev. Food Sci. Food Saf. 2023, 22, 4004–4029. [Google Scholar]
- Whittaker, J.; Baker, E.; Kumar, S.; Collingwood, R.; West, M.; Lee, P. Do variations in nasal irrigation recipes and storage effect the risk of bacterial contamination?–ERRATUM. J. Laryngol. Otol. 2023, 137, 942–944. [Google Scholar] [CrossRef]
- Kuzina, E.; Rafikova, G.; Mukhamatdyarova, S.; Sharipova, Y.Y.; Korshunova, T.Y. Biological Activity of Leached Chernozem under Oil and Sodium Chloride Pollution and the Effect of Treatment with Halotolerant Oil-Destructing Bacteria. Eurasian Soil Sci. 2023, 56, 75–86. [Google Scholar]
- Gavas, S.; Pawar, P.; Pandit, S.; Khanna, N.; Mathuriya, A.S.; Prasad, R. Role of Extremophiles in the Microbial Electrochemical Cell: Recent Advances. In Extremophiles; CRC Press: Oxfordshire, UK, 2023. [Google Scholar]
- Drissi Kaitouni, L.B.; Anissi, J.; Sendide, K.; El Hassouni, M. Diversity of hydrolase-producing halophilic bacteria and evaluation of their enzymatic activities in submerged cultures. Ann. Microbiol. 2020, 70, 1–15. [Google Scholar]
- Massaoudi, Y.; Anissi, J.; Lefter, R.; Lobiuc, A.; Sendide, K.; Ciobica, A.; Hassouni, M.E. Protective effects of two halophilic crude extracts from Pseudomonas zhaodongensis and Bacillus stratosphericus against memory deficits and anxiety-and depression-like behaviors in methionine-induced schizophrenia in mice focusing on oxidative stress status. Evid.-Based Complement. Altern. Med. 2020, 2020, 8852418. [Google Scholar]
- Ait Assou, S.; Anissi, J.; Sendide, K.; El Hassouni, M. Diversity and antimicrobial activities of actinobacteria isolated from mining soils in Midelt Region, Morocco. Sci. World J. 2023, 2023, 6106673. [Google Scholar] [CrossRef]
- De Simeis, D.; Serra, S. Actinomycetes: A never-ending source of bioactive compounds—An overview on antibiotics production. Antibiotics 2021, 10, 483. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Villegas, P.; Vigara, J.; Vila, M.; Varela, J.; Barreira, L.; Léon, R. Antioxidant, antimicrobial, and bioactive potential of two new haloarchaeal strains isolated from odiel salterns (Southwest Spain). Biology 2020, 9, 298. [Google Scholar] [CrossRef] [PubMed]
- Desai, C.; Patel, P.; Markande, A.; Kamala, K.; Sivaperumal, P. Exploration of haloarchaea for their potential applications in food industry. Int. J. Environ. Sci. Technol. 2020, 17, 4455–4464. [Google Scholar]
- Ma, Y.-C.; Gao, M.-R.; Yang, H.; Jiang, J.-Y.; Xie, W.; Su, W.-P.; Zhang, B.; Yeong, Y.-S.; Guo, W.-Y.; Sui, L.-Y. Optimization of C50 Carotenoids Production by Open Fermentation of Halorubrum sp. HRM-150. Appl. Biochem. Biotechnol. 2023, 195, 3628–3640. [Google Scholar]
- Adamski, J.C.; Roberts, J.A.; Goldstein, R.H. Entrapment of bacteria in fluid inclusions in laboratory-grown halite. Astrobiology 2006, 6, 552–562. [Google Scholar] [CrossRef]
- Pasteris, J.D.; Freeman, J.J.; Wopenka, B.; Qi, K.; Ma, Q.; Wooley, K.L. With a grain of salt: What halite has to offer to discussions on the origin of life. Astrobiology 2006, 6, 625–643. [Google Scholar]
- Vreeland, R.H.; Piselli, A.F., Jr.; McDonnough, S.; Meyers, S. Distribution and diversity of halophilic bacteria in a subsurface salt formation. Extremophiles 1998, 2, 321–331. [Google Scholar]
- Caton, T.M.; Caton, I.; Witte, L.R.; Schneegurt, M.A. Archaeal diversity at the Great Salt Plains of Oklahoma described by cultivation and molecular analyses. Microb. Ecol. 2009, 58, 519–528. [Google Scholar]
- Henriet, O.; Fourmentin, J.; Delincé, B.; Mahillon, J. Exploring the diversity of extremely halophilic archaea in food-grade salts. Int. J. Food Microbiol. 2014, 191, 36–44. [Google Scholar]
- Robinson, C.K.; Wierzchos, J.; Black, C.; Crits-Christoph, A.; Ma, B.; Ravel, J.; Ascaso, C.; Artieda, O.; Valea, S.; Roldán, M. Microbial diversity and the presence of algae in halite endolithic communities are correlated to atmospheric moisture in the hyper-arid zone of the A tacama D esert. Environ. Microbiol. 2015, 17, 299–315. [Google Scholar]
- Reang, L.; Bhatt, S.; Tomar, R.S.; Joshi, K.; Padhiyar, S.; Bhalani, H.; Kheni, J.; Vyas, U.; Parakhia, M. Extremozymes and compatible solute production potential of halophilic and halotolerant bacteria isolated from crop rhizospheric soils of Southwest Saurashtra Gujarat. Sci. Rep. 2024, 14, 15704. [Google Scholar] [CrossRef]
- Yao, H.; Liu, S.; Liu, T.; Ren, D.; Zhou, Z.; Yang, Q.; Mao, J. Microbial-derived salt-tolerant proteases and their applications in high-salt traditional soybean fermented foods: A review. Bioresour. Bioprocess. 2023, 10, 82. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Feng, S.; Luo, J.; Hao, B.; Diao, F.; Li, X.; Jia, B.; Wang, L.; Bao, Z.; Guo, W. Evaluation of microbial assemblages in various saline-alkaline soils driven by soluble salt ion components. J. Agric. Food Chem. 2021, 69, 3390–3400. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Akbar, A.; Permpornsakul, P.; Yanwisetpakdee, B.; Chen, X.; Anwar, M.; Ali, I. Molecular diversity of halophilic fungi isolated from mangroves ecosystem of miani hor, Balochistan, Pakistan. Pak. J. Bot. 2020, 52, 1823–1829. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Cong, P.; Kuang, S.; Tang, L.; Li, Y.; Dong, J.; Song, W. Long-term excessive application of K2SO4 fertilizer alters bacterial community and functional pathway of tobacco-planting soil. Front. Plant Sci. 2022, 13, 1005303. [Google Scholar] [CrossRef] [PubMed]
- Heinz, J.; Waajen, A.C.; Airo, A.; Alibrandi, A.; Schirmack, J.; Schulze-Makuch, D. Bacterial growth in chloride and perchlorate brines: Halotolerances and salt stress responses of Planococcus halocryophilus. Astrobiology 2019, 19, 1377–1387. [Google Scholar] [CrossRef]
- Vavourakis, C.D.; Ghai, R.; Rodriguez-Valera, F.; Sorokin, D.Y.; Tringe, S.G.; Hugenholtz, P.; Muyzer, G. Metagenomic insights into the uncultured diversity and physiology of microbes in four hypersaline soda lake brines. Front. Microbiol. 2016, 7, 211. [Google Scholar] [CrossRef]
- Papale, M.; Lo Giudice, A.; Conte, A.; Rizzo, C.; Rappazzo, A.C.; Maimone, G.; Caruso, G.; La Ferla, R.; Azzaro, M.; Gugliandolo, C. Microbial assemblages in pressurized Antarctic brine pockets (Tarn Flat, Northern Victoria Land): A hotspot of biodiversity and activity. Microorganisms 2019, 7, 333. [Google Scholar] [CrossRef]
- Lach, J.; Jęcz, P.; Strapagiel, D.; Matera-Witkiewicz, A.; Stączek, P. The methods of digging for “Gold” within the salt: Characterization of halophilic prokaryotes and identification of their valuable biological products using sequencing and genome mining tools. Genes 2021, 12, 1756. [Google Scholar] [CrossRef]
- Al-Daghistani, H.I.; Zein, S.; Abbas, M.A. Microbial communities in the Dead Sea and their potential biotechnological applications. Commun. Integr. Biol. 2024, 17, 2369782. [Google Scholar] [CrossRef]
- He, Y.; He, L.; Wang, Z.; Liang, T.; Sun, S.; Liu, X. Salinity Shapes the Microbial Communities in Surface Sediments of Salt Lakes on the Tibetan Plateau, China. Water 2022, 14, 4043. [Google Scholar] [CrossRef]
- Ali, I.; Prasongsuk, S.; Akbar, A.; Aslam, M.; Lotrakul, P.; Punnapayak, H.; Rakshit, S.K. Hypersaline habitats and halophilic microorganisms. Maejo Int. J. Sci. Technol. 2016, 10, 330–345. [Google Scholar]
- Al-Tarshi, M.; Dobretsov, S.; Al-Belushi, M. Bacterial Communities across Multiple Ecological Niches (Water, Sediment, Plastic, and Snail Gut) in Mangrove Habitats. Microorganisms 2024, 12, 1561. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, J.; Ge, A.-H.; Xie, W.; Yao, R.; Wang, X. Salinity drives niche differentiation of soil bacteria and archaea in Hetao Plain, China. J. Environ. Manag. 2024, 370, 122977. [Google Scholar]
- Kalwasińska, A.; Deja-Sikora, E.; Burkowska-But, A.; Szabó, A.; Felföldi, T.; Kosobucki, P.; Krawiec, A.; Walczak, M. Changes in bacterial and archaeal communities during the concentration of brine at the graduation towers in Ciechocinek spa (Poland). Extremophiles 2018, 22, 233–246. [Google Scholar]
- Ioannou, A.; Berkhout, M.D.; Geerlings, S.Y.; Belzer, C. Akkermansia muciniphila: Biology, microbial ecology, host interactions and therapeutic potential. Nat. Rev. Microbiol. 2025, 23, 162–177. [Google Scholar]
- Bhattarai, Y.; Muniz Pedrogo, D.A.; Kashyap, P.C. Irritable bowel syndrome: A gut microbiota-related disorder? Am. J. Physiol. -Gastrointest. Liver Physiol. 2017, 312, G52–G62. [Google Scholar]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Biswas, D.; Rahaman, S.O. Gut Microbiome and Its Impact on Health and Diseases; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Somda, N.; Tankoano, A.; Métuor-Dabiré, A.; Kaboré, D.; Bonkoungou, J.; Kpoda, D.; Sambe-Ba, B.; Dabiré, Y.; Saba, C.; Ouoba, I. A systematic review and meta-analysis of antibiotics resistance of foodborne pathogenic bacteria in West Africa between 2010 and 2020. J. Food Prot. 2023, 86, 100061. [Google Scholar]
- Wu, Z.; Li, M.; Qu, L.; Zhang, C.; Xie, W. Metagenomic insights into microbial adaptation to the salinity gradient of a typical short residence-time estuary. Microbiome 2024, 12, 115. [Google Scholar]


| Soil Samples | Loc. | pH | Salinity | Moisture Content | Conductivity | Na | K | Cl | Ca | Mg | Fe (mg/L) | SO4 | Nitrogen Content | Total Organic Carbon | Organic Matter |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1A | SWDKM | 7.9 | 30% | 26.59% | 95 | 27.2 | 0.34 | 42 | 0.018 | 0.053 | 9.51 | 3.2 | 0.11% | 0.29% | 1.01% |
| 2A | SSHWKM | 8.2 | 32% | 10% | 95.5 | 28.6 | 0.36 | 44.5 | 0.04 | 0.059 | 15.5 | 0.35 | 0.38% | 0.35% | 0.95% |
| 3A | SCFMKM | 8.65 | 36% | 8.27% | 99 | 33.5 | 0.42 | 49.4 | 0.041 | 0.065 | 16.8 | 0.32 | 0.97% | 0.39% | 1.05% |
| MEAN | 8.25 | 0.327 | 0.149533 | 96.5 | 29.8 | 0.39 | 45.3 | 0.033 | 0.059 | 13.9 | 1.29 | 0.0049 | 0.00343 | 0.010033 | |
| SD | 0.38 | 0.031 | 0.101147 | 2.1794 | 3.31 | 0.04 | 3.764 | 0.013 | 0.006 | 3.88 | 1.654 | 0.0044 | 0.0005 | 0.000503 |
| Brine Samples | Loc. | pH | Salinity | Conductivity | Na | K | Cl | Ca | Mg | Fe (mg/L) | SO4 | Nitrogen Content | Total Organic Carbon | Organic Matter |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SBK1 | P1KSM | 7.25 | 32.75% | 108 | 22.3 | 0.126 | 44.85 | 0.321 | 0.097 | 3.28 | 0.11 | 0.10% | 0.10% | 0.67% |
| SBK2 | P2KSM | 6.8 | 29.90% | 100 | 19.1 | 0.132 | 40.33 | 0.309 | 0.085 | 4.59 | 0.8 | 0.07% | 0.27% | 0.49% |
| SBK3 | P3KSM | 7.5 | 33.30% | 105 | 31.31 | 0.138 | 50.24 | 0.342 | 0.112 | 6.24 | 0.2 | 0.08% | 0.15% | 0.37% |
| Mean | 7.183 | 0.319833 | 104.3333 | 24.237 | 0.132 | 45.14 | 0.324 | 0.098 | 4.7033 | 0.37 | 0.000823 | 0.001733 | 0.0051 | |
| SD | 0.355 | 0.018251 | 4.041452 | 6.3312 | 0.006 | 4.9614 | 0.0167 | 0.014 | 1.4833 | 0.3751 | 0.000172 | 0.000874 | 0.00151 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malik, Y.; Ali, I.; Sajjad, A.; Jing, L.; Iqbal, I.; Rehman, A.u.; Azam, T.; Chen, X. Bacterial Diversity at Himalayan Pink Salt Extraction Site. Biology 2025, 14, 316. https://doi.org/10.3390/biology14030316
Malik Y, Ali I, Sajjad A, Jing L, Iqbal I, Rehman Au, Azam T, Chen X. Bacterial Diversity at Himalayan Pink Salt Extraction Site. Biology. 2025; 14(3):316. https://doi.org/10.3390/biology14030316
Chicago/Turabian StyleMalik, Yasmeen, Imran Ali, Ashif Sajjad, Luhuai Jing, Irfana Iqbal, Atiq ur Rehman, Toquier Azam, and Xiaoming Chen. 2025. "Bacterial Diversity at Himalayan Pink Salt Extraction Site" Biology 14, no. 3: 316. https://doi.org/10.3390/biology14030316
APA StyleMalik, Y., Ali, I., Sajjad, A., Jing, L., Iqbal, I., Rehman, A. u., Azam, T., & Chen, X. (2025). Bacterial Diversity at Himalayan Pink Salt Extraction Site. Biology, 14(3), 316. https://doi.org/10.3390/biology14030316






