Simple Summary
This study evaluates the efficacy of environmental DNA (eDNA) metabarcoding and underwater visual census (UVC) in assessing the diversity of subtidal macrobenthic communities. We compared water eDNA, sediment eDNA, and traditional UVC methods in the Nanji Islands, China. Sediment eDNA demonstrated superior performance in detecting key benthic phyla such as Annelida and Arthropoda, whereas UVC was more effective for large and active organisms. Integrating these methods provides a more comprehensive biodiversity assessment, highlighting the importance of combining molecular and traditional techniques for effective conservation and management strategies in marine ecosystems.
Abstract
The rapid advancement of environmental DNA (eDNA) technology has transformed ecological research, particularly in aquatic ecosystems. However, the optimal sampling matrix (e.g., water or sediment) and the potential for eDNA to replace or complement traditional underwater visual census (UVC) remain unclear. Here, we integrate water eDNA, sediment eDNA, and UVC approaches to systematically compare the diversity of benthic macrofauna in the subtidal zones of the Nanji Islands, China. Our results show that sediment eDNA samples exhibited the highest species richness, while UVC had the lowest. Each method revealed distinct species profiles, with relatively few shared taxa at the order level and below. Environmental eDNA showed significant advantages in detecting key phyla such as Annelida and Arthropoda. In contrast, traditional UVC was crucial for identifying certain taxa, such as Bryozoa, which were undetectable by eDNA methods. The low overlap in species detected by these methods underscores their complementary nature, highlighting the necessity of integrating multiple approaches to achieve a more comprehensive and accurate biodiversity assessment. Future research should focus on refining eDNA techniques, such as developing more universal primers, to further enhance their applicability in biodiversity monitoring.
1. Introduction
The subtidal zone, a critical interface between intertidal zones and shallow sea, usually harbors a rich diversity of benthic fauna, yet it is one of the areas most severely affected by human activities [1]. Traditional in situ observations through underwater visual census (UVC)—where trained SCUBA divers survey species richness and community assemblages in the field—have long been the predominant method for studying marine organisms [2]. However, this approach is limited by the need for specialized skills, suitable field conditions, and significant resources [3]. Moreover, the complex currents and topography of subtidal zones often pose substantial challenges for UVC surveys, restricting the comprehensive biomonitoring of these habitats [4].
Recent advancements in environmental DNA (eDNA) technology have revolutionized biodiversity assessments in aquatic ecosystems [5]. eDNA metabarcoding allows for the non-invasive detection of species through the analysis of genetic material present in environmental samples such as water and sediment [6]. This approach has been successfully applied to monitor fish and invertebrate communities in various aquatic habitats, including rivers, lakes, and marine environments [5,6,7,8]. However, the optimal sampling matrix (water vs. sediment) and the potential for eDNA to replace or complement the traditional UVC method to access the macrobenthos diversity remain uncertain.
The Nanji Islands, located in the eastern sea area of Zhejiang Province, China, is a biosphere reserve of the United Nations Educational, Scientific and Cultural Organization (UNESCO). This subtropical marine protected area is characterized by complex subtidal zones with high biodiversity [9]. The seabed terrain in this area slopes downwards from northwest to southeast, with a water depth generally between 15 and 25 m. There are two deep-water channels on the northeast and southwest sides of Nanji Island, with a depth of over 30 m and a maximum depth of 45 m. The subtidal areas here include several distinct habitat types, such as steep rocky cliffs, platforms, boulder fields, small area beaches, and sediments mainly composed of silty clay. Due to the fact that the subtidal zone with a depth less than 20 m is mainly composed of rocks, bottom trawl survey is impossible, and box-type mud collection is also limited. While the underwater visual census (UVC) is the remaining available traditional method, a systematic biodiversity survey is still lacking.
Generally, the Nanji Islands provide an ideal setting for comparing the effectiveness of different biodiversity assessment methods. In this study, we integrate water eDNA, sediment eDNA, and UVC methods to systematically compare their efficiency in detecting benthic macrofaunal diversity in the subtidal zone with a depth < 20 m of the Nanji Islands. Our objectives are to quantify the differences in species detection efficiency among the three methods, and explore the complementary nature of these methods across different taxonomic levels.
2. Materials and Methods
2.1. Study Area and Sample Location
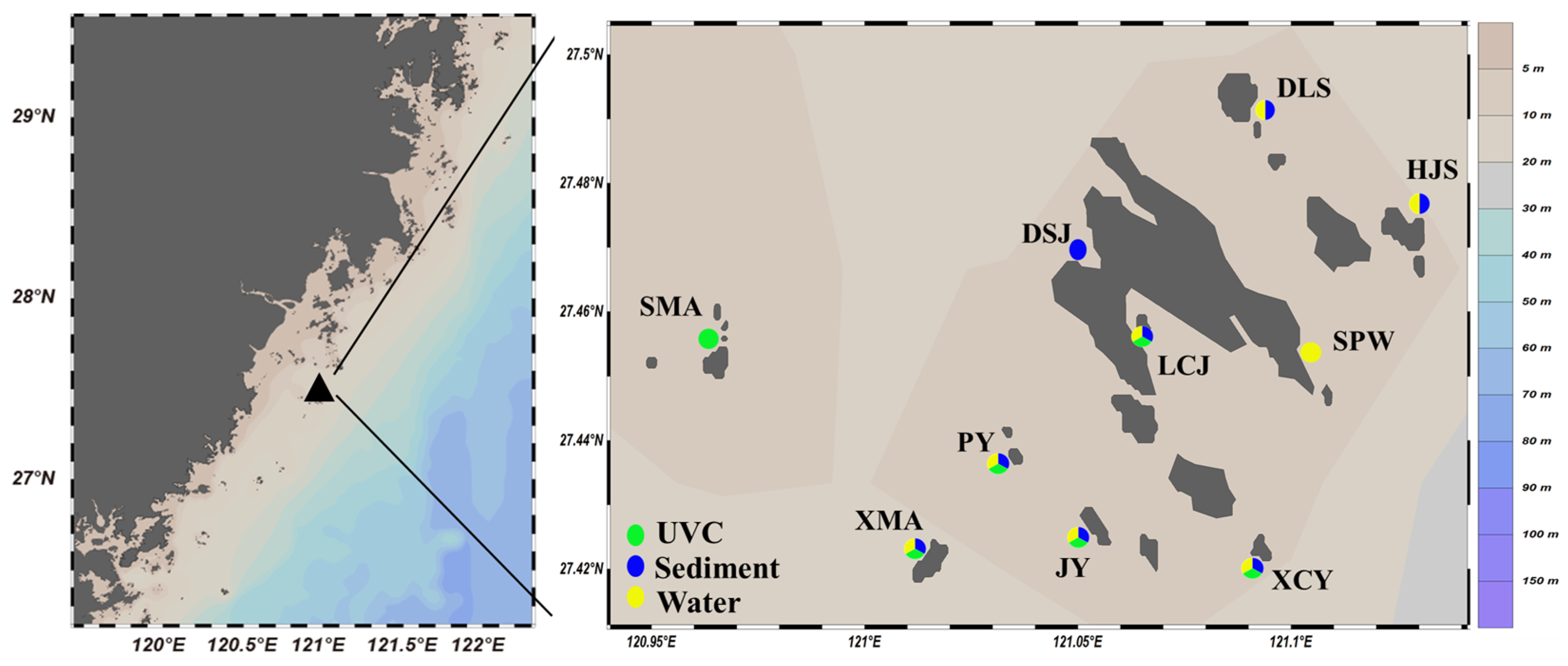
The Nanji Islands (27°27′43″ N–27°46′48″ N, 120°02′55″ E–121°07′58″ E) are located in the eastern sea area of Wenzhou City, Zhejiang Province, China. This subtropical marine protected area is characterized by mainly rocky inter- and sub-tidal zones with high biodiversity. Ten subtidal sites representing the broader subtidal environment were selected for this study based on accessibility by boat on 8–10 May 2024 (Figure 1).
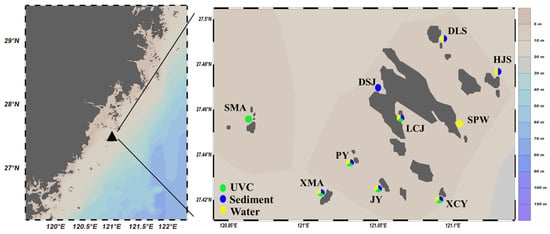
Figure 1.
Sampling sites in the subtidal zone of the Nanji Islands (The capitalized names in the right-side figure are abbreviations for each sub-island. “SMA” stands for “Shangmaan”, “XMA” for “Xiamaan”, “PY” for “Poyu”, “JY” for “Jianyu”, “XCY” for “Xiaochaiyu”, “LCJ” for “Longchuanjiao”, “DSJ” for “Dashanjiao”, “SPW” for “Sanpanwei”, “HJS” for “Houjishan”, and “DLS” for “Daleishan”).
2.2. Underwater Visual Census (UVC) and Photo Analysis
UVC survey conducted during high-tide period. The basic unit of UVC was a 50 m long transect line, with macrobenthos surveyed in two 1 m wide by 2 m high bands on either side of the transect line [10]. The proportions of the area examined in relation to the rocky subtidal area of each site ranged from approximately 10% to 30%. Digital photo-quadrats were taken at 2.5 m intervals along the transect line (i.e., 20 per 50 m transect). Due to the extremely low transparency of seawater (<1 m), the SMA, DSJ, HJS, and DLS sites were not suitable for diving. Consequently, the UVC units of only six stations with a depth range of 5–15 m were completely investigated (Figure 1, marked by green color). Two taxonomists (W. Huo and W. Chen) estimated the number of species from the visual data together, without prior knowledge of the eDNA outcomes. Due to the generally low transparency of seawater, many photo-quadrats were not very clear, and only 30 species could be identified to species level (Table A1).
2.3. Environmental DNA Sampling
Water and sediment samples were collected from ten stations in the subtidal zones of the Nanji Islands. Considering that the larvae of benthic organisms may float in surface water layers, 1 L of seawater was collected from both surface and bottom layers by divers to gather as much macrobenthos eDNA as possible. The samples were mixed thoroughly, and 1.5 L seawater was filtered [11] through a 0.22 µm mixed cellulose ester (MCE) membrane, and the filter paper was immediately transferred to a 1.5 mL cryopreservation tube and stored at −20 °C.
Sediment samples were collected using a grab sampler near the water sampling sites. At the sites XMA and XCY, however, the sediment could not be collected by the grab sampler due to the thin sediment layer. Instead, divers collected sediment using 50 mL centrifuge tubes in those sites. Approximately 20 g of surface sediment was placed into a sterile, enzyme-free sampling bag and stored at −20 °C until further analysis.
2.4. DNA Extraction and Sequencing
Environmental DNA was extracted from water sample filters using the DNeasy Power Water Kit (Qiagen, Hilden, Germany) and from sediment samples using the QIAamp PowerFecal Pro DNA Kit (Qiagen, Hilden, Germany), following standardized protocols to ensure high extraction efficiency and purity. The COI gene fragments were targeted and amplified using universal primers mlCOIintF (5′-GGWACWGGWTGAACWGTWTAYCCYCC-3′) [12] and jgHCO2198 (5′-TAIACYTCIGGRTGICCRAARAAYCA-3′) [13]. The PCR conditions were as follows: initial denaturation at 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 2 min, with a final extension at 72 °C for 5 min. Negative controls (DNA-free samples) were processed in parallel to ensure the reliability of the PCR. Sequence libraries were prepared using the TruSeq® DNA PCR-Free Kit (Illumina, San Diego, CA, USA) and quantified by Qubit and Q-PCR. The libraries were sequenced as 250 bp paired-end reads on a NovaSeq 6000 platform.
2.5. Bioinformatics Analysis
Raw sequencing data were processed using DADA2 (version 2.15.2) to remove primer sequences and low-quality reads [14]. Amplicon sequence variants (ASVs) were clustered at 97% similarity using VSEARCH, and ASVs with less than 0.01% abundance across all samples were filtered out [15,16]. Taxonomic annotations were performed against the NCBI database, focusing on eukaryotic groups. Non-marine groups (e.g., Arachnida, Insecta, and terrestrial Craniata) were manually excluded from the ASV table [17].
2.6. Statistical Analyses
To comprehensively assess the species diversity based on the three methods, species richness, Shannon–Wiener diversity, and Pielou’s evenness indices were employed. Species richness and the relative ratio of species number at class, order, and family level were compared among UVC, sediment eDNA, and water eDNA methods. Venn diagrams were generated to assess overlapping taxa detected by the different methods. As the species diversity data from sediment and water eDNA did not meet the normal distribution assumption (Shapiro–Wilk test, p < 0.05) or the homogeneity of variance assumption (Bartlett test, p < 0.05), a nonparametric Kruskal–Wallis test was used to analyze intergroup differences in species diversity. Post hoc Dunn tests with Holm–Bonferroni correction for p-values were further conducted to evaluate species diversity differences across different methods.
In the analysis of the community structure, Jaccard distances were calculated based on species composition matrices, and non-metric multidimensional scaling (NMDS) was used to visualize community differences among the three methods. The reliability of the ordination was evaluated by the stress value (the closer the value is to zero, the higher the degree of fit). The clustering of samples was observed in the NMDS plot, and the significance of community structure differences among groups was verified by ANOSIM tests (based on Jaccard distances). All statistical analyses and figures were performed using R version 4.0.3, with plots created using the ggplot2 package (version 3.3.3) [18].
3. Results
3.1. Sequencing and UVC Results
Only eight of ten water and sediment eDNA samples were successfully amplified even after repeating the PCR experiments several times, respectively (Figure 1, successful amplification stations marked by yellow and blue colors, respectively). A total of 657,161 and 752,136 raw reads were obtained from water and sediment eDNA samples, respectively. After bioinformatics processing, 1879 and 7487 amplicon sequence variants (ASVs) were identified for water and sediment eDNA, respectively. The majority of ASVs (79.30% for water eDNA and 64.90% for sediment eDNA) were either taxonomically unassigned or belonged to non-marine groups. The remaining ASVs represented a diverse range of marine taxa, including 11 phyla, 22 classes, 47 orders, 72 families, 72 genera, and 94 species for water eDNA; and 14 phyla, 28 classes, 64 orders, 114 families, 125 genera, and 166 species for sediment eDNA. In contrast, UVC identified a total of nine phyla, 16 classes, 18 orders, 29 families, 29 genera, and 39 species based on photo-quadrat analysis. Overall, the three methods collectively identified 15 phyla, 38 classes, 89 orders, 179 families, 206 genera, and 284 species (Table 1 and Table A1).

Table 1.
Species number of phyla using sediment and water eDNA and UVC methods.
3.2. Comparison of the Taxonomic Compositions Among the Three Methods
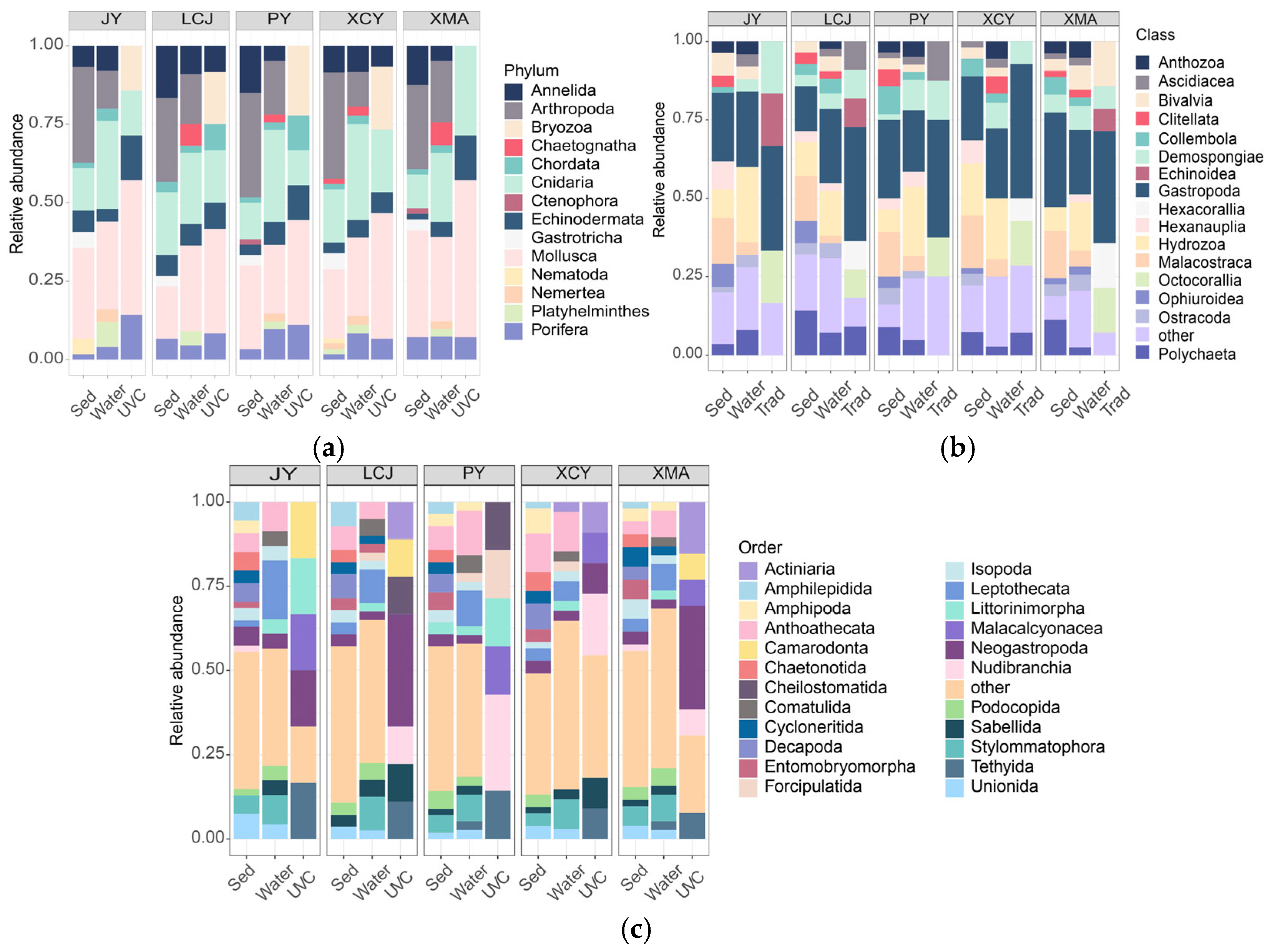
Taxonomic compositions were compared among the three methods at five common stations (Figure 2). Sediment eDNA detected the highest number of phyla (13), classes (27), and species (131), followed by water eDNA (10 phyla, 21 classes, 72 species) and UVC (seven phyla, 14 classes, 15 orders, 32 species). At the phylum level, sediment and water eDNA showed similar compositions, with Mollusca, Arthropoda, and Cnidaria being dominant (Table 1). In contrast, UVC identified Mollusca, Cnidaria, and Bryozoa as the most dominant group, and the Bryozoa was undetectable by eDNA methods (Figure 2a). At the class level, sediment eDNA was dominated by Gastropoda, followed by Malacostraca and Hydrozoa, while water eDNA showed a similar pattern with Gastropoda, Hydrozoa, and Demospongiae as dominant classes. UVC identified Gastropoda, Anthozoa, and Demospongiae as the dominant classes (Figure 2b). At the order level, sediment eDNA showed Anthomedusae, Decapoda, and Stylommatophora as dominant orders, while water eDNA was dominated by Leptothecata, Anthoathecata, and Stylommatophora. UVC identified Neogastropoda, Nudibranchia, and Demospongiae as dominant orders (Figure 2c).
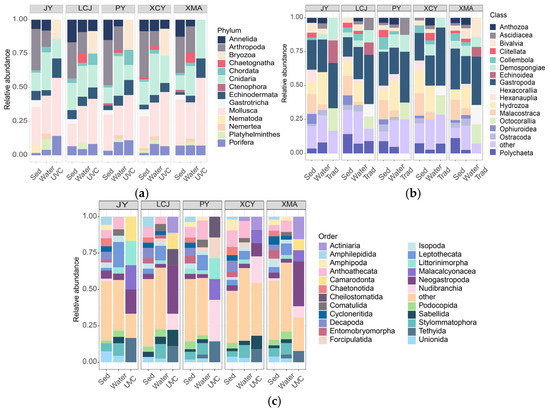
Figure 2.
Stacked bar charts showing the percentage of species richness at the phylum (a), class (b), and order (c) levels for underwater visual census (UVC) and sediment (Sed) and water eDNA methods.
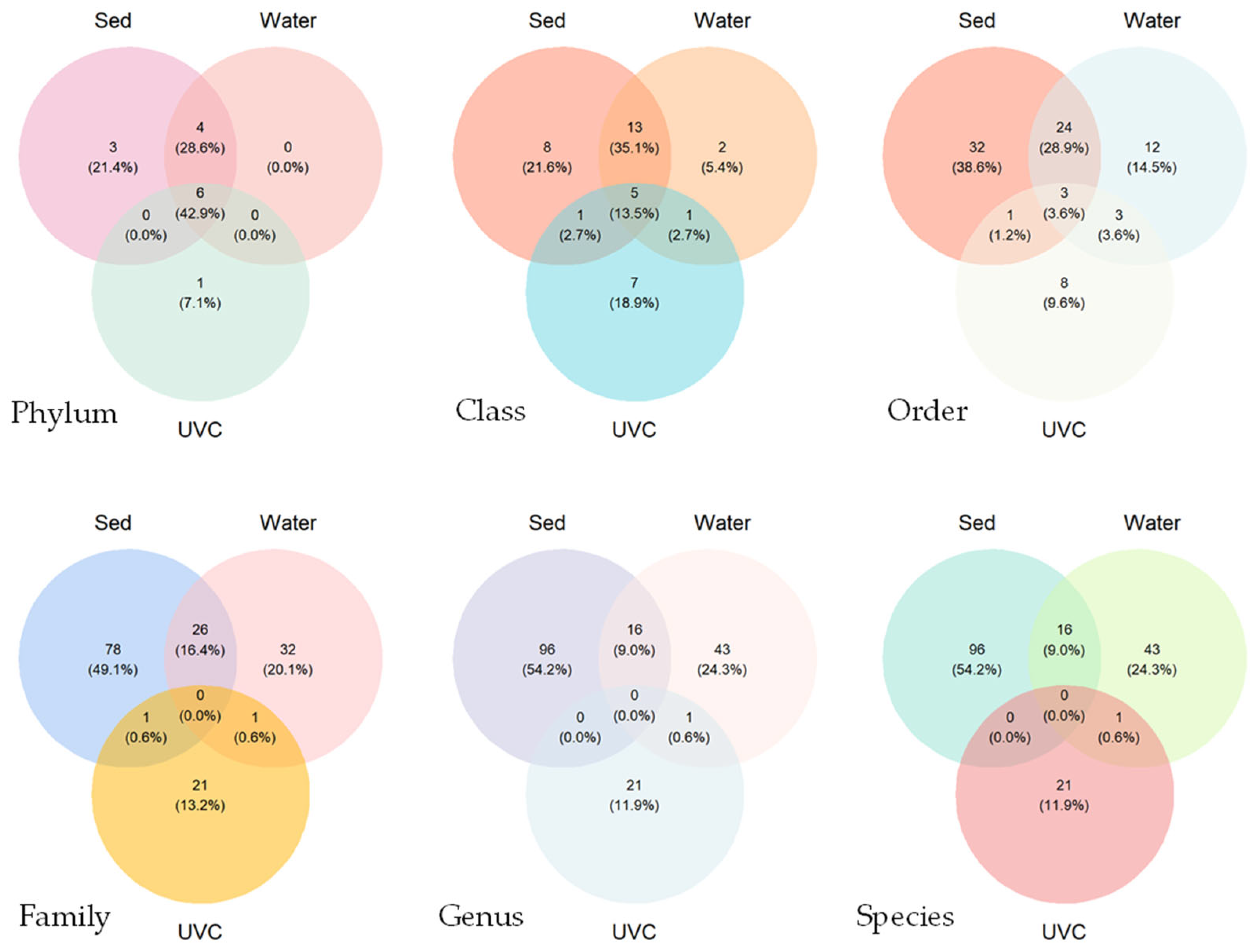
Venn diagrams revealed relatively high taxon overlaps at the phylum and class levels, and low overlaps at the order level and below among the taxonomic compositions from the three methods (Figure 3). Sediment eDNA detected the highest number of species overall, while traditional UVC methods detected fewer but unique species. No common species was found among the three methods. Only 18 common species were found between sediment and water eDNA samples, and only 1 common species was found between traditional UVC and sediment/water eDNA samples.
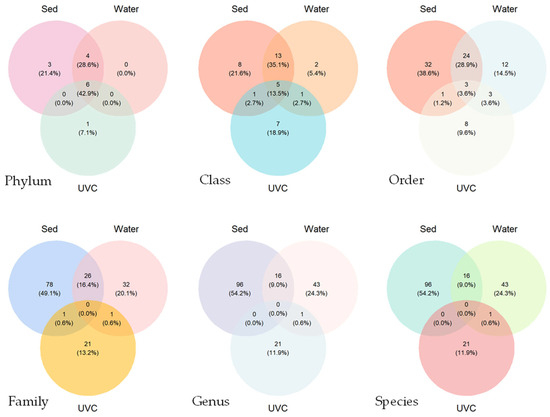
Figure 3.
Venn diagrams showing the species overlaps at the different taxonomic ranks detected by underwater visual census (UVC) and sediment (Sed) and water eDNA methods at the common stations.
3.3. Alpha and Beta Diversity Among the Three Methods
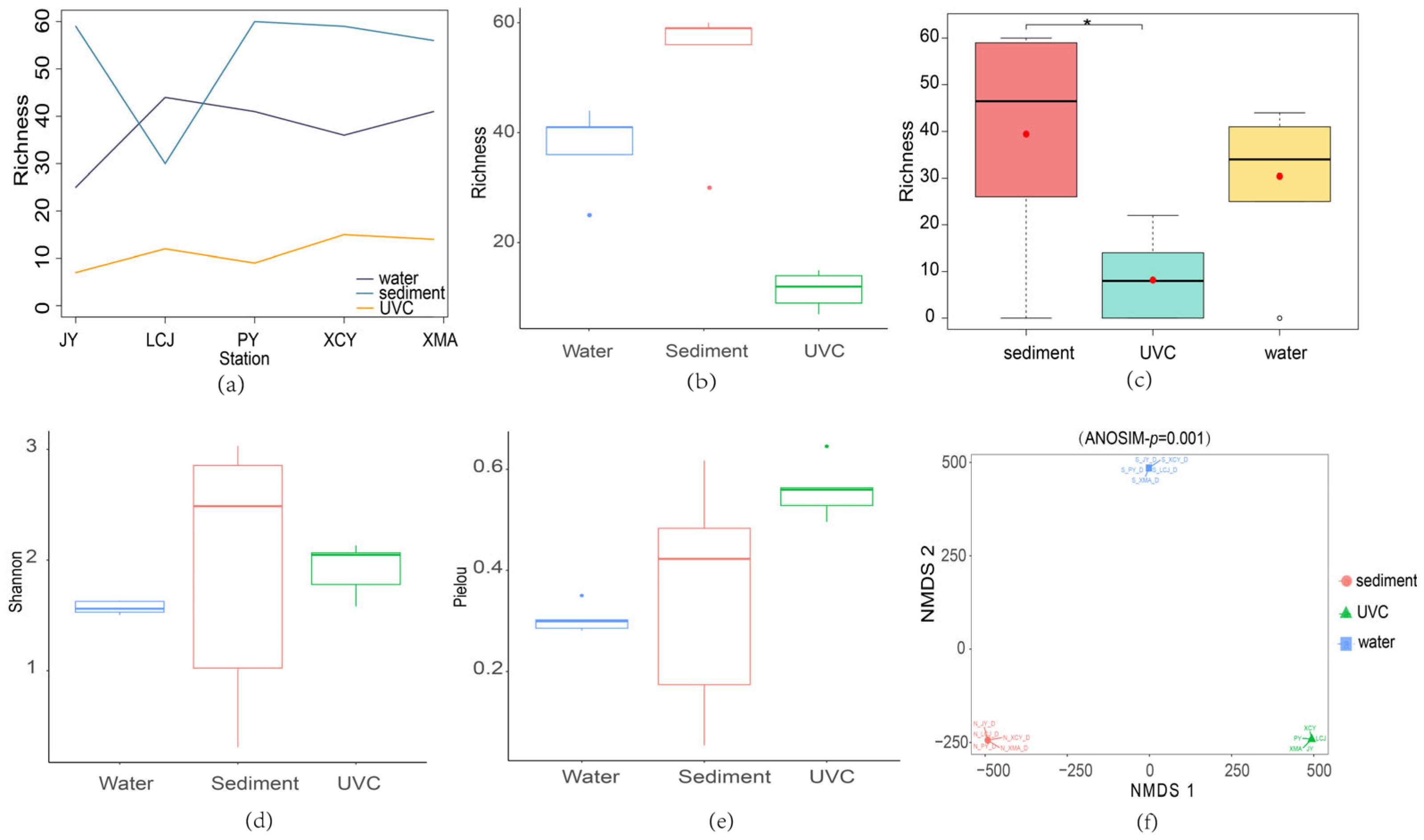
Species richness was highest in sediment eDNA samples across all stations except LCJ, with UVC showing the lowest species richness (Figure 4a). Overall, sediment eDNA samples exhibited the highest average species richness, while UVC had the lowest (Figure 4b). The Kruskal–Wallis test and post hoc Dunn test indicated significant differences in species richness between sediment eDNA and UVC methods (Figure 4c). Sediment eDNA also consistently showed higher Shannon and Pielou’s evenness indices compared to water eDNA, indicating superior performance in both species abundance and community evenness (Figure 4d,e).
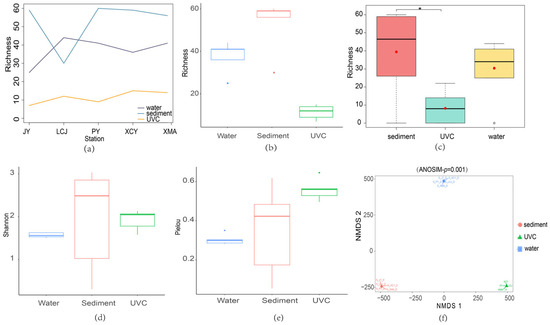
Figure 4.
Alpha and beta diversity analyses based on sediment eDNA, water eDNA, and underwater visual census (UVC) methods. (a) Species richness based on the sediment eDNA, water eDNA, and UVC at the common sampling stations (JY, LCJ, PY, XCY, XMA). (b) The total species richness based on the three methods from all the common stations. (c) Significant richness differences among the three methods conducted by post hoc Dunn tests. *, indicating significant differences. (d) The total Shannon index based on the three methods from all the common stations. (e) The total Pielou’s evenness index based on the three methods from all the common stations. (f) The clustering of samples with non-metric multidimensional scaling (NMDS) plot.
Non-metric multidimensional scaling (NMDS) ordination plots showed distinct clustering of samples from sediment eDNA, water eDNA, and UVC methods, with no overlap among the groups (Figure 4f). The stress value of zero and an ANOSIM test p-value of 0.001 confirmed the high reliability of the ordination results, indicating significant differences in community structure captured by the three methods. This suggests that each method provides unique insights into the biodiversity of the subtidal zone, highlighting the importance of integrating multiple approaches for comprehensive biodiversity assessment.
4. Discussion
4.1. Environmental DNA Is Essential for Biodiversity Assessment of Rocky Subtidal Zone
Traditional bottom trawling and grab sampling are unsuitable for biodiversity assessments in rocky subtidal zones. Underwater visual census (UVC) is often used for macrobenthic surveys, but its effectiveness is limited by low seawater transparency, as seen in this study. Compared to UVC, the present study demonstrates that eDNA is more effective for assessing benthic macrofaunal diversity, particularly for key phyla such as Annelida and Arthropoda (Table 1; Figure 3). The sediment eDNA detected a significantly higher species richness compared to the UVC method (Figure 4). Generally, both the sediment and water eDNA methods can uncover those relatively smaller benthos (e.g., Chaetognatha, Gastrotricha, Nemertea, Platyhelminthes, and most of Annelida and Arthropoda) that were missed by the UVC survey (Table 1 and Table A1). Furthermore, eDNA showed significant differences in community structure with the UVC (Figure 4f).
In terms of total number, the sediment eDNA detected more species than the water eDNA (166 vs. 94; Table 1). For all the phyla except Cnidaria and Chaetognatha, the sediment eDNA detected no less species than the water eDNA (Table 1). Notably, sediment eDNA failed to detect Octocorallia species, which were uncovered by water eDNA. Most octocorals prefer to live on hard substrates such as rocks, rather than sediments. Consequently, the octocoral eDNA in the sediment is too rare to amplify. In contrast, the eDNA concentration in water samples increases after filtration and concentration, which may contain more octocoral eDNA. For the planktonic Chaetognatha, it is not surprising that the water eDNA detected more species than the sediment eDNA (Table A1).
For the macrobenthos, the superior detection efficiency of sediment eDNA is likely due to the higher concentration and longer persistence of DNA in sediments compared to water samples, which allows for retrospective genetic monitoring and extends the seasonal window for species assessment [19,20,21,22,23,24,25]. For example, the decay rate of fish eDNA in sediment is significantly lower than that in water (0.033%/h vs. 1.9%/h), and eDNA is 8–1800 times more concentrated in sediment than in water. This suggests that sediment eDNA can capture a broader range of species, including those that may be temporarily absent or less active in the water column.
4.2. Limitations of Traditional UVC and eDNA Methods
While the traditional UVC method is effective for detecting large and highly active organisms, they have limited capacity for detecting small or cryptic species [2,26]. In the present study, this limitation is partly due to the difficulty in capturing and identifying relatively small organisms such as polychaetes (belonging to the phylum Annelida), Chaetognatha, Gastrotricha, Nemertea, Platyhelminthes, and Arthropoda, which often require further taxonomic examination under a microscope. Additionally, environmental factors such as water turbidity in the subtidal zones can further reduce the effectiveness of the UVC method. However, traditional UVC remains valuable for certain taxa that may not be well-represented in eDNA samples, such as Bryozoa, which was undetectable by eDNA in our study but was identified through the UVC method.
The missed detection of Bryozoa highlights the need for further refinement of eDNA techniques. The COI primers used in this study failed to detect Bryozoa, indicating potential limitations in primer universality or suboptimal annealing temperature for PCR reactions. Additionally, the incomplete nature of current DNA sequence databases limits the accuracy of species-level identifications. Future research should focus on developing more universal primers and optimal PCR reactions, and expanding reference databases to improve the reliability and resolution of eDNA-based biodiversity assessments.
4.3. Complementary Nature of the Methods and Implications for Biodiversity Monitoring
In general, taxonomic compositions from the three methods shared relatively few taxa at the order level and below, and no common species was found among these methods (Figure 2c and Figure 3). The low overlap in species detected by the three methods underscores their complementary nature. Sediment eDNA provided the most comprehensive species inventory, detecting a significantly higher number of species compared to water eDNA and traditional UVC methods. However, the unique detection of certain taxa by the UVC method highlights the importance of integrating both eDNA and UVC approaches for a more complete assessment of biodiversity. This integration can help overcome the limitations of each method, such as primer bias in eDNA techniques and the challenges of species identification in traditional methods. Future biodiversity monitoring efforts should consider the complementary nature of these methods and the potential for further technological advancements to enhance the accuracy and efficiency of biodiversity assessments. This integrated approach will be crucial for informing effective conservation and management strategies in complex aquatic environments.
5. Conclusions
The present study provides a comprehensive evaluation of environmental DNA (eDNA) metabarcoding of COI and traditional underwater visual census (UVC) methods for assessing benthic macrofaunal diversity in subtidal zones. While taxonomic compositions from the three methods show similar patterns at the phylum level, they share relatively few taxa at the order level and below. Sediment eDNA emerged as a highly effective tool, particularly for detecting key benthic phyla such as Annelida and Arthropoda, due to its enhanced preservation and detection capabilities. However, traditional UVC remains crucial for identifying certain taxa, such as Bryozoa, which were undetectable by eDNA methods. The low overlap in species detected by these methods underscores their complementary nature, highlighting the necessity of integrating multiple approaches to achieve a more comprehensive and accurate biodiversity assessment. Future research should focus on refining eDNA techniques, such as developing more universal primers and expanding reference databases, to further enhance their applicability in biodiversity monitoring.
Author Contributions
Conceptualization, Z.Z., Y.L., and K.X.; methodology, W.H., S.X., W.C., and X.L.; software, W.H. and Z.Z.; validation, Z.Z. and K.X.; formal analysis, W.H., S.X., Z.Z., and W.C.; investigation, X.L., Z.Z., S.X., and W.C.; writing—review and editing, Z.Z. and K.X.; visualization, W.H. and Z.Z.; supervision, Z.Z.; project administration, K.X.; funding acquisition, Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Key R&D Program of China, grant number 2022YFC2803800, the National Natural Science Foundation of China, grant number 42176128, the Strategic Priority Research Program of the Chinese Academy of Sciences, grant number XDB42000000, and Science and Technology Program of Nanji Islands National Marine Nature Reserve Administration, grant numbers and ZJYY-PYCG-2024092001 and JJZB-PYCG-2021112901. The APC was funded by No. 2022YFC2803800.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw sequence reads detected by eDNA are deposited in the NCBI Sequence Read Archive database with the BioProjects PRJNA1248872 and PRJNA1248901.
Acknowledgments
We thank Xiaoyu Zheng for help with photo collection.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A

Table A1.
Species composition based on the sediment and water eDNA and UVC methods.
Table A1.
Species composition based on the sediment and water eDNA and UVC methods.
| Species ID | Sediment | Water | UVC | Phylum | Class | Order | Family | Genus | Species |
|---|---|---|---|---|---|---|---|---|---|
| OTU_1958 | 1 | 1 | 0 | Annelida | Polychaeta | Polychaeta sp. NB-Po509 | |||
| OTU_1601 | 1 | 0 | 0 | Annelida | Clitellata | Enchytraeida | Enchytraeidae | Enchytronia | Enchytronia christenseni |
| OTU_4793 | 1 | 0 | 0 | Annelida | Polychaeta | Terebellida | Flabelligeridae | Treadwellius | Treadwellius bifidus |
| OTU_16546 | 1 | 0 | 0 | Annelida | Polychaeta | Maldanidae | Asychis | Asychis chilensis | |
| OTU_17049 | 1 | 0 | 0 | Annelida | Clitellata | Crassiclitellata | Megascolecidae | Perionyx | Perionyx rufulus |
| OTU_5756 | 1 | 0 | 0 | Annelida | Polychaeta | Phyllodocida | Nereididae | Neanthes | Neanthes sp. RP2011-N |
| OTU_7431 | 1 | 0 | 0 | Annelida | Polychaeta | Phyllodocida | Nereididae | Neanthes | Neanthes sp. 1 |
| OTU_4848 | 1 | 0 | 0 | Annelida | Polychaeta | Orbiniidae | Scoloplos | Scoloplos acmeceps | |
| OTU_2403 | 1 | 0 | 0 | Annelida | Phascolosomatiformes | Phascolosomatidae | Phascolosoma | Phascolosoma esculenta | |
| OTU_13606 | 1 | 0 | 0 | Annelida | Polychaeta | Sabellida | Serpulidae | Serpula | Serpula vermicularis |
| OTU_14440 | 1 | 0 | 0 | Annelida | Polychaeta | Sabellida | Serpulidae | Pileolaria | Pileolaria sp. 11BIOAK-0893 |
| OTU_3016 | 1 | 0 | 0 | Annelida | Polychaeta | Sabellida | Serpulidae | Crucigera | Crucigera sp. CMC01 |
| OTU_331 | 1 | 0 | 0 | Annelida | Polychaeta | Sabellida | Serpulidae | Serpula | Serpula columbiana |
| OTU_7545 | 1 | 0 | 0 | Annelida | Clitellata | Lumbriculida | Sparganophilidae | Sparganophilus | Sparganophilus sp. MAC_SRS_S115 |
| OTU_14076 | 1 | 0 | 0 | Annelida | Polychaeta | Spionida | Spionidae | Polydora | Polydora cornuta |
| OTU_2533 | 1 | 0 | 0 | Annelida | Polychaeta | Spionida | Spionidae | Paraprionospio | Paraprionospio patiens |
| OTU_16980 | 1 | 0 | 0 | Annelida | Polychaeta | Terebellida | Terebellidae | Thelepus | Thelepus sp. CMC01 |
| OTU_2402 | 1 | 0 | 0 | Annelida | Polychaeta | Terebellida | Terebellidae | Lysilla | Lysilla pacifica |
| OTU_15614 | 1 | 0 | 0 | Annelida | Polychaeta | Terebellida | Trichobranchidae | Terebellides | Terebellides stroemii |
| OTU_17438 | 1 | 0 | 0 | Annelida | Polychaeta | Xenopneusta | Urechidae | Urechis | Urechis unicinctus |
| OTU_13481 | 1 | 0 | 0 | Annelida | Polychaeta | Polychaeta sp. EBS61o-Po36 | |||
| OTU_3837 | 1 | 0 | 0 | Annelida | Polychaeta | Polychaeta sp. OPC-2015 | |||
| OTU_12617 | 0 | 1 | 0 | Annelida | Polychaeta | Sabellida | Serpulidae | Protolaeospira | Protolaeospira eximia |
| OTU_1436 | 0 | 1 | 0 | Annelida | Clitellata | Rhynchobdellida | Glossiphoniidae | Torix | Torix tukubana |
| OTU_1486 | 0 | 1 | 0 | Annelida | Clitellata | Crassiclitellata | Megascolecidae | Amynthas | Amynthas exiguus |
| OTU_2537 | 0 | 1 | 0 | Annelida | Polychaeta | Terebellida | Terebellidae | Loimia | Loimia arborea |
| OTU_3294 | 0 | 1 | 0 | Annelida | Polychaeta | Sabellida | Serpulidae | Serpula | Serpula vermicularis |
| OTU_518 | 0 | 1 | 0 | Annelida | Polychaeta | Spionida | Trochochaetidae | Trochochaeta | Trochochaeta multisetosa |
| OTU_6200 | 0 | 1 | 0 | Annelida | Polychaeta | Phyllodocida | Nereididae | Nereis | Nereis sp. CMC01 |
| UVC_01 | 0 | 0 | 1 | Annelida | Polychaeta | sabellida | Sabellidae | Sabellidae sp. | |
| UVC_02 | 0 | 0 | 1 | Arthropoda | Malacostraca | Decapoda | Paguridae | Pagurus | Pagurus hedleyi |
| OTU_2085 | 1 | 1 | 0 | Arthropoda | Collembola | Entomobryomorpha | Entomobryidae | Entomobrya | Entomobrya unostrigata |
| OTU_13808 | 1 | 0 | 0 | Arthropoda | Hexanauplia | Harpacticoida | Aegisthidae | Aegisthidae sp. DZMB479 | |
| OTU_8881 | 1 | 0 | 0 | Arthropoda | Hexanauplia | Harpacticoida | Aegisthidae | Aegisthidae sp. 1 | |
| OTU_14428 | 1 | 0 | 0 | Arthropoda | Malacostraca | Decapoda | Aeglidae | Aegla | Aegla longirostri complex sp. UFSM SG15 |
| OTU_7458 | 1 | 0 | 0 | Arthropoda | Hexanauplia | Sessilia | Archaeobalanidae | Conopea | Conopea basicuneata |
| OTU_11323 | 1 | 0 | 0 | Arthropoda | Ichthyostraca | Arguloida | Argulidae | Argulus | Argulus japonicus |
| OTU_13718 | 1 | 0 | 0 | Arthropoda | Malacostraca | Isopoda | Armadillidae | Spherillo | Spherillo sp. Kanzaki |
| OTU_5774 | 1 | 0 | 0 | Arthropoda | Branchiopoda | Anostraca | Branchipodidae | Parartemia | Parartemia serventyi |
| OTU_8686 | 1 | 0 | 0 | Arthropoda | Branchiopoda | Anostraca | Branchipodidae | Parartemia | Parartemia sp. 1 |
| OTU_6592 | 1 | 0 | 0 | Arthropoda | Malacostraca | Decapoda | Cambaridae | Faxonius | Faxonius validus |
| OTU_14411 | 1 | 0 | 0 | Arthropoda | Malacostraca | Amphipoda | Chiltoniidae | Chiltoniidae sp. CU1 | |
| OTU_5711 | 1 | 0 | 0 | Arthropoda | Hexanauplia | Sessilia | Chthamalidae | Chthamalus | Chthamalus panamensis |
| OTU_13431 | 1 | 0 | 0 | Arthropoda | Ostracoda | Podocopida | Cyprididae | Strandesia | Strandesia velhoi |
| OTU_14178 | 1 | 0 | 0 | Arthropoda | Ostracoda | Podocopida | Cyprididae | Cyprideis | Cyprideis torosa |
| OTU_9379 | 1 | 0 | 0 | Arthropoda | Ostracoda | Podocopida | Darwinulidae | Darwinula | Darwinula stevensoni |
| OTU_14387 | 1 | 0 | 0 | Arthropoda | Hexanauplia | Calanoida | Diaptomidae | Hesperodiaptomus | Hesperodiaptomus arcticus |
| OTU_4874 | 1 | 0 | 0 | Arthropoda | Collembola | Symphypleona | Dicyrtomidae | Dicyrtomidae sp. BIOUG21696-E03 | |
| OTU_14442 | 1 | 0 | 0 | Arthropoda | Collembola | Entomobryomorpha | Entomobryidae | Entomobrya | Entomobrya decemfasciata |
| OTU_5455 | 1 | 0 | 0 | Arthropoda | Collembola | Entomobryomorpha | Entomobryidae | Lepidocyrtus | Lepidocyrtus sp. COLNO287-09 |
| OTU_8097 | 1 | 0 | 0 | Arthropoda | Collembola | Entomobryomorpha | Entomobryidae | Entomobrya | Entomobrya sp. HB0852_3 |
| OTU_4780 | 1 | 0 | 0 | Arthropoda | Malacostraca | Amphipoda | Eusiridae | Eusiridae sp. 06 MK-2021 | |
| OTU_4668 | 1 | 0 | 0 | Arthropoda | Malacostraca | Decapoda | Grapsidae | Grapsus | Grapsus albolineatus |
| OTU_2814 | 1 | 0 | 0 | Arthropoda | Malacostraca | Isopoda | Haploniscidae | Mastigoniscus | Mastigoniscus sp. NB-Iso41 |
| OTU_14314 | 1 | 0 | 0 | Arthropoda | Collembola | Poduromorpha | Hypogastruridae | Hypogastruridae sp. BIOUG23106-B05 | |
| OTU_7317 | 1 | 0 | 0 | Arthropoda | Malacostraca | Isopoda | Ischnomesidae | Ischnomesus | Ischnomesus sp. lineage 7d JB-2020 |
| OTU_7631 | 1 | 0 | 0 | Arthropoda | Ostracoda | Podocopida | Loxoconchidae | Microloxoconcha | Microloxoconcha ikeyai |
| OTU_13364 | 1 | 0 | 0 | Arthropoda | Malacostraca | Amphipoda | Lysianassidae | Eurythenes | Eurythenes gryllus |
| OTU_4425 | 1 | 0 | 0 | Arthropoda | Malacostraca | Amphipoda | Lysianassidae | Eurythenes | Eurythenes sp. 1 |
| OTU_4697 | 1 | 0 | 0 | Arthropoda | Malacostraca | Amphipoda | Lysianassidae | Eurythenes | Eurythenes sp. 2A2 HR-2015 |
| OTU_17634 | 1 | 0 | 0 | Arthropoda | Hexanauplia | Harpacticoida | Miraciidae | Psammotopa | Psammotopa sp. n. SR-2019 |
| OTU_6939 | 1 | 0 | 0 | Arthropoda | Malacostraca | Mysida | Mysidae | Paramesopodopsis | Paramesopodopsis rufa |
| OTU_1240 | 1 | 0 | 0 | Arthropoda | Collembola | Poduromorpha | Neanuridae | Morulina | Morulina mackenziana |
| OTU_2412 | 1 | 0 | 0 | Arthropoda | Malacostraca | Decapoda | Oregoniidae | Pleistacantha | Pleistacantha kannu |
| OTU_1556 | 1 | 0 | 0 | Arthropoda | Malacostraca | Decapoda | Palaemonidae | Macrobrachium | Macrobrachium naiyanetri |
| OTU_5707 | 1 | 0 | 0 | Arthropoda | Malacostraca | Decapoda | Pandalidae | Plesionika | Plesionika semilaevis |
| OTU_9864 | 1 | 0 | 0 | Arthropoda | Malacostraca | Decapoda | Pandalidae | Pandalus | Pandalus montagui |
| OTU_13528 | 1 | 0 | 0 | Arthropoda | Diplopoda | Polyxenida | Polyxenidae | Polyxenidae sp. Biologic-POLX001 | |
| OTU_12157 | 1 | 0 | 0 | Arthropoda | Malacostraca | Decapoda | Porcellanidae | Petrolisthes | Petrolisthes rufescens |
| OTU_13725 | 1 | 0 | 0 | Arthropoda | Malacostraca | Decapoda | Portunidae | Portunus | Portunus ventralis |
| OTU_1312 | 1 | 0 | 0 | Arthropoda | Collembola | Entomobryomorpha | Tomoceridae | Tomocerus | Tomocerus ocreatus |
| OTU_1352 | 1 | 0 | 0 | Arthropoda | Malacostraca | Isopoda | Tylidae | Tylos | Tylos ponticus |
| OTU_17543 | 1 | 0 | 0 | Arthropoda | Malacostraca | Amphipoda | Typhlogammaridae | Typhlogammarus | Typhlogammarus sp. ZH-2014 |
| OTU_14472 | 1 | 0 | 0 | Arthropoda | Hexanauplia | Sessilia | Verrucidae | Metaverruca | Metaverruca recta |
| OTU_13506 | 1 | 0 | 0 | Arthropoda | Malacostraca | Amphipoda | Amphipoda sp. LPdivOTU157 | ||
| OTU_17242 | 1 | 0 | 0 | Arthropoda | Hexanauplia | Copepoda | Copepoda environmental sample | ||
| OTU_2669 | 1 | 0 | 0 | Arthropoda | Hexanauplia | Copepoda | Copepoda sp. RR-2014 | ||
| OTU_6511 | 1 | 0 | 0 | Arthropoda | Arthropoda environmental sample | ||||
| OTU_7414 | 1 | 0 | 0 | Arthropoda | Hexanauplia | Harpacticoida | Harpacticoida sp. 10BIOBC-00681 | ||
| OTU_11071 | 0 | 1 | 0 | Arthropoda | Hexanauplia | Calanoida | Diaptomidae | Allodiaptomus | Allodiaptomus mirabilipes |
| OTU_11156 | 0 | 1 | 0 | Arthropoda | Ostracoda | Podocopida | Cyprididae | Cypretta | Cypretta campechensis |
| OTU_15398 | 0 | 1 | 0 | Arthropoda | Arthropoda sp. NZAC 03011539 | ||||
| OTU_15775 | 0 | 1 | 0 | Arthropoda | Ostracoda | Podocopida | Candonidae | Pseudocandona | Pseudocandona hartwigi |
| OTU_17132 | 0 | 1 | 0 | Arthropoda | Malacostraca | Euphausiacea | Euphausiidae | Pseudeuphausia | Pseudeuphausia sinica |
| OTU_1993 | 0 | 1 | 0 | Arthropoda | Collembola | Symphypleona | Bourletiellidae | Heterosminthurus | Heterosminthurus sp. 2 JRD-2015 |
| OTU_4032 | 0 | 1 | 0 | Arthropoda | Malacostraca | Amphipoda | Stenothoidae | Metopa | Metopa boeckii |
| OTU_4984 | 0 | 1 | 0 | Arthropoda | Hexanauplia | Siphonostomatoida | Siphonostomatoida sp. DZMB115 | ||
| OTU_6169 | 0 | 1 | 0 | Arthropoda | Branchiopoda | Diplostraca | Limnadiidae | Limnadopsis | Limnadopsis cf. parvispinus |
| OTU_6178 | 0 | 1 | 0 | Arthropoda | Ostracoda | Podocopida | Cyprididae | Ngarawa | Ngarawa sp. 950.5 |
| OTU_6301 | 0 | 1 | 0 | Arthropoda | Malacostraca | Isopoda | Desmosomatidae | Desmosomatidae sp. NB-Iso234 | |
| OTU_8282 | 0 | 1 | 0 | Arthropoda | Malacostraca | Isopoda | Ligiidae | Ligia | Ligia baudiniana |
| OTU_8283 | 0 | 1 | 0 | Arthropoda | Malacostraca | Amphipoda | Niphargidae | Niphargus | Niphargus parenzani |
| UVC_03 | 0 | 0 | 1 | Bryozoa | Gymnolaemata | Cheilostomatida | Membraniporidae | Biflustra | Biflustra grandicella |
| UVC_04 | 0 | 0 | 1 | Bryozoa | Bryozoa sp. | ||||
| UVC_05 | 0 | 0 | 1 | Bryozoa | Gymnolaemata | Cheilostomatida | Bitectiporidae | entapora | entapora fascialis |
| UVC_31 | 0 | 0 | 1 | Bryozoa | Stenolaemata | Cyclostomatida | Crisiidae | Crisia | Crisia elongata |
| UVC_33 | 0 | 0 | 1 | Bryozoa | Cheilostomatida | Candidae | Caberea | Caberea lata | |
| OTU_10240 | 1 | 1 | 0 | Chaetognatha | Sagittoidea | Aphragmophora | Sagittidae | Sagitta | Sagitta sp. ZP304 |
| OTU_36 | 0 | 1 | 0 | Chaetognatha | Sagittoidea | Aphragmophora | Sagittidae | Zonosagitta | Zonosagitta nagae |
| OTU_93 | 0 | 1 | 0 | Chaetognatha | Chaetognatha environmental sample | ||||
| OTU_971 | 0 | 1 | 0 | Chaetognatha | Chaetognatha environmental sample | ||||
| OTU_14459 | 1 | 0 | 0 | Chordata | Ascidiacea | Aplousobranchia | Didemnidae | Didemnum | Didemnum candidum |
| OTU_13738 | 1 | 0 | 0 | Chordata | Thaliacea | Pyrosomata | Pyrosomatidae | Pyrostremma | Pyrostremma spinosum |
| OTU_13999 | 1 | 0 | 0 | Chordata | Thaliacea | Salpida | Salpidae | Cyclosalpa | Cyclosalpa sp. USNM IZ 1449879 |
| OTU_13033 | 1 | 0 | 0 | Chordata | Ascidiacea | Stolidobranchia | Styelidae | Cnemidocarpa | Cnemidocarpa verrucosa |
| OTU_2807 | 1 | 0 | 0 | Chordata | Ascidiacea | Stolidobranchia | Styelidae | Cnemidocarpa | Cnemidocarpa verrucosa |
| OTU_121 | 0 | 1 | 0 | Chordata | Ascidiacea | Stolidobranchia | Styelidae | Cnemidocarpa | Cnemidocarpa sp. 1 |
| UVC_06 | 0 | 0 | 1 | Chordata | Ascidiacea | Ascidiacea sp. | |||
| UVC_30 | 0 | 0 | 0 | Chordata | Ascidiacea | Stolidobranchia | Styelidae | styela | styela canopus |
| OTU_8074 | 1 | 1 | 0 | Cnidaria | Hydrozoa | Leptothecata | Aequoreidae | Aequorea | Aequorea sp. USNM IZ 1450497 |
| OTU_17794 | 1 | 1 | 0 | Cnidaria | Hydrozoa | Anthoathecata | Candelabridae | Candelabrum | Candelabrum cocksii |
| OTU_1173 | 1 | 1 | 0 | Cnidaria | Hydrozoa | Leptothecata | Clytiidae | Clytia | Clytia paulensis |
| OTU_1784 | 1 | 1 | 0 | Cnidaria | Hydrozoa | Leptothecata | Clytiidae | Clytia | Clytia gracilis |
| OTU_16831 | 1 | 1 | 0 | Cnidaria | Hydrozoa | Anthoathecata | Corynidae | Coryne | Coryne sp. JRH-2014 |
| OTU_17154 | 1 | 0 | 0 | Cnidaria | Anthozoa | Scleractinia | Acroporidae | Acropora | Acropora sp. IP0357 |
| OTU_17856 | 1 | 0 | 0 | Cnidaria | Hydrozoa | Anthoathecata | Bougainvilliidae | Bougainvillia | Bougainvillia muscus |
| OTU_2568 | 1 | 0 | 0 | Cnidaria | Anthozoa | Scleractinia | Caryophylliidae | Dasmosmilia | Dasmosmilia cf. lymani MVK-2010 |
| OTU_2663 | 1 | 0 | 0 | Cnidaria | Anthozoa | Scleractinia | Caryophylliidae | Rhizosmilia | Rhizosmilia robusta |
| OTU_9057 | 1 | 0 | 0 | Cnidaria | Scyphozoa | Rhizostomeae | Cassiopeidae | Cassiopea | Cassiopea sp. AUQLCBC |
| OTU_3574 | 1 | 0 | 0 | Cnidaria | Hydrozoa | Anthoathecata | Corynidae | Stauridiosarsia | Stauridiosarsia cliffordi |
| OTU_13582 | 1 | 0 | 0 | Cnidaria | Staurozoa | Stauromedusae | Craterolophidae | Craterolophus | Craterolophus convolvulus |
| OTU_3839 | 1 | 0 | 0 | Cnidaria | Hydrozoa | Anthoathecata | Eudendriidae | Eudendrium | Eudendrium carneum |
| OTU_5607 | 1 | 0 | 0 | Cnidaria | Hydrozoa | Leptothecata | Lafoeidae | Lafoea | Lafoea dumosa |
| OTU_10300 | 1 | 0 | 0 | Cnidaria | Hydrozoa | Leptothecata | Laodiceidae | Laodicea | Laodicea undulata |
| OTU_13976 | 1 | 0 | 0 | Cnidaria | Staurozoa | Stauromedusae | Lucernariidae | Lucernaria | Lucernaria quadricornis |
| OTU_17088 | 1 | 0 | 0 | Cnidaria | Staurozoa | Stauromedusae | Lucernariidae | Lucernariidae sp. CCSMA208-10 | |
| OTU_10965 | 1 | 0 | 0 | Cnidaria | Hydrozoa | Leptothecata | Obeliidae | Obelia | Obelia dichotoma |
| OTU_13515 | 1 | 0 | 0 | Cnidaria | Hydrozoa | Leptothecata | Obeliidae | Obelia | Obelia sp. DNZ116 |
| OTU_3127 | 1 | 0 | 0 | Cnidaria | Hydrozoa | Anthoathecata | Oceaniidae | Turritopsis | Turritopsis lata |
| OTU_14409 | 1 | 0 | 0 | Cnidaria | Hydrozoa | Anthoathecata | Pandeidae | Leuckartiara | Leuckartiara sp. PS-2018 |
| OTU_13700 | 1 | 0 | 0 | Cnidaria | Hydrozoa | Siphonophorae | Rhodaliidae | Dendrogramma | Dendrogramma enigmatica |
| OTU_4766 | 1 | 0 | 0 | Cnidaria | Hydrozoa | Leptothecata | Sertulariidae | Fraseroscyphus | Fraseroscyphus hozawai |
| OTU_3775 | 1 | 0 | 0 | Cnidaria | Anthozoa | Scleractinia | Siderastreidae | Psammocora | Psammocora profundacella |
| OTU_11341 | 1 | 0 | 0 | Cnidaria | Scyphozoa | Semaeostomeae | Ulmaridae | Aurelia | Aurelia sp. IDPAJGB |
| OTU_8311 | 1 | 0 | 0 | Cnidaria | Hydrozoa | Anthoathecata | Zancleidae | Zanclea | Zanclea sango |
| OTU_10172 | 0 | 1 | 0 | Cnidaria | Hydrozoa | Leptothecata | Eirenidae | Helgicirrha | Helgicirrha malayensis |
| OTU_104 | 0 | 1 | 0 | Cnidaria | Hydrozoa | Leptothecata | Clytiidae | Clytia | Clytia hemisphaerica |
| OTU_10944 | 0 | 1 | 0 | Cnidaria | Hydrozoa | Anthoathecata | Corymorphidae | Euphysa | Euphysa aurata |
| OTU_11207 | 0 | 1 | 0 | Cnidaria | Anthozoa | Actiniaria | Phymanthidae | Phymanthus | Phymanthus sp. IP0414 |
| OTU_117 | 0 | 1 | 0 | Cnidaria | Anthozoa | Alcyonacea | Plexauridae | Euplexaura | Euplexaura sp. A CSM-2013 |
| OTU_15386 | 0 | 1 | 0 | Cnidaria | Staurozoa | Stauromedusae | Haliclystidae | Haliclystus | Haliclystus inabai |
| OTU_15496 | 0 | 1 | 0 | Cnidaria | Hydrozoa | Anthoathecata | Corynidae | Corynidae sp. BCnid2010-002 | |
| OTU_15730 | 0 | 1 | 0 | Cnidaria | Hydrozoa | Anthoathecata | Asyncorinidae | Asyncoryne | Asyncoryne ryniensis |
| OTU_15819 | 0 | 1 | 0 | Cnidaria | Hydrozoa | Siphonophorae | Prayidae | Praya | Praya reticulata |
| OTU_15894 | 0 | 1 | 0 | Cnidaria | Scyphozoa | Rhizostomeae | Rhizostomatidae | Rhopilema | Rhopilema hispidum |
| OTU_15897 | 0 | 1 | 0 | Cnidaria | Hydrozoa | Anthoathecata | Pandeidae | Amphinema | Amphinema sp. JRH-2012 |
| OTU_16864 | 0 | 1 | 0 | Cnidaria | Hydrozoa | Leptothecata | Plumulariidae | Plumularia | Plumularia virginiae |
| OTU_16960 | 0 | 1 | 0 | Cnidaria | Hydrozoa | Anthoathecata | Bougainvilliidae | Bougainvillia | Bougainvillia muscus |
| OTU_17210 | 0 | 1 | 0 | Cnidaria | Hydrozoa | Siphonophorae | Agalmatidae | Marrus | Marrus sp. BO-2021 |
| OTU_17266 | 0 | 1 | 0 | Cnidaria | Hydrozoa | Anthoathecata | Corynidae | Coryne | Coryne sp. JRH-2014 |
| OTU_1954 | 0 | 1 | 0 | Cnidaria | Hydrozoa | Leptothecata | Aglaopheniidae | Aglaophenia | Aglaophenia tubulifera |
| OTU_1962 | 0 | 1 | 0 | Cnidaria | Hydrozoa | Leptothecata | Obeliidae | Obelia | Obelia sp. BFHL 2016 |
| OTU_2471 | 0 | 1 | 0 | Cnidaria | Hydrozoa | Anthoathecata | Pandeidae | Amphinema | Amphinema sp. PS-2017 |
| OTU_2502 | 0 | 1 | 0 | Cnidaria | Hydrozoa | Anthoathecata | Zancleidae | Zanclea | Zanclea giancarloi |
| OTU_3269 | 0 | 1 | 0 | Cnidaria | Scyphozoa | Semaeostomeae | Cyaneidae | Cyanea | Cyanea sp. RUNNPEC |
| OTU_473 | 0 | 1 | 0 | Cnidaria | Hydrozoa | Anthoathecata | Corynidae | Coryne | Coryne sp. JRH-2014 |
| OTU_5038 | 0 | 1 | 0 | Cnidaria | Hydrozoa | Leptothecata | Obeliidae | Obelia | Obelia bidentata |
| OTU_8072 | 0 | 1 | 0 | Cnidaria | Anthozoa | Alcyonacea | Victorgorgiidae | Victorgorgia | Victorgorgia sp. 2 KMM-2015 |
| OTU_8128 | 0 | 1 | 0 | Cnidaria | Anthozoa | Scleractinia | Acroporidae | Astreopora | Astreopora myriophthalma |
| OTU_8132 | 0 | 1 | 0 | Cnidaria | Scyphozoa | Semaeostomeae | Pelagiidae | Chrysaora | Chrysaora lactea |
| OTU_8177 | 0 | 1 | 0 | Cnidaria | Scyphozoa | Coronatae | Nausithoidae | Nausithoe | Nausithoe sp. NHM_353 |
| OTU_969 | 0 | 1 | 0 | Cnidaria | Hydrozoa | Leptothecata | Obeliidae | Obelia | Obelia sp. MZUSP 3356 |
| UVC_36 | 0 | 0 | 1 | Cnidaria | Hexacorallia | Actiniaria | Actiniidae | Paracondylactis | Paracondylactis sinensis |
| UVC_07 | 0 | 0 | 1 | Cnidaria | Octocorallia | Malacalcyonacea | Melithaeidae | Melithaea | Melithaea formosa |
| UVC_08 | 0 | 0 | 1 | Cnidaria | Hexacorallia | Ceriantharia | Cerianthidae | Cerianthidae sp. | |
| UVC_09 | 0 | 0 | 1 | Cnidaria | Octocorallia | malacalcyonacea | Isididae | Hicksonella | Hicksonella guishanensis |
| UVC_10 | 0 | 0 | 1 | Cnidaria | Octocorallia | Octocorallia sp. | |||
| UVC_11 | 0 | 0 | 1 | Cnidaria | Hexacorallia | Actiniaria | Actiniidae | Anthopleura | Anthopleura japonica |
| UVC_12 | 0 | 0 | 1 | Cnidaria | Hexacorallia | Actiniaria | Actiniidae | Anthopleura | Anthopleura inornata |
| OTU_13644 | 1 | 0 | 1 | Ctenophora | Nuda | Beroida | Beroidae | Beroe | Beroe ovata |
| OTU_14325 | 1 | 0 | 0 | Ctenophora | Nuda | Beroida | Beroidae | Beroe | Beroe sp. 2 |
| UVC_34 | 0 | 0 | 1 | Echinodermata | Holothuroidea | Dendrochirotida | Cucumariidae | Actinia | Actinia equina |
| OTU_8133 | 0 | 0 | 1 | Echinodermata | Asteroidea | Forcipulatida | Asteriidae | Stichaster | Stichaster striatus |
| UVC_13 | 0 | 0 | 1 | Echinodermata | Echinoidea | Camarodonta | Echinometridae | Anthocidaris | Anthocidaris crassispina |
| UVC_14 | 0 | 0 | 1 | Echinodermata | Echinoidea | Camarodonta | Temnopleuridae | Temnopleurus | Temnopleurus hardwickii |
| OTU_4816 | 1 | 0 | 0 | Echinodermata | Ophiuroidea | Euryalida | Gorgonocephalidae | Astrodendrum | Astrodendrum sp. NSMT:E-6273 |
| OTU_324 | 1 | 0 | 0 | Echinodermata | Ophiuroidea | Amphilepidida | Ophiactidae | Ophiactis | Ophiactis perplexa |
| OTU_2974 | 1 | 0 | 0 | Echinodermata | Ophiuroidea | Amphilepidida | Ophiolepididae | Ophiolepis | Ophiolepis irregularis |
| OTU_5418 | 1 | 0 | 0 | Echinodermata | Ophiuroidea | Amphilepidida | Ophiotrichidae | Ophiothrix | Ophiothrix spiculata |
| OTU_14449 | 1 | 0 | 0 | Echinodermata | Crinoidea | Cyrtocrinida | Sclerocrinidae | Neogymnocrinus | Neogymnocrinus richeri |
| OTU_11136 | 0 | 1 | 0 | Echinodermata | Crinoidea | Comatulida | Antedonidae | Antedon | Antedon loveni |
| OTU_16102 | 0 | 1 | 0 | Echinodermata | Ophiuroidea | Euryalida | Gorgonocephalidae | Astrothrombus | Astrothrombus chrysanthi |
| OTU_466 | 0 | 1 | 0 | Echinodermata | Crinoidea | Comatulida | Antedonidae | Antedon | Antedon loveni |
| OTU_8133 | 0 | 1 | 0 | Echinodermata | Asteroidea | Forcipulatida | Asteriidae | Stichaster | Stichaster striatus |
| OTU_17372 | 1 | 0 | 0 | Gastrotricha | Chaetonotida | Chaetonotidae | Chaetonotus | Chaetonotus aff. gelidus MK-2019 | |
| OTU_3701 | 1 | 0 | 0 | Gastrotricha | Chaetonotida | Chaetonotidae | Aspidiophorus | Aspidiophorus sp. n. MK-2019 | |
| OTU_4126 | 1 | 0 | 0 | Gastrotricha | Chaetonotida | Chaetonotidae | Aspidiophorus | Aspidiophorus sp. 1 | |
| OTU_4761 | 1 | 0 | 0 | Gastrotricha | Chaetonotida | Chaetonotidae | Chaetonotus | Chaetonotus sp. 2 MK-2019 | |
| OTU_6356 | 1 | 0 | 0 | Gastrotricha | Chaetonotida | Chaetonotidae | Chaetonotus | Chaetonotus sp. 1 | |
| OTU_7757 | 1 | 0 | 0 | Gastrotricha | Chaetonotida | Chaetonotidae | Aspidiophorus | Aspidiophorus sp. n. MK-2019 | |
| OTU_15575 | 0 | 1 | 0 | Gastrotricha | Macrodasyida | Thaumastodermatidae | Tetranchyroderma | Tetranchyroderma sp. 3 TK-2011 | |
| OTU_1292 | 1 | 1 | 0 | Mollusca | Gastropoda | Cycloneritida | Neritidae | Neritina | Neritina sp. Suji |
| OTU_1469 | 1 | 1 | 0 | Mollusca | Gastropoda | Systellommatophora | Onchidiidae | Melayonchis | Melayonchis tillieri |
| OTU_3647 | 1 | 1 | 0 | Mollusca | Gastropoda | Pachychilidae | Sulcospira | Sulcospira paludiformis | |
| OTU_94 | 1 | 1 | 0 | Mollusca | Gastropoda | Planorbidae | Biomphalaria | Biomphalaria straminea | |
| OTU_6172 | 1 | 1 | 0 | Mollusca | Gastropoda | Neogastropoda | Terebridae | Myurella | Myurella affinis |
| OTU_10351 | 1 | 0 | 0 | Mollusca | Gastropoda | Architaenioglossa | Ampullariidae | Pomacea | Pomacea catemacensis |
| OTU_14521 | 1 | 0 | 0 | Mollusca | Gastropoda | Architaenioglossa | Ampullariidae | Pomacea | Pomacea canaliculata |
| OTU_7625 | 1 | 0 | 0 | Mollusca | Gastropoda | Aplysiida | Aplysiidae | Aplysia | Aplysia fasciata |
| OTU_13638 | 1 | 0 | 0 | Mollusca | Gastropoda | Trochida | Calliostomatidae | Calliostoma | Calliostoma iridium |
| OTU_14338 | 1 | 0 | 0 | Mollusca | Gastropoda | Stylommatophora | Camaenidae | Xanthomelon | Xanthomelon interpositum |
| OTU_14515 | 1 | 0 | 0 | Mollusca | Gastropoda | Stylommatophora | Camaenidae | Satsuma | Satsuma jacobii |
| OTU_2134 | 1 | 0 | 0 | Mollusca | Gastropoda | Stylommatophora | Camaenidae | Satsuma | Satsuma hemihelvus |
| OTU_5901 | 1 | 0 | 0 | Mollusca | Gastropoda | Stylommatophora | Camaenidae | Satsuma | Satsuma sp. SPW-2007 |
| OTU_9949 | 1 | 0 | 0 | Mollusca | Gastropoda | Stylommatophora | Clausiliidae | Montenegrina | Montenegrina helvola |
| OTU_736 | 1 | 0 | 0 | Mollusca | Gastropoda | Neogastropoda | Costellariidae | Pusia | Pusia savignyi |
| OTU_6464 | 1 | 0 | 0 | Mollusca | Gastropoda | Nudibranchia | Cuthonellidae | Cuthonella | Cuthonella punicea |
| OTU_4617 | 1 | 0 | 0 | Mollusca | Gastropoda | Architaenioglossa | Diplommatinidae | Diplommatina | Diplommatina sp. nov. AG MS-2010 |
| OTU_6514 | 1 | 0 | 0 | Mollusca | Gastropoda | Nudibranchia | Dotidae | Doto | Doto coronata |
| OTU_10095 | 1 | 0 | 0 | Mollusca | Gastropoda | Ellobiida | Ellobiidae | Ellobium | Ellobium scheepmakeri |
| OTU_3788 | 1 | 0 | 0 | Mollusca | Gastropoda | Lepetellida | Fissurellidae | Fissurella | Fissurella sp. SWA-2011 |
| OTU_2686 | 1 | 0 | 0 | Mollusca | Gastropoda | Stylommatophora | Geomitridae | Xerocrassa | Xerocrassa ponsi |
| OTU_4632 | 1 | 0 | 0 | Mollusca | Gastropoda | Littorinimorpha | Hydrobiidae | Marstonia | Marstonia lustrica |
| OTU_14429 | 1 | 0 | 0 | Mollusca | Gastropoda | Neogastropoda | Muricidae | Concholepas | Concholepas concholepas |
| OTU_9579 | 1 | 0 | 0 | Mollusca | Gastropoda | Neogastropoda | Muricidae | Drupa | Drupa morum |
| OTU_14543 | 1 | 0 | 0 | Mollusca | Gastropoda | Cycloneritida | Neritidae | Nerita | Nerita atramentosa |
| OTU_1864 | 1 | 0 | 0 | Mollusca | Gastropoda | Cycloneritida | Neritidae | Neritina | Neritina dilatata |
| OTU_5511 | 1 | 0 | 0 | Mollusca | Cephalopoda | Teuthida | Ommastrephidae | Dosidicus | Dosidicus gigas |
| OTU_10283 | 1 | 0 | 0 | Mollusca | Gastropoda | Systellommatophora | Onchidiidae | Paromoionchis | Paromoionchis daemelii |
| OTU_13627 | 1 | 0 | 0 | Mollusca | Gastropoda | Systellommatophora | Onchidiidae | Onchidella | Onchidella marginata |
| OTU_14456 | 1 | 0 | 0 | Mollusca | Gastropoda | Pachychilidae | Sulcospira | Sulcospira paludiformis | |
| OTU_1546 | 1 | 0 | 0 | Mollusca | Bivalvia | Unionida | Unionidae | Middendorffinaia | Middendorffinaia mongolica |
| OTU_4294 | 1 | 0 | 0 | Mollusca | Bivalvia | Unionida | Unionidae | Pleuronaia | Pleuronaia barnesiana |
| OTU_4892 | 1 | 0 | 0 | Mollusca | Bivalvia | Unionida | Unionidae | Toxolasma | Toxolasma lividus |
| OTU_7039 | 1 | 0 | 0 | Mollusca | Bivalvia | Unionida | Unionidae | Aculamprotula | Aculamprotula fibrosa |
| OTU_10224 | 1 | 0 | 0 | Mollusca | Gastropoda | Littorinimorpha | Vermetidae | Dendropoma | Dendropoma gregarium |
| OTU_5629 | 1 | 0 | 0 | Mollusca | Gastropoda | Littorinimorpha | Vermetidae | Dendropoma | Dendropoma platypus |
| OTU_14128 | 1 | 0 | 0 | Mollusca | Bivalvia | Bivalvia environmental sample | |||
| OTU_12851 | 0 | 1 | 0 | Mollusca | Gastropoda | Stylommatophora | Camaenidae | Satsuma | Satsuma hemihelvus |
| OTU_15803 | 0 | 1 | 0 | Mollusca | Bivalvia | Galeommatida | Galeommatidae | Galeomma | Galeomma turtoni |
| OTU_1980 | 0 | 1 | 0 | Mollusca | Gastropoda | Stylommatophora | Geomitridae | Candidula | Candidula sp. AR_BC7338 |
| OTU_3248 | 0 | 1 | 0 | Mollusca | Gastropoda | Neogastropoda | Terebridae | Hastula | Hastula matheroniana |
| OTU_3959 | 0 | 1 | 0 | Mollusca | Gastropoda | Physidae | Physella | Physella acuta | |
| OTU_4408 | 0 | 1 | 0 | Mollusca | Gastropoda | Stylommatophora | Helicidae | Eobania | Eobania vermiculata |
| OTU_470 | 0 | 1 | 0 | Mollusca | Gastropoda | Plakobranchidae | Thuridilla | Thuridilla vatae | |
| OTU_6184 | 0 | 1 | 0 | Mollusca | Gastropoda | Cycloneritida | Phenacolepadidae | Plesiothyreus | Plesiothyreus newtoni |
| OTU_6194 | 0 | 1 | 0 | Mollusca | Bivalvia | Unionida | Unionidae | Hyriopsis | Hyriopsis bialatus complex sp. 2 PP-2017 |
| OTU_6526 | 0 | 1 | 0 | Mollusca | Gastropoda | Stylommatophora | Clausiliidae | Montenegrina | Montenegrina umbilicata |
| OTU_7189 | 0 | 1 | 0 | Mollusca | Gastropoda | Stylommatophora | Camaenidae | Rhagada | Rhagada angulata |
| OTU_8077 | 0 | 1 | 0 | Mollusca | Gastropoda | Siphonariida | Siphonariidae | Siphonaria | Siphonaria lessonii |
| OTU_86 | 0 | 1 | 0 | Mollusca | Gastropoda | Littorinimorpha | Hydrobiidae | Hauffenia | Hauffenia subpiscinalis |
| OTU_965 | 0 | 1 | 0 | Mollusca | Bivalvia | Galeommatida | Galeommatidae | Galeomma | Galeomma turtoni |
| UVC_37 | 0 | 0 | 1 | Mollusca | Bivalvia | Mytilida | Mytilidae | Perna | Perna viridis |
| UVC_38 | 0 | 0 | 1 | Mollusca | Bivalvia | Mytilida | Modiolidae | Vignadula | Vignadula atrata |
| UVC_15 | 0 | 0 | 1 | Mollusca | Cephalopoda | Sepiida | Sepiidae | Sepiella | Sepiella maindroni de Rochebrune |
| UVC_16 | 0 | 0 | 1 | Mollusca | Gastropoda | Trochidae | Calliostomatidae | Tristichotrochus | Tristichotrochus unicum |
| UVC_17 | 0 | 0 | 1 | Mollusca | Gastropoda | Trochidae | Trochidae | Monodonta | Monodonta labio |
| UVC_18 | 0 | 0 | 1 | Mollusca | Gastropoda | Littorinimorpha | Ovulidae | Ovulidae sp. | |
| UVC_19 | 0 | 0 | 1 | Mollusca | Gastropoda | Littorinimorpha | Ovulidae | Cuspivolva | Cuspivolva formosa |
| UVC_20 | 0 | 0 | 1 | Mollusca | Gastropoda | Neogastropoda | Buccinidae | Cantharus | Cantharus cecillei |
| UVC_21 | 0 | 0 | 1 | Mollusca | Gastropoda | Neogastropoda | Cymatiidae | Gyrineum | Gyrineum natator |
| UVC_22 | 0 | 0 | 1 | Mollusca | Gastropoda | Neogastropoda | Muricidae | Chicoreus | Chicoreus asianus |
| UVC_23 | 0 | 0 | 1 | Mollusca | Gastropoda | Neogastropoda | Muricidae | Reishia | Reishia clavigera |
| UVC_24 | 0 | 0 | 1 | Mollusca | Gastropoda | Neogastropoda | Muricidae | Reishia | Reishia luteostoma |
| UVC_25 | 0 | 0 | 1 | Mollusca | Gastropoda | Neogastropoda | Nassariidae | Nassarius | Nassarius hepaticus |
| UVC_26 | 0 | 0 | 1 | Mollusca | Gastropoda | Nudibranchia | Nudibranchia sp. | ||
| UVC_27 | 0 | 0 | 1 | Mollusca | Gastropoda | Nudibranchia | Facelinidae | Sakuraeolis | Sakuraeolis sakuracea |
| UVC_28 | 0 | 0 | 1 | Mollusca | Gastropoda | Nudibranchia | Dendrodorididae | Doriopsilla | Doriopsilla sp. |
| UVC_32 | 0 | 0 | 1 | Mollusca | Gastropoda | Pyramidellidae | Odostomia | Odostomia subangulata | |
| OTU_7430 | 1 | 0 | 0 | Nematoda | Chromadorea | Strongylida | Ancylostomatidae | Ancylostoma | Ancylostoma ceylanicum |
| OTU_7275 | 1 | 0 | 0 | Nematoda | Chromadorea | Desmodorida | Desmodoridae | Metachromadora | Metachromadora sp. 2AMA |
| OTU_13801 | 1 | 0 | 0 | Nematoda | Chromadorea | Rhabditida | Rhabditidae | Caenorhabditis | Caenorhabditis inopinata |
| OTU_8772 | 1 | 0 | 0 | Nemertea | Pilidiophora | Heteronemertea | Baseodiscidae | Baseodiscus | Baseodiscus sp. 2 SA-2011 |
| OTU_2470 | 1 | 0 | 0 | Nemertea | Palaeonemertea | Tubulanidae | Tubulanus | Tubulanus polymorphus | |
| OTU_17313 | 0 | 1 | 0 | Nemertea | Enopla | Monostilifera | Monostilifera sp. MCZ IZ 45646 | ||
| OTU_502 | 0 | 1 | 0 | Nemertea | Enopla | Monostilifera | Amphiporidae | Amphiporus | Amphiporus cruentatus |
| OTU_17155 | 1 | 0 | 0 | Phoronida | Phoroniformea sp. larval OTU P1 | ||||
| OTU_435 | 1 | 1 | 0 | Platyhelminthes | Trematoda | Diplostomida | Diplostomidae | Crassiphiala | Crassiphiala sp. Lineage 2 |
| OTU_16856 | 0 | 1 | 0 | Platyhelminthes | Rhabditophora | Tricladida | Geoplanidae | Microplana | Microplana sp. MOTU37 |
| OTU_897 | 1 | 0 | 0 | Porifera | Demospongiae | Bubarida | Dictyonellidae | Scopalina | Scopalina lophyropoda |
| OTU_7359 | 1 | 0 | 0 | Porifera | Demospongiae | Suberitida | Halichondriidae | Ciocalypta | Ciocalypta sp. DE-2012 |
| OTU_8918 | 1 | 0 | 0 | Porifera | Demospongiae | Haplosclerida | Niphatidae | Amphimedon | Amphimedon queenslandica |
| OTU_7393 | 1 | 0 | 0 | Porifera | Homoscleromorpha | Homosclerophorida | Plakinidae | Plakina | Plakina coerulea |
| OTU_9873 | 1 | 0 | 0 | Porifera | Homoscleromorpha | Homosclerophorida | Plakinidae | Plakina | Plakina sp. 1 |
| OTU_7613 | 1 | 0 | 0 | Porifera | Demospongiae | Axinellida | Raspailiidae | Raspailiidae sp. Po.25592 | |
| OTU_9606 | 1 | 0 | 0 | Porifera | Demospongiae | Tetractinellida | Tetillidae | Cinachyrella | Cinachyrella alloclada |
| OTU_10893 | 0 | 1 | 0 | Porifera | Demospongiae | Suberitida | Suberitidae | Aaptos | Aaptos nuda |
| OTU_15892 | 0 | 1 | 0 | Porifera | Demospongiae | Suberitida | Halichondriidae | Halichondriidae sp. UCMPWC1015 | |
| OTU_16058 | 0 | 1 | 0 | Porifera | Demospongiae | Poecilosclerida | Mycalidae | Mycale | Mycale mirabilis |
| OTU_16093 | 0 | 1 | 0 | Porifera | Porifera sp. Sp08 | ||||
| OTU_17151 | 0 | 1 | 0 | Porifera | Demospongiae | Tetractinellida | Tetillidae | Cinachyrella | Cinachyrella australiensis |
| OTU_2563 | 0 | 1 | 0 | Porifera | Demospongiae | Tethyida | Hemiasterellidae | Adreus | Adreus fascicularis |
| OTU_5032 | 0 | 1 | 0 | Porifera | Demospongiae | Haplosclerida | Chalinidae | Haliclona | Haliclona sp. P2MK |
| UVC_35 | 0 | 0 | 1 | Porifera | Demospongiae | Tethyida | Tethyidae | Tethya | Tethya aurantium |
| UVC_29 | 0 | 0 | 1 | Porifera | Demospongiae | Tethyida | Tethyidae | Tethya | Tethya sp. 1 |
References
- Halpern, B.S.; Frazier, M.; Afflerbach, J.; Lowndes, J.S.; Micheli, F.; O’Hara, C.; Scarborough, C.; Selkoe, K.A. Recent Pace of Change in Human Impact on the World’s Ocean. Sci. Rep. 2019, 9, 11609. [Google Scholar] [CrossRef] [PubMed]
- Chung, A.; Kam, Y.C.; Shea, S.K.H.; Schunter, C. Detecting Fish Diversity in Urban-Impacted Ecosystems: A Comparative Approach of eDNA Metabarcoding and UVC. Environ. DNA 2024, 6, e70048. [Google Scholar] [CrossRef]
- Stat, M.; Huggett, M.J.; Bernasconi, R.; DiBattista, J.D.; Berry, T.E.; Newman, S.J.; Harvey, E.S.; Bunce, M. Ecosystem Biomonitoring with eDNA: Metabarcoding across the Tree of Life in a Tropical Marine Environment. Sci. Rep. 2017, 7, 12240. [Google Scholar] [CrossRef]
- Jardim, V.L.; Grall, J.; Barros-Barreto, M.B.; Bizien, A.; Benoit, T.; Braga, J.C.; Brodie, J.; Burel, T.; Cabrito, A.; Diaz-Pulido, G.; et al. Common Terminology to Unify Research and Conservation of Coralline Algae and the Habitats They Create. Aquat. Conserv. 2025, 35, e70121. [Google Scholar] [CrossRef]
- Deiner, K.; Bik, H.M.; Mächler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; De Vere, N.; et al. Environmental DNA Metabarcoding: Transforming How We Survey Animal and Plant Communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Brochmann, C.; Willerslev, E. Towards Next-generation Biodiversity Assessment Using DNA Metabarcoding. Mol. Ecol. 2012, 21, 2045–2050. [Google Scholar] [CrossRef]
- Djurhuus, A.; Closek, C.J.; Kelly, R.P.; Pitz, K.J.; Michisaki, R.P.; Starks, H.A.; Walz, K.R.; Andruszkiewicz, E.A.; Olesin, E.; Hubbard, K.; et al. Environmental DNA Reveals Seasonal Shifts and Potential Interactions in a Marine Community. Nat. Commun. 2020, 11, 254. [Google Scholar] [CrossRef]
- West, K.M.; Stat, M.; Harvey, E.S.; Skepper, C.L.; DiBattista, J.D.; Richards, Z.T.; Travers, M.J.; Newman, S.J.; Bunce, M. eDNA Metabarcoding Survey Reveals Fine-scale Coral Reef Community Variation across a Remote, Tropical Island Ecosystem. Mol. Ecol. 2020, 29, 1069–1086. [Google Scholar] [CrossRef]
- Cai, H. Brief Introduction to Nanji Islands National Marine Nature Reserve; China Ocean Press: Beijing, China, 2021; pp. 1–19. [Google Scholar]
- Jessop, S.A.; Saunders, B.J.; Goetze, J.S.; Harvey, E.S. A comparison of underwater visual census, baited, diver operated and remotely operated stereo-video for sampling shallow water reef fishes. Estuar. Coast. Shelf Sci. 2022, 276, 108017. [Google Scholar] [CrossRef]
- Suominen, S.; Provoost, P.; Principe, S.; Boulanger, E.; Burrows, M.T.; Campoy, A.N.; Costello, M.J.; Earl, C.; Gante, H.F.; Gillard, E.; et al. Engaging Communities to Safeguard Ocean Life: UNESCO Environmental DNA Expeditions; United Nations Educational, Scientific and Cultural Organization: Paris, France, 2024; p. 9. [Google Scholar]
- Leray, M.; Yang, J.Y.; Meyer, C.P.; Mills, S.C.; Agudelo, N.; Ranwez, V.; Boehm, J.T.; Machida, R.J. A New Versatile Primer Set Targeting a Short Fragment of the Mitochondrial COI Region for Metabarcoding Metazoan Diversity: Application for Characterizing Coral Reef Fish Gut Contents. Front. Zool. 2013, 10, 34. [Google Scholar] [CrossRef]
- Geller, J.; Meyer, C.; Parker, M.; Hawk, H. Redesign of PCR Primers for Mitochondrial Cytochrome c Oxidase Subunit I for Marine Invertebrates and Application in All-Taxa Biotic Surveys. Mol. Ecol. Resour. 2013, 13, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Beermann, A.J.; Zizka, V.M.A.; Elbrecht, V.; Baranov, V.; Leese, F. DNA Metabarcoding Reveals the Complex and Hidden Responses of Chironomids to Multiple Stressors. Environ. Sci. Eur. 2018, 30, 26. [Google Scholar] [CrossRef]
- Steinke, D.; Braukmann, T.W.; Manerus, L.; Woodhouse, A.; Elbrecht, V. Effects of Malaise Trap Spacing on Species Richness and Composition of Terrestrial Arthropod Bulk Samples. Metabarcoding Metagenom. 2021, 5, e59201. [Google Scholar] [CrossRef]
- Di Capua, I.; Piredda, R.; Mazzocchi, M.G.; Zingone, A. Metazoan Diversity and Seasonality through eDNA Metabarcoding at a Mediterranean Long-Term Ecological Research Site. ICES J. Mar. Sci. 2021, 78, 3303–3316. [Google Scholar] [CrossRef]
- Ginestet, C. Ggplot2: Elegant Graphics for Data Analysis. J. R. Stat. Soc. Ser. A Stat. Soc. 2011, 174, 245–246. [Google Scholar] [CrossRef]
- Turner, C.R.; Uy, K.L.; Everhart, R.C. Fish Environmental DNA Is More Concentrated in Aquatic Sediments than Surface Water. Biol. Conserv. 2015, 183, 93–102. [Google Scholar] [CrossRef]
- Buxton, A.S.; Groombridge, J.J.; Griffiths, R.A. Seasonal Variation in Environmental DNA Detection in Sediment and Water Samples. PLoS ONE 2018, 13, e0191737. [Google Scholar] [CrossRef]
- Collins, R.A.; Wangensteen, O.S.; O’Gorman, E.J.; Mariani, S.; Sims, D.W.; Genner, M.J. Persistence of Environmental DNA in Marine Systems. Commun. Biol. 2018, 1, 185. [Google Scholar] [CrossRef]
- Nevers, M.B.; Przybyla-Kelly, K.; Shively, D.; Morris, C.C.; Dickey, J.; Byappanahalli, M.N. Influence of Sediment and Stream Transport on Detecting a Source of Environmental DNA. PLoS ONE 2020, 15, e0244086. [Google Scholar] [CrossRef]
- Brandt, M.I.; Pradillon, F.; Trouche, B.; Henry, N.; Liautard-Haag, C.; Cambon-Bonavita, M.-A.; Cueff-Gauchard, V.; Wincker, P.; Belser, C.; Poulain, J.; et al. Evaluating Sediment and Water Sampling Methods for the Estimation of Deep-Sea Biodiversity Using Environmental DNA. Sci. Rep. 2021, 11, 7856. [Google Scholar] [CrossRef] [PubMed]
- Ogata, M.; Masuda, R.; Harino, H.; Sakata, M.K.; Hatakeyama, M.; Yokoyama, K.; Yamashita, Y.; Minamoto, T. Environmental DNA Preserved in Marine Sediment for Detecting Jellyfish Blooms after a Tsunami. Sci. Rep. 2021, 11, 16830. [Google Scholar] [CrossRef] [PubMed]
- Suzzi, A.L.; Huggett, M.J.; Gaston, T.F.; MacFarlane, G.R.; Alam, M.R.; Gibb, J.; Stat, M. eDNA Metabarcoding Reveals Shifts in Sediment Eukaryote Communities in a Metal Contaminated Estuary. Mar. Pollut. Bull. 2023, 191, 114896. [Google Scholar] [CrossRef] [PubMed]
- Roblet, S.; Priouzeau, F.; Gambini, G.; Cottalorda, J.-M.; Gastaldi, J.M.; Pey, A.; Raybaud, V.; Suarez, G.R.; Serre, C.; Sabourault, C.; et al. From Sight to Sequence: Underwater Visual Census vs Environmental DNA Metabarcoding for the Monitoring of Taxonomic and Functional Fish Diversity. Sci. Total Environ. 2024, 956, 177250. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).