Abstract
PRDM (PRDI-BF1 and RIZ homology domain containing) protein family members are characterized by the presence of a PR domain and a variable number of Zn-finger repeats. Experimental evidence has shown that the PRDM proteins play an important role in gene expression regulation, modifying the chromatin structure either directly, through the intrinsic methyltransferase activity, or indirectly through the recruitment of chromatin remodeling complexes. PRDM proteins have a dual action: they mediate the effect induced by different cell signals like steroid hormones and control the expression of growth factors. PRDM proteins therefore have a pivotal role in the transduction of signals that control cell proliferation and differentiation and consequently neoplastic transformation. In this review, we describe pathways in which PRDM proteins are involved and the molecular mechanism of their transcriptional regulation.
1. Structure of PRDM Proteins and Their Alternative Gene Products
The PRDM (PRDI-BF1 and RIZ homology domain containing) protein family is characterized by the presence of an N-terminal PR (PRDI-BF1 and RIZ1 homology) domain. The PR domain shares high homology with the catalytic SET (Suppressor of variegation 3–9, Enhancer of zeste and Trithorax) domain that defines a group of histone methyltransferases [1]. In the human genome there are 17 genes encoding for proteins with a PR/SET and all of them but PRDM11 have a variable number of Zn-finger domains [2]. PRDM proteins have a pivotal role in the transduction of signals that control cell proliferation and differentiation and consequently neoplastic transformation [3]. A common characteristic of PRDM family genes is the expression of different molecular forms by alternative splicing or by the action of different promoters. Furthermore, some genes of this family are expressed as two alternative forms, one lacking the PR domain (PR-minus) but otherwise identical to the other PR-containing product (PR-plus) (PRDM1, PRDM2, PRDM3, PRDM16) [4,5,6,7]. Others genes encode for proteins that differ for the presence or absence of Zn-finger domains (PRDM6, PRDM9) [8,9]. Recent reviews presented schematic diagrams showing the main PRDM gene products [2,3,10].
1.1. Alternative Promoters
PRDM1 and PRDM2, initially identified as Blimp-1 (B lymphocyte-induced maturation protein-1) and RIZ (Retinoblastoma interacting zinc finger protein) respectively, have two promoters that encode for a PR-plus and a PR-minus isoform. PRDM1 promoters are localized upstream of exon 1 and exon 4 respectively. These transcriptional start sites at two promoters guide: PRDI-BF1 (Positive regulatory domain I-binding factor 1) α (PR-plus) e PRDI-BF1β (PR-minus) that differ only by the PR domain presence [4,11]. One promoter of PRDM2 is located upstream of the open reading frame in a region including exon 1a and a second promoter is located within intron 5 and exon 6 [6]. Similarly to PRDM1, PRDM2 expresses two proteins, PRDM2a/RIZ1 (PR-plus) and PRDM2b/RIZ2 (PR-minus), by differential transcription initiated by the two promoters.
PRDM16 encodes a Zn-finger protein (MEL1) that shares 63% sequence similarity to PRDM3/MECOM (MDS1 and EVI1 complex locus, also known as EVI1, MDS1). Like PRDM3, two mRNAs coding for PR-plus and PR-minus protein are transcribed from this locus: PRDM16/MEL1 (MDS1/EVI1-like gene 1), the PR-plus form, with the PR domain coded from codon ATC91 (exon 2) to codon CCC223 (exon 5) and PRDM16/MEL1S, the PR-minus form, initiated from an internal codon ATG599 (exon 9) [12,13].
1.2. Alternative Splicing
PRDM1 encodes also for an alternatively spliced transcript lacking exon 7; this variant (Blimp-1Δexon7) lacks DNA binding activity and fails to bind G9a or HDAC1/2, but retains the ability to interact with Prmt5 (protein methyltransferase 5) [14]. This evidence suggests that the expression of PRDM1 alternative splicing variants is regulated during development by chromatin structure modification and fine-tunes PRDM1’s functional capabilities [14].
PRDM3/MECOM is a complex locus containing EV1 and MDS1 genes, located on chromosome 3q26. This complex locus encodes for different gene products generated by alternative splicing or by intragenic splicing [15].
The major and most studied protein, EVI1 (Ecotropic virus integration site 1 protein homolog), also named MECOM (E) is a 1051 aminoacid protein [16], that consists of an N-terminal seven-zinc finger domain, a central transcription repression domain, a second zinc finger domain with three finger motifs and a C-terminal acidic region. One EVI1 mRNA splice variant, identified as PRDM3/EVI1/Δ324, is a protein that lacks zinc fingers 6 and 7 as well as part of the transcription repression domain. The PRDM3/EVI1-Rp9 variant is abundant both in humans and mice and lacks 9 amino acids in the repression domain. The variant, PRDM3/EVI1/Δ105, is a protein truncated of 105 aminoacids at its C-terminus and is detected only in murine but not in human cells.
EVI1 may form a fusion transcript with the MDS1 gene located upstream. The use of alternative transcriptional start sites generates mRNA combining sequences derived from the MDS1 (Myelodysplasia syndrome-associated protein 1) gene, which is located upstream of EVI1, and the EVI1 sequences starting from exon 2. The derived protein, called MDS1/EVI1 or MECOM (ME), from this mRNA contains a 188 amino acid extension encoding a PR domain at its N-terminus, but is otherwise identical to the EVI1 protein [5,15,17].
In mice, Prdm6 encodes for four isoforms referred to as Prdm6/4#, 3#, 33# and 36#, produced by alternative splicing. Prdm6/4# has a PR/SET domain in the central region and four Zn-finger domains at its C-terminal region. Prdm6/3# and Prdm6/33# have an additional sequence of 31 residues produced by retention of the first intron, absent in the Prdm6/4# transcript. Similarly, Prdm6/36# has a single amino acid insertion if compared to Prdm6/4#, derived from the recognition of an alternative splicing site 3 bp upstream of the intron 1/exon 2 boundary. Prdm6/33# is a PR-minus isoform, obtained by an alternative splicing event that results in the deletion of exons 3–5 of transcript encoding for Prdm6/4#. Prdm6/36#, missing the fourth Zn-finger domain, derives from an alternative splicing in intron 7 of Prdm6/4# transcript that includes an in-frame stop codon [9].
The Prdm9 murine gene encodes for three isoforms generated by alternative splicing: one isoform has a PR domain in its N-terminal region and a Zn-finger motif in its C-terminal portion. The other two isoforms generated by alternative splicing lack the Zn-finger domain responsible of the nuclear localization [8].
In 2002 Siegel, analyzing the protein extracted from mouse brain by Western blot with an antibody to the C-terminal region of Prdm10/tristanin, identified two molecular forms of this protein of 50 kDa and 25 kDa respectively. This finding corroborates the hypothesis that there are also different molecular variants encoded by the gene Prdm10 [18].
The functional relevance of the different variants has not yet been elucidated. Table 1 summarizes the information relative to PRDM proteins obtained from Uniprot [19] and National Center for Biotechnology Information protein database [20].

Table 1.
PRDM (PRDI-BF1 and RIZ homology domain containing) proteins derived by alternative promoters activity or alternative splicing.
| Human | ||||||||
|---|---|---|---|---|---|---|---|---|
| gene name | protein name | localization | molecular forms | alternative promoter (UNIPROT entry) | splicing variants (UNIPROT entry) | length (aa) | PR domain | HMT activity |
| PRDM1 (BLIMP1) | PR domain zinc finger protein 1 (BLIMP1) | nucleuscytoplasm | 3 | Isoform 1 'canonical' sequence (O75626-1) | Isoform 2 | 825 | aa 85-205 | no |
| 1-36: missing | ||||||||
| (Beta-interferon gene positive regulatory domain I-binding factor) | (O75626-2) | |||||||
| Isoform 3 | partially missing | no | ||||||
| 1-3: MLD → MEK | ||||||||
| (PR domain-containing protein 1) | 4-137: missing | |||||||
| Positive regulatory domain I-binding factor 1) | (O75626-3) | |||||||
| PRDM2 (KMT8, RIZ) | PR domain zinc finger protein 2 | nucleus | 3 | Isoform 1 (RIZ1) 'canonical' sequence (Q13029-1) | Isoform 2 (MTB-Zf) | 1,718 | aa 27-145 | H3K9 |
| (GATA-3-binding protein G3B) | 1679-1682: SYSL → RNFL | |||||||
| 1683-1718: missing * | ||||||||
| (Lysine N - methyltransferase 8) | (Q13029-2) | |||||||
| (MTB-ZF) | Isoform 3 (RIZ2) 1-201: missing (13029-3) | no | no | |||||
| (MTE-binding protein) | ||||||||
| (PR domain-containing protein 2) | ||||||||
| (RIZ, Retinoblastoma protein-interacting zinc finger protein) | ||||||||
| PRDM3/MECOM (EVI1) | MDS1 and EVI1 complex locus protein EVI1 (Ecotropic virus integration site 1 protein homolog-EVI-1) | nucleus | 6 | Isoform 1 (Evi-1a) 'canonical' sequence | 1,051 | aa 79-194 | H3K9me1 | |
| (Q03112-1) | ||||||||
| Isoform 2 (Evi-1c) (Mds1/Evi1) | ||||||||
| (Q03112-3) | ||||||||
| 1-1: M → MRSKGRARKL...APGEELLL-FM | ||||||||
| Contains an additional SET domain at positions 79-194 | ||||||||
| Isoform 3 (Mds1) | ||||||||
| (Q13465-1) | ||||||||
| Isoform 4 | ||||||||
| 1-1: M → MILDEFYNVKFCIDASQPD-VGSWLKYIRFAGCYDQHNLVACQINDQIFYRVVADIAPGEELLLFM | ||||||||
| 138-138: K → KQ | ||||||||
| (Q03112-4) | ||||||||
| Isoform 5 | ||||||||
| 672-680: missing | ||||||||
| (Q03112-5) | ||||||||
| Isoform 6 | ||||||||
| 138-138: K → KQ | ||||||||
| 672-680: missing | ||||||||
| (Q03112-6) | ||||||||
| PRDM4 (PFM1) | PR domain zinc finger protein 4 (PR domain-containing protein 4) | nucleus | 1 | 801 | aa 412-533 | no | ||
| PRDM5 (PFM2) | PR domain zinc finger protein 5 (PR domain-containing protein 5) | nucleus | 3 | Isoform 1 'canonical' sequence | 630 | aa 8-128 | no | |
| (Q9NQX1-1) | ||||||||
| Isoform 2 | ||||||||
| 218-248: missing | ||||||||
| (Q9NQX1-2) | ||||||||
| Isoform 3 (Q9NQX1-3) | ||||||||
| 101-111: EGENIFYLAVE → DKNLGP-AEWRG | ||||||||
| 112-630: missing | ||||||||
| PRDM6 (PFM3) | Putative histone-lysine N -methyltransferase PRDM6 | nucleus | 3 | Isoform 1 (Q9NQX0-3) | 595 | aa 247-369 | H4K20 | |
| 'canonical' sequence | ||||||||
| (PR domain zinc finger protein 6) | Isoform 2 (B) | |||||||
| 1-182: missing | ||||||||
| (PR domain-containing protein 6) | ||||||||
| 314-595: missing | ||||||||
| (Q9NQX0-2) | ||||||||
| Isoform 3 (A) | ||||||||
| 1-182: missing | ||||||||
| (Q9NQX0-1) | ||||||||
| PRDM7 (PFM4) | Probable histone-lysine
N
- methyltransferase PRDM7 (PR domain zinc finger protein 7) (PR domain-containing protein 7) | nucleus | 3 | Isoform 1 'canonical' sequence | 492 | aa 246-362 | no | |
| (Q9NQW5-3) | ||||||||
| Isoform 2 (B) | ||||||||
| 1-206: missing | ||||||||
| 318-377: YVNCARDDEE...RSSIEPAESL → TKARDPSMSL...RGSESGaaIF | ||||||||
| 378-492: missing | ||||||||
| (Q9NQW5-2) | ||||||||
| Isoform 3 (A) | ||||||||
| 1-206: missing | ||||||||
| 368-492: RSSIEPAESL...VKRSKKGPNS → KWGSKWKKEL...GEAPVCRKDE | ||||||||
| (Q9NQW5-1) | ||||||||
| PRDM8 (PFM5) | PR domain zinc finger protein 8 | nucleus | 2 | Isoform 1 'canonical' sequence | aa 8-135 | H3K9 | ||
| (Q9NQV8-1) | ||||||||
| Isoform 2 | ||||||||
| (PR domain-containing protein 8) | 332-334: GRG → aaL | |||||||
| 335-689: missing | ||||||||
| (Q9NQV8-2) | ||||||||
| PRDM9 ( PFM6 ) | Histone-lysine N -methyltransferase PRDM9 | nucleus | 1 | (Q9NQV7) | 894 | aa 246-362 | H3K4me3 | |
| (PR domain zinc finger protein 9; | ||||||||
| PR domain-containing protein 9) | ||||||||
| PRDM10 (KIaa 1231; PFM7; TRIS) | PR domain zinc finger protein 10 (PR domain-containing protein 10) (Tristanin) | nucleus | 6 | Isoform 3 'canonical' sequence | 1,147 | aa 206-330 | no | |
| (Q9NQV6-3) | ||||||||
| Isoform 2 | ||||||||
| 1-97: MDSKDESSHV...AYVQQDATAQ → MSAYSVPSTFA | ||||||||
| 511-514: missing | ||||||||
| 952-985: missing | ||||||||
| (Q9NQV6-2 | ||||||||
| Isoform 1 | ||||||||
| 1-97: MDSKDESSHV...AYVQQDATAQ → MSAYSVPSTFA | ||||||||
| (Q9NQV6-1) | ||||||||
| Isoform 4 | ||||||||
| 511-514: missing | ||||||||
| 984-984: I → IQVSEPTASAPSSA * | ||||||||
| (Q9NQV6-4) | ||||||||
| Isoform 5 | ||||||||
| 1-97: MDSKDESSHV...AYVQQDATAQ → MSAYSVPSTFA | ||||||||
| 984-984: I → IQVSEPTASAPSSA * | ||||||||
| (Q9NQV6-5) | ||||||||
| Isoform 6 | ||||||||
| 511-514: missing | ||||||||
| 984-984: I → IQVSEPTASAPSSA | ||||||||
| 1132-1147: TTTNGNGSSEVHITKP → AGSKVIQNEF...IVFKRISKRI * | ||||||||
| (Q9NQV6-6) | ||||||||
| PRDM11 (PFM8) | PR domain-containing protein 11 | 2 | Isoform 1 'canonical' sequence | 511 | aa 149-264 | no | ||
| (Q9NQV5-1) | ||||||||
| Isoform 2 | ||||||||
| 1-34: missing * | ||||||||
| (Q9NQV5-2) | ||||||||
| PRDM12 (PFM9) | PR domain zinc finger protein 12 | nucleus | 1 | (Q9H4Q4) | 367 | aa 87-207 | no | |
| (PR domain-containing protein 12) | ||||||||
| PRDM13 ( PFM10 ) | PR domain zinc finger protein 13 | nucleus | 1 | (Q9H4Q3) | 707 | aa 1-116 | no | |
| (PR domain-containing protein 13) | ||||||||
| PRDM14 | PR domain zinc finger protein 14 | nucleus | 1 | (Q9GZV8) | 571 | aa 253-371 | no | |
| (PR domain-containing protein 14) | ||||||||
| PRDM15 (C21orf83; ZNF298) | PR domain zinc finger protein 15 | nucleus | 1 | (P57O71) | 1,507 | aa 406-529 | no | |
| (PR domain-containing protein 15)(Zinc finger protein 298) | ||||||||
| PRDM16 (KIaa 1675; MEL1; PFM13) | PR domain zinc finger protein 16 | nucleus | 4 | Isoform 1 'canonical' sequence | 1,276 | aa 83-215 | H3K9me1 | |
| (Q9HAZ2-1) | ||||||||
| Isoform 2 (MEL1L) | ||||||||
| (PR domain-containing protein 16) | 1233-1251: missing * | |||||||
| (Q9HAZ2-2) | ||||||||
| Isoform 3 | ||||||||
| 191-191: Q → QV | ||||||||
| (Transcription factor MEL1) | 868-868: missing * | |||||||
| (Q9HAZ2-3) | ||||||||
| Isoform 4 | ||||||||
| Also known as: | ||||||||
| MEL1S | ||||||||
| 1-184: missing | ||||||||
| (Q9HAZ2-4) | ||||||||
| ZNF408 (PFM14; PRDM17) | Zinc finger protein 408 | nucleus | (Q9H9D4) | 720 | ||||
| (PR domain zinc finger protein 17) | ||||||||
| Mouse | ||||||||
| gene name | protein name | localization | molecular forms | alternative promoter(UNIPROT entry) | splicing variants(UNIPROT entry) | length (aa) | PR domain | HMT activity |
| Prdm1 (Blimp1) | PR domain zinc finger protein 1(B lymphocyte-induced maturation protein 1-Blimp1) | nucleuscytoplasm | 5 | Isoform 1 'canonical' sequence(Q60636-1) | Isoform 2 | 856 | aa 118-237 | no |
| Also known as: 1A | ||||||||
| 1-47: MREAYLRCWIFSWKNVWVRP-CQRLHFKTVLLQGSLLYTALDSYSTVQ → MLDLLLEKRVGTTL | ||||||||
| (Q60636-2) | ||||||||
| Isoform 3 Also known as: 1B 1-67: missing (Q60636-3) | Isoform 4 (1C) | |||||||
| 1-47: MREAYLRCWI...TALDSYSTVQ → MTPGVPGHRTQQRPQHISALSDK-AKDCSK | ||||||||
| (Q60636-4) | ||||||||
| Isoform 5 | ||||||||
| Also known as: delta exon 7; | ||||||||
| (Beta-interferon gene positive regulatory domain I-binding factor)(PR domain-containing protein 1) | ||||||||
| 624-666: missing | ||||||||
| (Q60636-5) | ||||||||
| Prdm2 (KMT; Riz1 ; Znfpr1c1 ) | Prdm2 protein | nucleus | 1 | 1,670 | aa 34-144 | H3K9 | ||
| PRDM3 / Mecom ( Evi1 ) | MDS1 and EVI1 complex locus protein EVI1 (Ecotropic virus integration site 1 protein-EVI-1) | nucleus | 2 | Isoform 1 'canonical' sequence | 1,042 | aa 81-196 | H3K9me1 | |
| (P14404-1) | ||||||||
| Isoform 2 | ||||||||
| (Q9Z1L8-1) | ||||||||
| Prdm4 | PR domain zinc finger protein 4 | nucleus | 1 | (Q80V63) | 803 | aa 415-536 | no | |
| (PR domain-containing protein 4) | ||||||||
| Prdm5 | PR domain zinc finger protein 5 | nucleus | 1 | (Q9CXE0) | 599 | aa 8-128 | no | |
| (PR domain-containing protein 5) | ||||||||
| Prdm6 ( Gm92 ; Prism ) | Putative histone-lysine
N
-methyltransferase PRDM6 (PR domain zinc finger protein 6) (PR domain-containing protein 6) | Isoform 1 'canonical' sequence | 596 | aa 248-370 | H4K20 | |||
| (Q3UZD5-1) | ||||||||
| Isoform 2 | ||||||||
| 1-201: missing | ||||||||
| (Q3UZD5-2) | ||||||||
| Isoform 3 | ||||||||
| 1-392: missing | ||||||||
| (Q3UZD5-3) | ||||||||
| Isoform 4 | ||||||||
| 28-58: missing | ||||||||
| (Q3UZD5-4) | ||||||||
| Prdm8 | PR domain zinc finger protein 8 | nucleus | (Q8BZ97) | 687 | aa 8-135 | H3K9 | ||
| (PR domain-containing protein 8) | ||||||||
| Prdm9 ( Hst1 ; Meisetz ) | Histone-lysine N -methyltransferase PRDM9 | nucleus | 4 | Isoform 1 'canonical' (Meisetz) | 843 | aa 246-362 | H3K4me3 | |
| (Q96EQ9-1) | ||||||||
| Isoform 2 (Meisetz-S1) | ||||||||
| (Hybrid sterility protein 1) | 382-404: ELRTEIHPCLLCSLAFSSQKFLT → GGHYYDSLKKKEKREFSLRIFIF | |||||||
| (Meiosis-induced factor containing a PR/SET domain and zinc-finger motif) | 405-843: missing | |||||||
| (Q96EQ9-2) | ||||||||
| Isoform 3 (Meisetz-S2) | ||||||||
| 382-418: ELRTEIHPCLLCSLAFSSQKFL-TQHMEWNHRTEIFPG → DLFIIICKYT-VAVFRHTRRGSQILLRMVVSHHVVAGI | ||||||||
| (PR domain zinc finger protein 9) | ||||||||
| 419-843: missing | ||||||||
| (PR domain-containing protein 9) | ||||||||
| (Q96EQ9-3) | ||||||||
| Isoform 4 | ||||||||
| 1-121: missing | ||||||||
| 382-404: ELRTEIHPCLLCSLAFSSQKFLT → GGHYYDSLKKKEKREFSLRIFIF | ||||||||
| 405-843: missing | ||||||||
| (Q96EQ9-4) | ||||||||
| Prdm10 ( Gm1112 , Tris ) | PR domain zinc finger protein 10 | nucleus | 2 | Isoform 1 'canonical' sequence | 1,184 | aa 200-324 | no | |
| (Q3UTQ7-1) | ||||||||
| Isoform 2 | ||||||||
| 318-341: WYaaSYAEFVNQKIHDISEEE-RKV → QNWIHSCLPARVMIRALSY-KRILP | ||||||||
| (PR domain-containing protein 10) | ||||||||
| 342-1184: missing | ||||||||
| (Tristanin) | ||||||||
| (Q3UTQ7-2) | ||||||||
| Prdm11 | PR domain-containing protein 11 | nucleus | 1 | (A2AGX3) | 565 | aa 115-230 | no | |
| Prdm12 (Gm998) | PR domain zinc finger protein 12 | nucleus | 1 | (A2AJ77) | 365 | aa 87-207 | no | |
| (PR domain-containing protein 12) | ||||||||
| Prdm13 | PR domain zinc finger protein 13 | nucleus | 2 | Isoform 1 'canonical' sequence; | 754 | aa 5-164 | no | |
| (E9PZZ1-1) | ||||||||
| Isoform 2 | ||||||||
| (PR domain-containing protein 13) | ||||||||
| 1-48: missing | ||||||||
| (E9PZZ1-2) | ||||||||
| Prdm14 | PR domain zinc finger protein 14 | nucleus | 1 | (E9Q3T6) | 561 | aa 243-360 | no | |
| (PR domain-containing protein 14) | ||||||||
| PRDM15 (C21orf83; E130018M06Rik; ORF62; Zfp298) | PR domain containing 15 | nucleus | 1 | 1,174 | aa 76-191 | no | ||
| PRDM16 (Kiaa; 1675; Mel1) | PR domain zinc finger protein 16 | nucleus | 3 | Isoform 1 'canonical' sequence | 1,275 | aa 83-215 | H3K9me1 | |
| (A2A935-1) | ||||||||
| Isoform 2 | ||||||||
| 129-129: E → EQ | ||||||||
| (PR domain-containing protein 16) | ||||||||
| 868-868: Y → YS | ||||||||
| 1174-1176: CVE → HMQ | ||||||||
| 1177-1275: missing * | ||||||||
| (Transcription factor MEL1) | (A2A935-2) | |||||||
| Isoform 3 | ||||||||
| 868-868: Y → YS * | ||||||||
| (A2A935-3) | ||||||||
* No experimental confirmation available.
2. PRDM Proteins in Signal Transduction and Transcription Control
PRDM protein are involved in the transduction of many signals that are responsible for proliferation and differentiation control. PRDM proteins, through the formation of chromatin remodeling complexes, regulate gene expression acting generally as transcription repressors [21,22,23,24]. Some members of the PRDM family show an intrinsic methyltransferase activity [8,25] while others act indirectly, recruiting chromatin remodeling enzymes [22,26,27].
2.1. Nuclear Receptor Superfamily Signal Transduction
Nuclear receptors act as ligand-dependent transcription factors, modulating gene expression by direct interaction with well conserved consensus sequences of target genes: cis-acting hormone-regulatory elements [28].
Several findings suggest that the PRDM2 gene product PRDM2a/RIZ1 is a downstream effector of estrogen action and is related to estrogen-regulated cell proliferation in classical estrogen target tissues. PRDM2 proteins interact with estrogen receptor (ER) through a LXXLL motif and their interaction is dependent on 17β-estradiol treatment [29,30,31]. PRDM2a has in vitro histone H3K9 methyltransferase activity and is a weak activator or a repressor of transcription [25,32,33]. It acts as co-activator of estrogen-dependent gene transcription when its methyltransferase activity is inhibited by estradiol (Figure 1) [30,34]. Medici et al. in fact demonstrated that PRDM2a is able to bestow estrogen inducibility to a promoter containing an incomplete ERE and a G/C TTGGC motif [29].
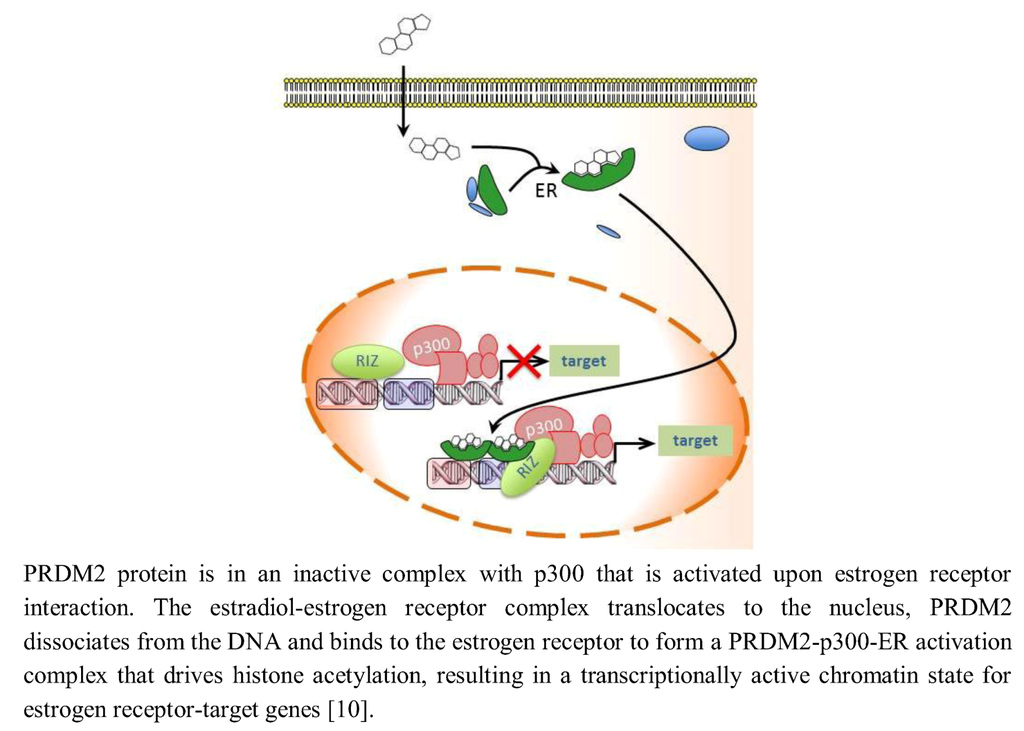
Figure 1.
PRDM2 is an estrogen receptor co-activator.
Figure 1.
PRDM2 is an estrogen receptor co-activator.
Garcia Bassets et al. have shown the fundamental role of histone methyltransferase (HMT), including PRDM2a, in maintaining in off-state the promoters regulated by nuclear receptors, such as ERα or androgen receptor (AR). However, the H3-K9 methylation-mediated down-regulation allows the action of lysine-specific demethylase 1 (LSD1) molecules recruited by steroid nuclear receptors ligand, complexed to the same genes [35,36]. Based on this, the opening of regulated genes could involve two crucial events leading to the enhancer effect of the nuclear receptor to promote the DNA unwinding in transcription: the recruitment of topoisomerase β [37] and of OGG1 (8-oxoguanine DNA glycosylase), due to the oxygen radicals produced by the LSD1 action in the removal of the methyl group of dimethyl H3K9 with production of monomethyl H3K9 [38]. This would explain why the PRDM2 (PR-minus) form is unable to produce enhancer effects in presence of estradiol, as observed with ERE-Luc reporter assay experiments in vitro [39], despite the presence of domains for the recruitment of p300 and p160 co-activators [34]. It might be expected that its full function as co-activator would be due to the presence of the PR domain. In this way, PRDM2a would provide the substratum to histone demethylase near the ERE sequences, thereby supporting and stabilizing the binding of the receptor to DNA, for its ability to recognize flanking sequences and to interact with the AF-2 core sequence in the ER hormone binding domain [29,30].
Moreover, PRDM2 gene products are endowed with DNA-binding as well as transcription factor-binding activities. In fact PRDM2 was independently isolated as a retinoblastoma-binding protein (RIZ) [40], a DNA-binding protein (MTB-Zf), or as a GATA3 transcription factor binding protein (G3B) [41]. MTB-Zf (essentially identical to PRDM2b) binds to the MTE DNA element GTCATATGAC of human hemeoxygenase-1 gene and can weakly activate transcription [32]. G3B (PRDM2) interacts with the transcription factor GATA-3, regulating the expression of several genes critical for T-cell function and development [42].
Nuclear receptor ligands modulate expression of several PRDM genes. In breast cancer cells (MCF-7 cell line), 17β-estradiol stimulation specifically modulates expression of PRDM2 gene products (PRDM2a and PRDM2b), inducing a shift in the balance of their intracellular concentrations; in particular 17β-estradiol induced a selective decrease in PRDM2a transcript and an increase in total PRDM2 mRNA, accounted by an increase in the PRDM2b form [31]. In fact it was recently demonstrated that the promoter 2 of the PRDM2 gene contains an estrogen responsive element (ERE) endowed with enhancer activity that is recognized by ERα [43]. Moreover, with the innovative DNA-picked chromatin (DPC) assay, it was possible to observe that estradiol treatment induces a preferential interaction between hormone-responsive PRDM2 promoter (promoter 2) and the polyadenylation site. Formation of loops has been implicated not only in bringing together far upstream or downstream regions with regulatory or transcribed gene regions, but also in establishing contacts between the 5' and 3' ends of genes, [44,45], in agreement with the now prevalent hypothesis that 3' end-processing factors interact with components of the transcriptional machinery [46]. In the DPC assay, estradiol treatment increased by 60–70% the amount of molecules from exons 9a and 10 (where alternative polyA addition occurs) specifically associated to the captured estradiol-sensitive PRDM2 promoter 2, whereas the recovery of those captured by the estradiol-insensitive PRDM2 promoter (promoter 1) was decreased. The 17β-estradiol remodels the chromatin architecture of PRDM2 gene locus to create a loop for the mRNA transcription with poliA-exon 9a, leading to the production of oncogenic variants [47]. In non-primary target tissues, however, 17β-estradiol could have an opposite effect, inducing a shift in the PRDM2a/PRDM2b molar ratio in favor of PRDM2a. In fact, in other cell types the hormone stimulation did not affect PRDM2a expression, as in the EPN (epithelial cell line derived from normal human prostate) cell line, or increased it, as SAOS2 (osteosarcoma) cells; serum treatment produced the same effect [48,49].
PRDM2 proteins might also be mediators of androgen effects. In EPN cells, 5α-dihydrotestosterone (DHT) induced a slight increase in cell growth, related to a sharp increase of PRDM2a mRNA and protein concentration. Further investigation could confirm whether PRDM2 is an androgen responsive gene, because there is an androgen responsive element (ARE) at -361 bp, in the upstream regulatory region of the promoter 1 of PRDM2 gene [49,50].
PRDM2 proteins might also be active for retinoid action. In fact, in a human promyelocytic leukemia cell line (HL60) treatment with retinoic acid induced a selective expression of PRDM2a and a redistribution of the protein within the nucleus, correlated to the granulocytic differentiation. In HL60 cells, PRDM2a expression was also induced by activation of a retinoid receptor-independent maturation pathway based on retinoid X receptor agonist and protein kinase A synergism [51].
Similarly to PRDM2 acting as co-activator of ERα, PRDM16 stimulates adipogenesis by binding and co-activating, in a ligand-dependent manner, the peroxisome-proliferator-activated receptor γ (PPAR-γ) [52]. PRDM16 also is able to stimulate the function of PGC-1 (Peroxisome proliferator-activated receptor-γ co-activator) α and β in the brown-white fat switch. PRDM16 probably also has a transcriptional repressor activity because the fusion proteins PRDM16/MEL1 or PRDM16/MEL1S-GAL4 DNA-binding domain negatively regulates transcription [7].
2.2. Luteinizing Hormone (LH) Signaling
LH stimulates testosterone synthesis in Leydig cells inducing the expression of cytochrome P450 enzymes, 3β-hydroxysteroid dehydrogenase and LH receptor. Prdm8 is a transcriptional repressor that specifically methylates lysine 9 of histone H3. The overexpression of Prdm8 wild-type protein or its mutant deletion, lacking the PR domain, induced a reduction in the expression levels of the steroidogenic enzyme gene p450c17c coding for a component of cytochrome P450 family, and of Luteinizing Hormone Receptor gene when steroidogenesis was induced in mouse Leydig cells (TM3 cell line) by LH treatment [53]. This evidence suggests that Prdm8 could negatively control steroidogenesis.
2.3. Insulin-Like Growth Factor-1 (IGF-1) Signaling
PRDM2a acts as a repressor of a subgroup of genes involved in IGF-1 signaling. A chromatin immunoprecipitation (ChIP) assay showed that PRDM2a down-regulates IGF-1 expression through a direct binding to its promoter, increasing histone H3K9 methylation. PRDM2a also positively controls insulin-like growth factor-binding protein 2 (IGFBP-2) and SPARC expression [54]. Moreover, PRDM2a is involved in IGF-1R activation and signal transduction. In fact, forced PRDM2a expression in chronic myelogenous leukemia-blast crisis (CML-BC) cell lines decreases activation of IGF-1 receptor and of the downstream signaling components ERK 1/2 and AKT.
2.4. NGF Signaling
Neurotrophins influence a wide number of functions in the nervous system, including neuronal cell survival, cell differentiation and apoptosis, synaptic plasticity, control of axonal guidance and dendrite growth [55,56]. These actions are mediated by neurotrophin binding to two separate receptor classes, the Trk family of tyrosine kinase receptors and the p75 neurotrophin receptor, a member of the tumor necrosis factor receptor superfamily. SC-1 (Schwann Cell factor 1), the Prdm4 gene product binds to the p75 neurotrophin receptor and provides a downstream transducer for the effects of nerve growth factor (NGF) through this receptor. In fact, NGF treatment of the monkey kidney fibroblast-like cell line (COS) induces a translocation of Prdm4/SC-1 from the cytoplasm to the nucleus that is related to a reduction in bromodeoxyuridine (BrdU) incorporation. The translocation of Prdm4/SC-1 to the nucleus was specific for p75, as NGF binding to the TrkA receptor prevented the nuclear localization of Prdm4/SC-1 (Figure 2) [57]. On the contrary, both TrkA and p75NTR are able to enhance the repressive transcriptional activity of Prdm4/SC-1, implying the role of Prdm4/SC1 as a transducer of NGF signaling by these two receptors [58]. Prdm4/SC-1 acts as a transcriptional repressor forming complexes with trichostatin A (TSA)-sensitive histone deacetylases HDAC1, 2 and 3 and negatively controls cell cycle progression down-regulating cyclin E expression, essential for the G1-S phase transition [58]. In mice cortical neural stem cells (NSCs), Prdm4/SC-1 recruit the chromatin modifier Prmt5 via its N-terminus and partly via the PR/SET domain, probably as part of an epigenetic regulatory complex that maintains the “stem-like” cell state of the NSCs by preserving their proliferative capacity and modulating their cell cycle progression [59].
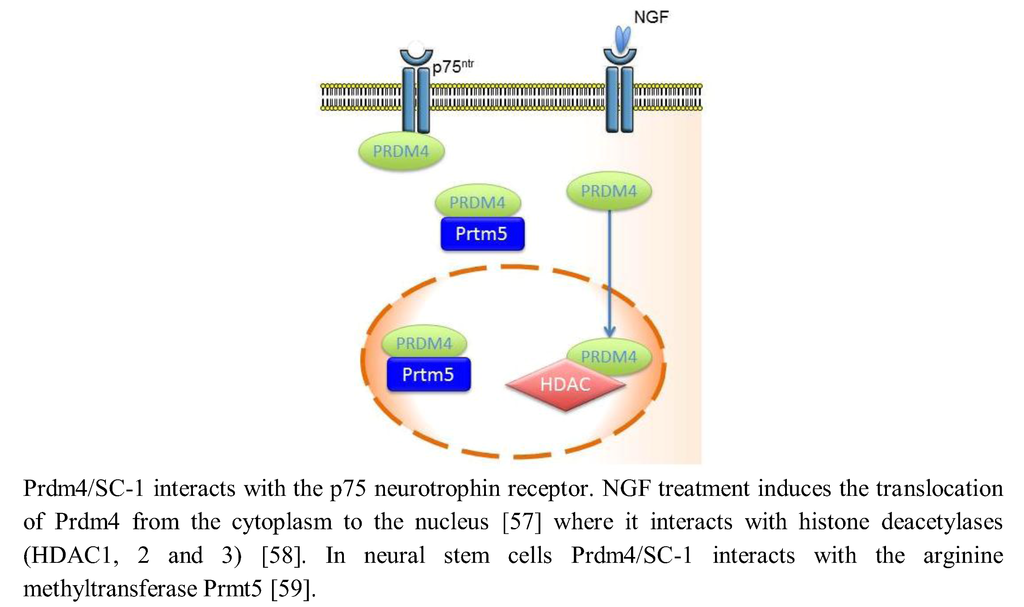
Figure 2.
Prdm4/SC-1 (Schwann Cell factor 1) provides a downstream transducer for the effects of nerve growth factor (NGF) through the p75 neurotrophin receptor and forms an epigenetic regulatory complex with Prmt5 that probably maintains the “stem-like” cellular state of a NSC.
Figure 2.
Prdm4/SC-1 (Schwann Cell factor 1) provides a downstream transducer for the effects of nerve growth factor (NGF) through the p75 neurotrophin receptor and forms an epigenetic regulatory complex with Prmt5 that probably maintains the “stem-like” cellular state of a NSC.
2.5. Wnt/β-Catenin and BMP/SMAD Signaling
Wnt signaling is involved in many aspects of embryonic development, such as morphogenetic movements, cell type specification, and patterning. In addition, Wnt/β-catenin regulates pluripotency and differentiation in various stem cell systems, including Embryonic Stem (ES) cells [60]. In murine ES cells, derived from inner cell mass of blastocyst prior the formation of epiblast, the activation of Wnt/β-catenin signaling, through the inhibition of glycogen synthase kinase-3β (GSK3β), leads to the maintaining of pluripotency, induced by bone morphogenetic protein (BMP) or fibroblast growth factor (FGF) via PI3K⁄AKT activation [61,62,63].
PRDM14 is essential for the maintenance of the pluripotent state of human and, potentially, murine ESC, but not for the murine epiSCs (derived from post-implantation epiblast cells), and enhances epigenetic reprogramming of human and murine somatic cells to induced pluripotent stem cells (iPSC) [64].
Co-expression of PRDM1 and PRDM14 is obligatory for the establishment of germ cell lineage [65]. In mammals, the PGCs, the first germ lineage cells are specified in the proximal epiblast [66] and their normal proliferation is ensured by the GSK-3-mediated suppression of Wnt/β-catenin signaling. In PGCs, activation of the Wnt/β-catenin signaling is involved in nuclear reprogramming in culture and nevertheless its aberrant activation leads to germ cell deficiency due to the delay of the cell cycle progression [67]. Wnt signaling alone however, is not sufficient for PGC formation in the absence of BMP. Wnt3, expressed in the epiblast at around E5.5 [68], is a key factor in conferring Bmp4 responsiveness to the epiblasts, giving them the competence to form PGC-like cells. Therefore, Wnt signaling facilitates the response of the epiblast to BMP but itself is not sufficient to induce the PGCs [69]. In the proximal epiblast, BMP/Smad signals induce PRDM1 [70], essential for specification of PGCs [71,72]. PRDM1 complexed with arginine methyltransferase Prtm5, regulates epigenetic reprogramming in germ cell lineages, resulting in high levels of H2A/H4 R3 methylation [26]. Prmt5, a class II arginine methyltransferase, is responsible for the monomethylation of arginine (Rme1) [73] and it has been shown that it methylates cytoplasmic R3 of H2A rather than H4, and that it might be involved in the repression of differentiation genes [74].
Other epigenetic changes, associated with PRDM1 expression, allow PGC to escape the somatic pathway: PGCs show low levels of DNA methylation and H3K9me2 histone marks while acquiring high levels of H3K27me3 modifications [75]. The expression of somatic genes, as Hoxa1 and Hoxb1, is repressed [76] at the same time as the expression of pluripotent marks (Sox2, Pousf1 and Nanog) is re-activated [77].
In vitro, the ES cells are capable of differentiating in germ cells [78] and these are at least equivalent to the PGCs that migrate into the fetal gonad and have the potential to undergo meiosis and produce sperm [79]. In embryonic cells fated to become PGCs, PRDM14 is co-expressed with PRDM1 and is critical to the reacquisition of potential pluripotency and successful epigenetic reprogramming [80]. In these cells, PRDM14 expression is regulated by BMP and SMAD signaling and is involved in the establishment of germ cell lineage. The loss of PRDM14 causes defects in genome-wide epigenetic reprogramming with a shift of H3K9me2/H3K27me3 ratio caused by increased expression of the G9a-Like Protein 1 (GLP1, Euchromatic Histone N-Methyltransferase 1, and failure to upregulate Sox2 expression [81]. PRDM1 is therefore not required for the derivation or the maintenance of murine ESCs while it is obligatory for PGC specification and is critical for the maintenance of unipotent germ cells [82].
Aberrant activation of Wnt/β-catenin signaling is also frequently involved in cancers, accompanied with elevated levels of active β-catenin. In addition to genetic defects, epigenetic silencing of Wnt/β-catenin antagonists also leads to aberrant Wnt/β-catenin signaling in tumors [83]. PRDM5 antagonizes the Wnt/β-catenin signaling in normal cells and in cancer cells. By TOPFlash luciferase reporter assay, it was demonstrated that PRDM5 significantly inhibits the T Cell Factor (TCF)/Lymphoid enhancer-binding factor (LEF)-dependent transcription thus hypothesizing that PRDM5 forms a complex with the transcriptional factor TCF (Figure 3) [84]. In agreement with this evidence, the promoter reporter activity of cyclin D1 (CCND1), a Wnt/β-catenin downstream target gene whose product binds CDK4, was markedly decreased when PRDM5 was overexpressed [84]. By ChIP assay it was demonstrated that PRDM5 directly binds the promoters of several oncogenes, such as CDK4 and TWIST1 and PRDM5 expression resulted in significantly decreased levels of active transcription marks H3K4me3 and acetyl-histone H4 in CDK4 and TWIST1 promoters.
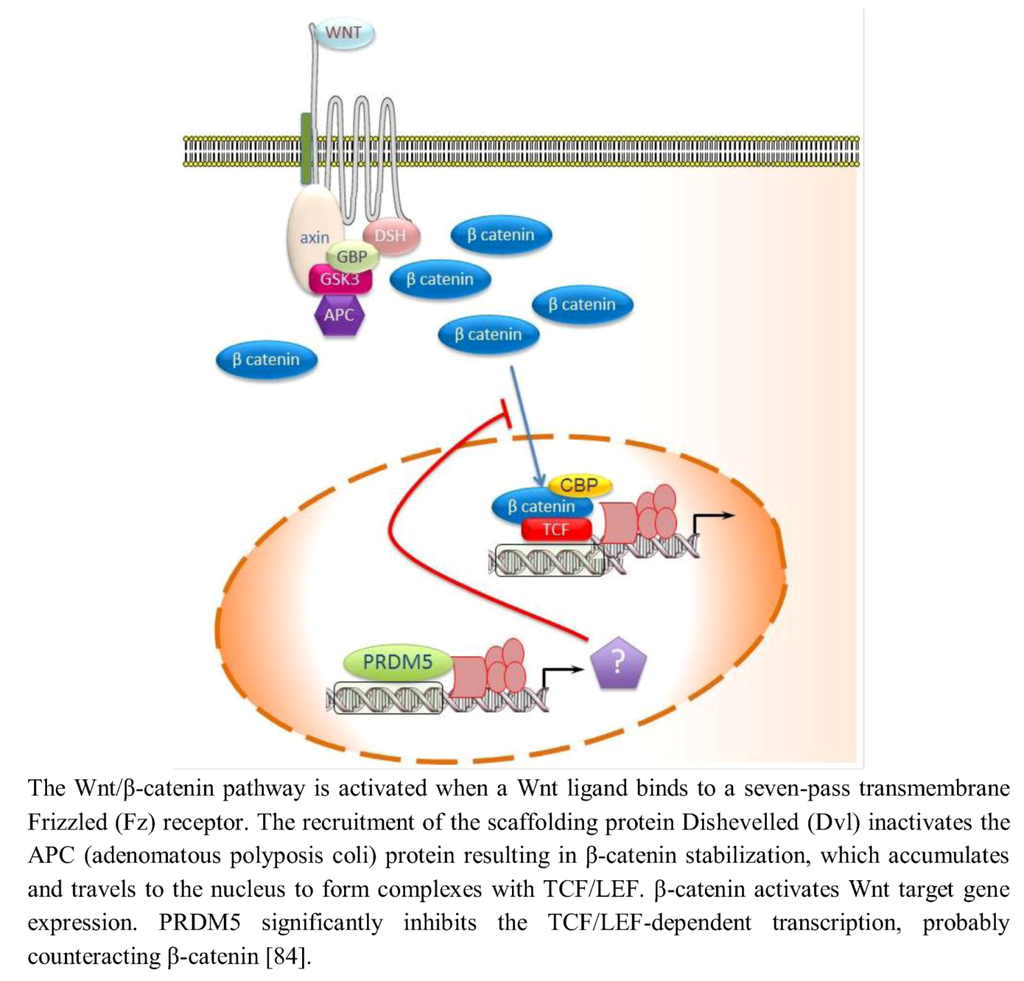
Figure 3.
PRDM5 regulates the Wnt/β-catenin signaling in normal cells and in cancer cells.
Figure 3.
PRDM5 regulates the Wnt/β-catenin signaling in normal cells and in cancer cells.
2.6. Neural Progenitor Maintenance and Differentiation
The nervous system of mammals contains a large number of neurons in a diverse array of neuron classes. Transcription factors play central roles in generating this complexity by controlling neural progenitor cell proliferation, patterning, and defining neuron fate [85,86]. One family that has emerged as important in this regard is the basic helix-loop-helix (bHLH) containing transcription factors [87,88]. For example, evolutionary conserved basic Helix-Loop-Helix (bHLH) transcription factor cascades downstream of Notch signaling is necessary for both the maintenance of neural progenitor cell character and the progression of neurogenesis, while Bhlhb5 olig-related transcription factors Bhlhb5 (also known as Bhlhe22) function predominantly as transcriptional repressors. Bhlhb5 expression is almost exclusively limited to post-mitotic neurons rather than proliferating neural progenitors, hinting at the possibility that Bhlhb5 regulates later aspects of neuronal differentiation [89,90,91].
2.7. Notch Signaling
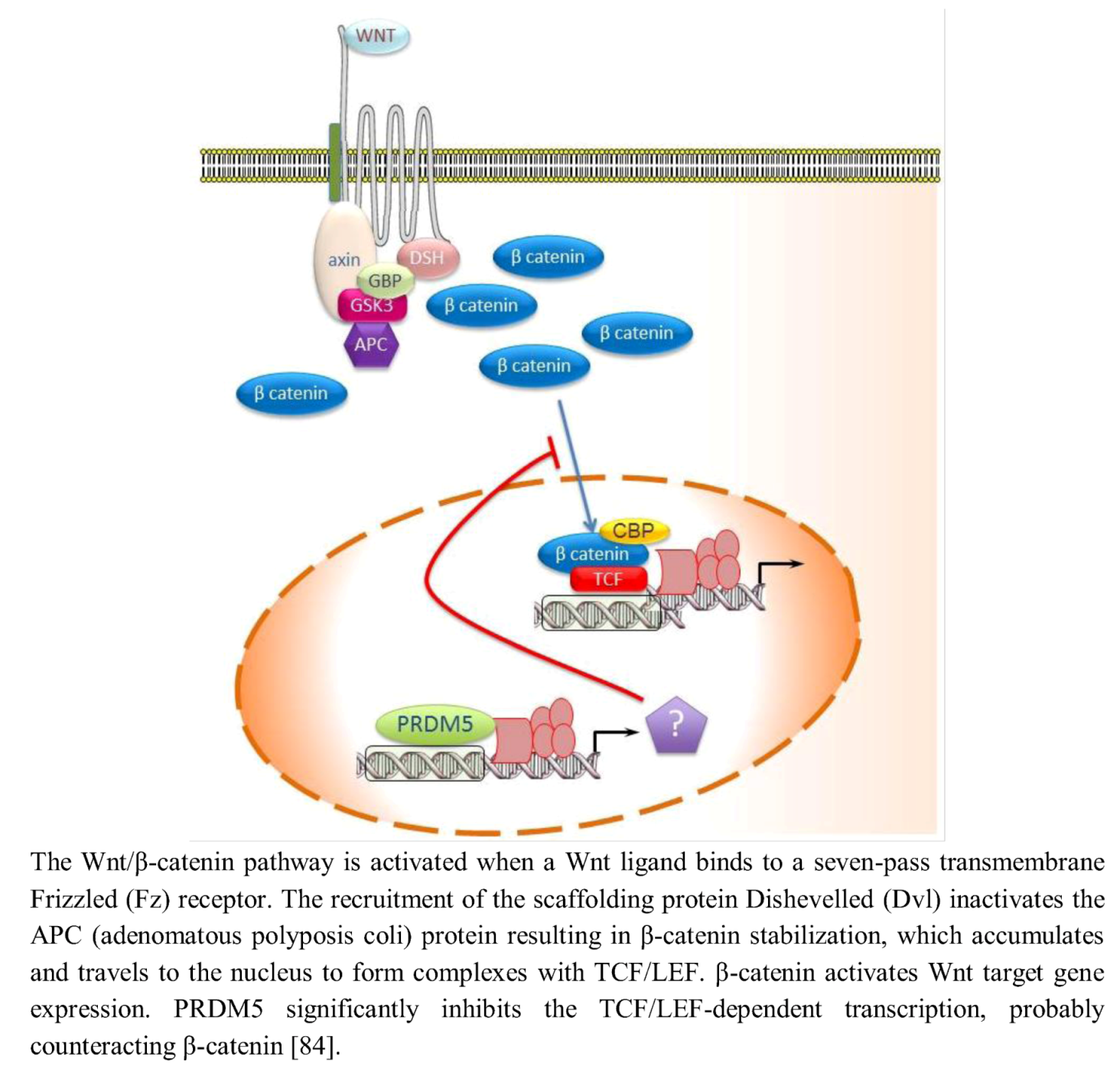
In mammals, Notch activity maintains neural progenitors through an effector pathway consisting of the bHLH Hairy and enhancer of split homologue transcription factors Hes1 and Hes5. Notch up-regulates the transcription of Hes factors that then function as DNA-binding repressors and antagonize the expression of proneural bHLH genes [92]. Hence, low Notch activity reduces Hes activity and leads to up-regulation of proneural bHLH factors such as Neurogenin2 (Ngn2) and Mammalian achaetescute homolog1 (Mash1); these factors then repress neural progenitor cell maintenance and promote neuronal differentiation [93]. Evidence has revealed an involvement of PRDM protein in the transcriptional regulation mediated by Notch signaling. Hamlet (Ham), the Drosophila homolog of mammalian Prdm3/Evi1 and Prdm16, controls olfactory receptor neuron (ORN) development fate by modifying the cellular response to the Notch signals. Ham up-regulating H3K27me3 and down-regulating H3K4me3 directs chromatin-modification events at specific Notch targets, altering the accessibility for Su(H) binding at the enhancer. In nascent ORNs, Ham activity erased the Notch state that was inherited from the parental pNa intermediate precursor cell. This permitted a new and modified response of Notch targets in the subsequent round of Notch signaling [94]. mRNA in situ hybridization analysis showed that in the developing murine telencephalon, Prdm family genes are expressed at high level in a spatially and temporally restricted manner. The Notch-Hes pathway controls their expression: in particular Hes positively or negatively regulated expression of Prdm16 and Prdm8, respectively. In fact, in Hes-null telencephalon neural differentiation is enhanced, Prdm8 expression is up-regulated, and Prdm16 expression is down-regulated. Conversely, electroporation of Hes1 into the developing telencephalon in utero up-regulates Prdm16 expression (Figure 4) implying that Prdm16 is positively regulated by Hes1 during neurogenesis and expressed in the neural progenitor cell population. As Hes1 protein is believed to act as a transcriptional repressor, positive regulation of Prdm16 by Hes1 may not be direct; it is possible that Hes1 acts by repressing a repressor of Prdm16 expression. Moreover, Prdm16 tags neuronal progenitor cells while Prdm8 does it in the post-mitotic neurons [95].
2.8. Neural Circuit Formation
Bhlhb5 binds specific DNA sequence elements and then recruits Prdm8 to inhibit expression of target genes that must be repressed to permit correct development of neural circuits. Mice lacking either Bhlhb5 or Prdm8 have strikingly similar cellular and behavioral abnormalities including axonal mistargeting by neurons of the dorsal telencephalon and abnormal itch-like behavior [96], suggesting that Bhlhb5 and Prdm8 are required partners for key aspects of neuronal development. One important target of the Prdm8/Bhlhb5 repressor complex is Cadherin-11 (Cdh11), a cell-cell adhesion molecule involved in neural circuit assembly.
Prdm8 and Prdm16 gene products represent therefore, strong new candidates as regulators of neural progenitor cell proliferation and neural differentiation in mammals’ central nervous system (CNS).
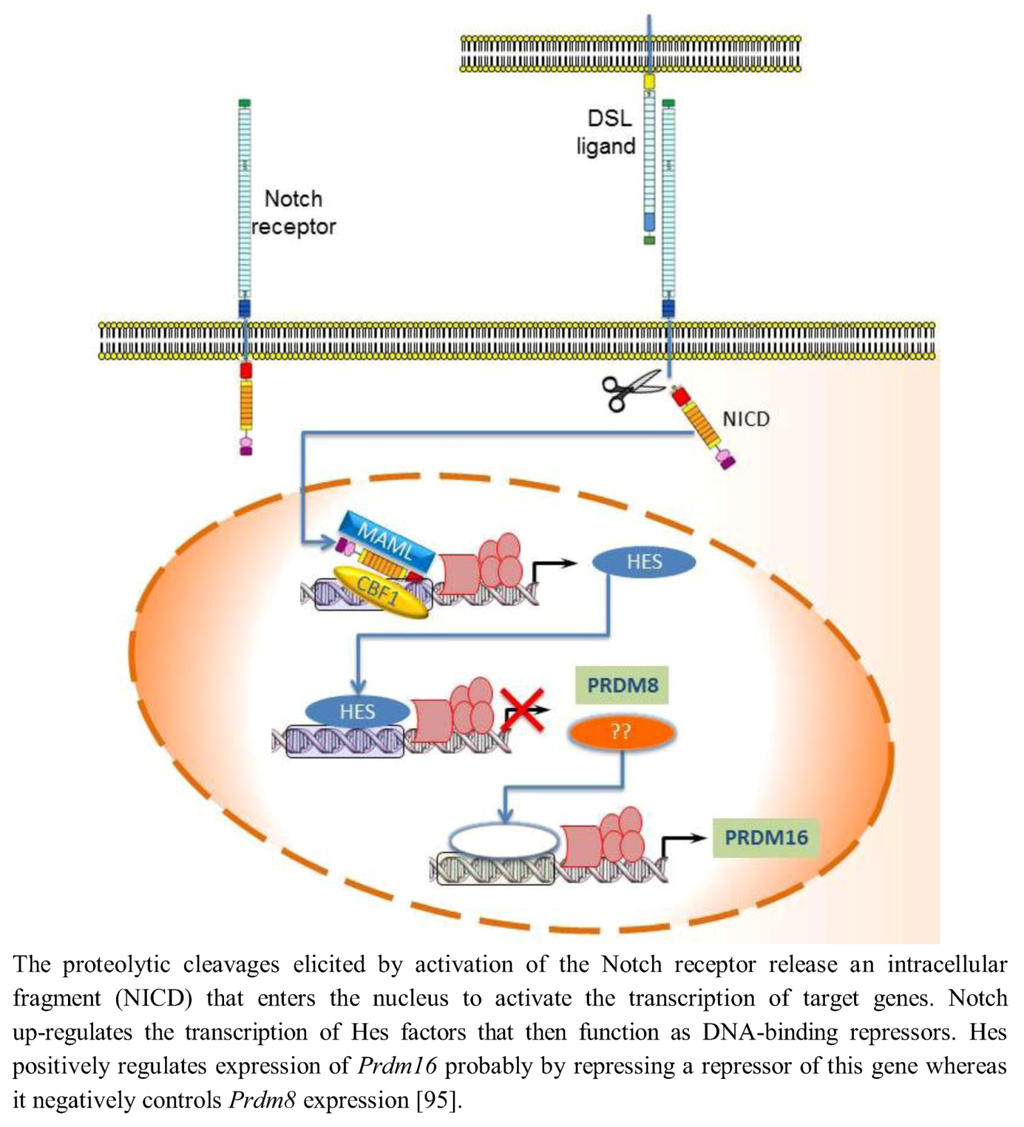
Figure 4.
Notch-Hes pathway controls the expression of Prdm8 and Prdm16.
Figure 4.
Notch-Hes pathway controls the expression of Prdm8 and Prdm16.
2.9. TGF-β Signaling
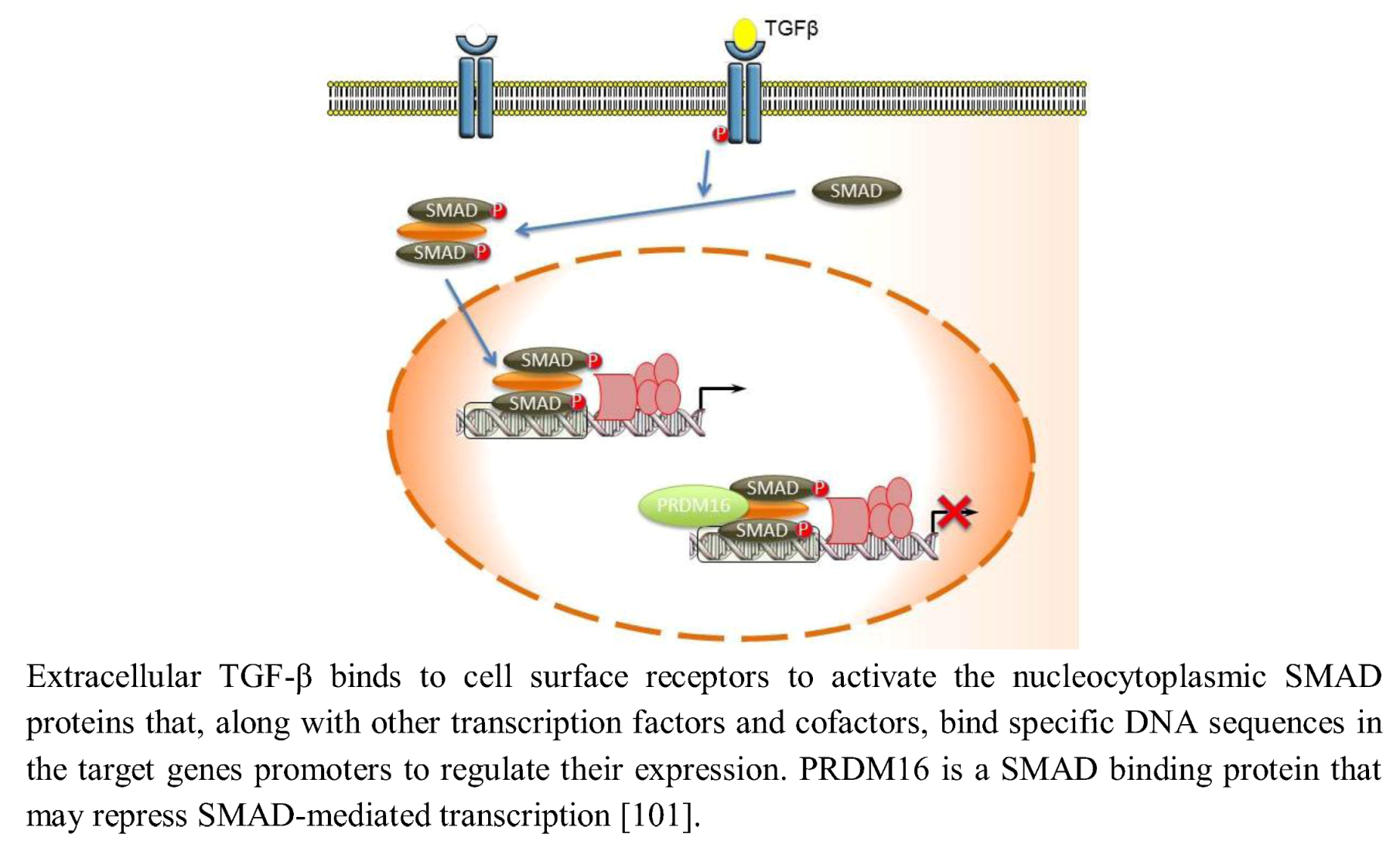
The Transforming growth factor β (TGF-β) signaling regulates central cell processes, such as proliferation and extracellular matrix production during development of the orofacial region [97,98,99,100]. Extracellular TGF-β binds to cell surface receptors to activate the nucleocytoplasmic SMAD proteins that, along with other transcription factors and cofactors, bind specific DNA sequences in the target genes’ promoters to regulate their expression. PRDM16 is a SMAD-binding protein that can bind a number of different SMADs, including TGF-β and BMP-regulated SMADs, and may modulate their signaling via the TGF-β pathway (Figure 5) [101]. PRDM16 is similar in structure to PRDM3, which has been previously demonstrated to bind and thereby inactivate SMAD3 proteins through its DNA binding domain-1 (Zn-finger domain-1) and repress TGF-β cell growth-inhibitory signaling [102]. PRDM3 and PRDM16, however, bind SMADs and recruit CtBP, which in turn join histone deacetylases (HDACs) to deacetylate histones and repress SMAD mediated transcription [10,21,102,103,104,105].
Prdm16 is expressed in the murine embryonic secondary palate [101] where it plays a downstream regulatory role in mediating TGF-β signaling, affecting embryonic craniofacial development. Indeed, Prdm16 knockout murine embryos display a completely penetrant cleft palate [103]. In Prdm16−/− fetuses, chromatin immunoprecipitation-promoter microarray analysis (ChIP-Chip) has revealed a gene expression change of markers for bone (Opn) and muscle (Myf-4) development. The expression of Opn, [106], linked to human cases of orofacial clefting, was significantly reduced, while that of Myf-4 was significantly increased, allowing to assume a role for Prdm16 to myo-, chondro- and/ or osteogenesis in the developing orofacial region, in addition to regulating other processes of normal development. Prdm16 knockout could cause an abnormal muscle and/or bone development leading to altered morphogenesis of the nascent palatal processes with the failure of reorientation and subsequent separation of the oral and nasal cavities [107].
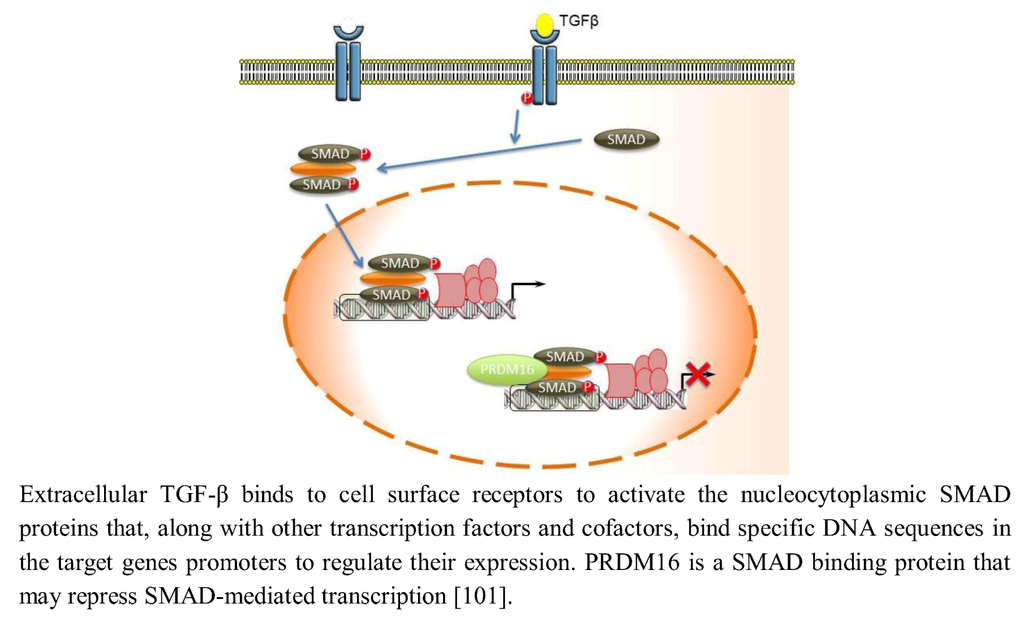
Figure 5.
PRDM16 modulates TGF-β signaling.
Figure 5.
PRDM16 modulates TGF-β signaling.
3. PRDM Proteins in the Host Defence
PRDM1 is a transcription repressor that plays a critical role in terminal differentiation of B cells into antibody-secreting plasma cells [11]. PRDM1/Blimp-1 modifies the architecture of chromatin through the interaction with several proteins. The Pro/Ser rich domain interacts with the Groucho family proteins [108], LSD1 (Lysine-Specific Demethylase-1), and the HDAC 2 [24].
Interleukin 21-producing T helper lymphocytes are central to humoral immune response because this cytokine is required for the antibody production induced by IL-6. In B cells, IL-21-treatment induces the expression of signal transducer and activator of transcription 3 (STAT3), required for optimal immunoglobulin production and an up-regulation of PRDM1, the master plasma cell factor [109].
PRDM1 plays also a crucial role in controlling T cell homeostasis [110,111]. In activated T cells, PRDM1 is induced by IL-2 signaling and inhibits IL-2 production in a negative feedback loop [112]. In naive T helper cells, IL-4 promotes the TH2 differentiation and inhibits the TH1 differentiation, which induces the down-regulation of IL-2. PRDM1 is an IL-4 responsive gene that potentiates the IL-2 inhibition. In fact, IL-4-mediated IL-2 suppression was less pronounced in activated, PRDM1- deficient T helper cells [113].
Recent studies revealed that the PRDM1 expression level and, consequently, the secretion of pro-inflammatory cytokines was regulated not only at transcriptional level by activation of T helper cells but also at post-transcriptional level, by enhanced miR-9 expression. The miR-9 is particularly abundant in activated human T helper cells and controls expression of PRDM1 and B cell lymphoma-6 protein (Bcl-6). In fact, suppression of miR-9 led to increased expression of PRDM1 and Bcl-6, which subsequently resulted in diminished secretion of IL-2 and IFN-γ [114].
Another microRNA gene cluster is repressed, in T follicular helper cell (TFH cells), by Bcl-6 to maintain the expression of several TFH genes implicated in lineage commitment [115]. Bcl-6 is a transcriptional repressor that is, at low concentration, recruited by T-bet, a TH1-specific T box transcription factor, to maintain the TH1 gene-expression profile [116,117]. PRDM1 is directly targeted by Bcl-6 and is responsible for the repression of a subset of TFH signature gene in TH1 cells [118]. Oestreich and colleagues hypothesized a flexibility between TH1 and TFH-like gene-expression regulated by T-bet-Bcl-6 complex, through the activation or repression of PRDM1. In TH1 cells, the variations of the ratio between Bcl-6 and T-bet are regulated by the low or high concentration of IL-2. In this way, low concentration of IL-2 enables the Foxo transcription factor to activate Bcl-6 transcription; Bcl-6 in turn represses the PRDM1 expression, promoting the expression of TFH signature genes [118].
In addition to controlling the fate of effector T helper cells, PRDM1 cooperates with transcription factor IRF4 for the differentiation of natural Treg cells. Expression of IL-10 is essential for this particular effector function and PRDM1is responsible for the remodeling of active chromatin at the locus Il10 via trimethylation of histone H3 at Lys27 [119]. In these cells, the PRDM1 gene is a target for the transcription factor FOXP3, which regulates also the expression of IRF4 [120,121]. These transcription factors directly regulate PRDM1 expression in Treg cells by binding two sites in the 3' region and between exons 5 and 6 of PRDM1 (conserved noncoding sequence 9) [122,123].
PRDM2 is involved in the regulation of inflammatory response in host defense and might play an important role in inflammatory diseases. In the murine leukemic monocyte macrophage cell line (RAW 267.4) PRDM2 is a lipopolysaccharide (LPS)-responsive gene that increases the production of TNF-α (Tumor necrosis factor α) and IL-6 by nuclear factor-κB (NF-κB) activation. LPS, in fact, augments the PRDM2a expression via the activation of PI3K/Akt/NF-κB pathway. In turn, PRDM2a increases TNF-α expression level and IL-6 cytokine enhancing NF-κB activation. PRDM2a knock-down by RNA interference led, in fact, to the inactivation of NF-κB in response to LPS. TNF-α induced PRDM2 expression by the activation of NF-κB and AKT signaling. PRDM2a negatively regulates the proliferative activity of TNF-α-treated human monocytic leukemia cells via activation of p53. In fact, PRDM2a forced expression produces an increase of p53 protein expression and silencing of RIZ1 prevented it. On the other hand, a p53 inhibitor enhanced the TNF-α-induced PRDM2a expression [124,125].
PRDM5 is probably involved with the regulation of hematopoiesis. PRDM5 is in fact able to interact with Growth factor independent 1 (Gfi1) transcription factor, essential for hematopoiesis [126], whose inactivation impaired blood cell formation, causing neutropenia and lymphopenia and release from bone marrow of immature cells [127,128,129]. At molecular level, PRDM5 acts as a sequence-specific DNA binding transcription factor interacting with Gfi1 and recruiting the histone methyltransferase G9a, histone deacetylases HDAC1, 2 and 3 to its target gene promoters [130] to repress transcription.
PRDM5 can also activate some target genes, such as NOTCH2, IL6R, MYB and c-MYC, whose transcriptional regulation is also controlled by Gfi1, suggesting that Gfi1-PRDM5 interaction activates rather than represses transcription. Neutropenia-associated PRDM5 sequence variants interfere with its transcriptional activity.
5. Conclusions and Perspectives
PRDM gene family has a pivotal role in the control of the proliferation/differentiation switch and expression of its member is relevant during tumorigenesis, when some PRDM genes are frequently silenced by genetic or epigenetic mechanisms. Several members of the family express forms containing the SET/PR domain closely involved in cell differentiation and forms without this domain have an oncogenic potential (e.g., PRDM2, PRDM3 and PRDM16 gene variants) [13,136]. An imbalance in the amounts of the two products frequently occurs in tumor progression through either disruption or underexpression of the PR-plus form or overexpression of the PR-minus one. Nevertheless, expression of forms missing the PR domain is not only limited to neoplastic transformation and tumor progression. Actually, the significance of the balance between the different forms and the mechanism controlling the ratio is unknown.
PRDM family expanded in vertebrates in parallel with the increased complexity of the genome in higher organisms. PRDM genes are grouped in five subfamilies and the genes lying in sister branches of the tree maintain similar gene organization, splicing patterns, and functions. For example, PRDM2 and PRDM5, belonging to the same subfamily (composed of PRDM2, PRDM5, PRDM3, and PRDM16), have histone methyltransferase activity and are involved in cell cycle progression regulation [29,30].
By comparing the evolutionary features of PRDM genes with their expression in human tissues, it is evident that the newer genes have a lower expression than the older genes and acquire tissue specificity, suggesting a progressive specialization and/or a tighter regulation of their functions. Could the concomitant expression of old and new genes in a tissue suggest a cooperation in the establishment of the phenotype? This behavior is shown by PRDM1 and PRDM14, cooperating during germ cell development, and by PRDM3 and PRDM16, participating to maintain mammalian heterochromatin integrity. We hypothesize that the cooperation is a common characteristic of the PRDM gene family. Moreover, we observed (data non published) that PRDM2 gene siRNA silencing did not produce major phenotypic changes but increased the expression level of other PRDM-family proteins, suggesting that these could have a vicarious role.
PRDM proteins are localized in the nucleus where they participate in the transcriptional regulation of gene expression. However, the function of the PRDM protein in the cytosolic compartment is not completely clarified. Recently it has been demonstrated that PRDM3 and PRDM16 methylate H3K9me1 in the cytosol. Moreover, PRDM2a and Prdm4/SC-1 translocate from the cytosol to the nucleus after retinoic acid and NGF treatment respectively. We hypothesize that other than the role in histone code PRDM proteins targets other cytosolic proteins and control their function. PRDM protein as PRDM2 and PRDM16 are co-activators of the nuclear receptor superfamily and participate in the steroid genomic pathway. No clues are available about the involvement of PRDM proteins in the steroid non genomic pathway.
Acknowledgements
This investigation was supported by grants from the Italian Ministry for University and Scientific and Technological Research (PRIN).
References and Notes
- Xiao, B.; Wilson, J.R.; Gamblin, S.J. SET domains and histone methylation. Curr. Opin. Struct. Biol. 2003, 13, 699–705. [Google Scholar] [CrossRef]
- Fumasoni, I.; Meani, N.; Rambaldi, D.; Scafetta, G.; Alcalay, M.; Ciccarelli, F. Family expansion and gene rearrangements contributed to the functional specialization of PRDM genes in vertebrates. BMC Evol. Biol. 2007, 7, 187. [Google Scholar]
- Fog, C.K.; Galli, G.G.; Lund, A.H. PRDM proteins: Important players in differentiation and disease. BioEssays 2011, 34, 50–60. [Google Scholar]
- Gyory, I.; Fejer, G.; Ghosh, N.; Seto, E.; Wright, K.L. Identification of a functionally impaired positive regulatory domain I binding factor 1 transcription repressor in myeloma cell lines. J. Immunol. 2003, 170, 3125–3133. [Google Scholar]
- Hirai, H. The transcription factor Evi-1. Int. J. Biochem. Cell Biol. 1999, 31, 1367–1371. [Google Scholar] [CrossRef]
- Liu, L.; Shao, G.; Steele-Perkins, G.; Huang, S. The retinoblastoma interacting zinc finger gene RIZ produces a PR domain-lacking product through an internal promoter. J. Biol. Chem. 1997, 272, 2984–2991. [Google Scholar] [CrossRef]
- Lahortiga, I.; Agirre, X.; Belloni, E.; Vazquez, I.; Larrayoz, M.J.; Gasparini, P.; Lo Coco, F.; Pelicci, P.G.; Calasanz, M.J.; Odero, M.D. Molecular characterization of a t(1;3)(p36;q21) in a patient with MDS. MEL1 is widely expressed in normal tissues, including bone marrow, and it is not overexpressed in the t(1;3) cells. Oncogene 2004, 23, 311–316. [Google Scholar]
- Hayashi, K.; Yoshida, K.; Matsui, Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 2005, 438, 374–378. [Google Scholar] [CrossRef]
- Wu, Y.; Ferguson Iii, J.E.; Wang, H.; Kelley, R.; Ren, R.; McDonough, H.; Meeker, J.; Charles, P.C.; Wang, H.; Patterson, C. PRDM6 is enriched in vascular precursors during development and inhibits endothelial cell proliferation, survival, and differentiation. J. Mol. Cell. Cardiol. 2008, 44, 47–58. [Google Scholar] [CrossRef]
- Hohenauer, T.; Moore, A.W. The Prdm family: Expanding roles in stem cells and development. Development 2012, 139, 2267–2282. [Google Scholar]
- Turner, C.A., Jr.; Mack, D.H.; Davis, M.M. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell 1994, 77, 297–306. [Google Scholar] [CrossRef]
- Mochizuki, N.; Shimizu, S.; Nagasawa, T.; Tanaka, H.; Taniwaki, M.; Yokota, J.; Morishita, K. A novel gene, MEL1, mapped to 1p36.3 is highly homologous to the MDS1/EVI1 gene and is transcriptionally activated in t(1;3)(p36;q21)-positive leukemia cells. Blood 2000, 96, 3209–3214. [Google Scholar]
- Nishikata, I.; Sasaki, H.; Iga, M.; Tateno, Y.; Imayoshi, S.; Asou, N.; Nakamura, T.; Morishita, K. A novel EVI1 gene family, MEL1, lacking a PR domain (MEL1S) is expressed mainly in t(1;3)(p36;q21)-positive AML and blocks G-CSF-induced myeloid differentiation. Blood 2003, 102, 3323–3332. [Google Scholar] [CrossRef]
- Morgan, M.A.J.; Mould, A.W.; Li, L.; Robertson, E.J.; Bikoff, E.K. Alternative splicing regulates Prdm1/Blimp-1 DNA binding activities and co-repressor interactions. Mol. Cell. Biol. 2012, 32, 3403–3413. [Google Scholar]
- Wieser, R. The oncogene and developmental regulator EVI1: Expression, biochemical properties, and biological functions. Gene 2007, 396, 346–357. [Google Scholar] [CrossRef]
- Morishita, K.; Parganas, E.; William, C.L.; Whittaker, M.H.; Drabkin, H.; Oval, J.; Taetle, R.; Valentine, M.B.; Ihle, J.N. Activation of EVI1 gene expression in human acute myelogenous leukemias by translocations spanning 300–400 kilobases on chromosome band 3q26. Proc. Natl. Acad. Sci. USA 1992, 89, 3937–3941. [Google Scholar]
- Fears, S.; Mathieu, C.; Zeleznik-Le, N.; Huang, S.; Rowley, J.D.; Nucifora, G. Intergenic splicing of MDS1 and EVI1 occurs in normal tissues as well as in myeloid leukemia and produces a new member of the PR domain family. Proc. Natl. Acad. Sci. USA 1996, 93, 1642–1647. [Google Scholar] [CrossRef]
- Siegel, D.A.; Huang, M.K.; Becker, S.F. Ectopic dendrite initiation: CNS pathogenesis as a model of CNS development. Int. J. Dev. Neurosci. 2002, 20, 373–389. [Google Scholar] [CrossRef]
- UniProt. Available online: http://www.uniprot.org/ (accessed on 05 October 2012).
- National Center for Biotechnology Information Protein Database. Available online: http://www.ncbi.nlm.nih.gov/protein/ (accessed on 05 October 2012).
- Izutsu, K.; Kurokawa, M.; Imai, Y.; Maki, K.; Mitani, K.; Hirai, H. The corepressor CtBP interacts with Evi-1 to repress transforming growth factor-β signaling. Blood 2001, 97, 2815–2822. [Google Scholar]
- Davis, C.A.; Haberland, M.; Arnold, M.A.; Sutherland, L.B.; McDonald, O.G.; Richardson, J.A.; Childs, G.; Harris, S.; Owens, G.K.; Olson, E.N. PRISM/PRDM6, a Transcriptional Repressor That Promotes the Proliferative Gene Program in Smooth Muscle Cells. Mol. Cell. Biol. 2006, 26, 2626–2636. [Google Scholar] [CrossRef]
- Kajimura, S.; Seale, P.; Tomaru, T.; Erdjument-Bromage, H.; Cooper, M.P.; Ruas, J.L.; Chin, S.; Tempst, P.; Lazar, M.A.; Spiegelman, B.M. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 2008, 22, 1397–1409. [Google Scholar] [CrossRef]
- Yu, J.; Angelin-Duclos, C.; Greenwood, J.; Liao, J.; Calame, K. Transcriptional Repression by Blimp-1 (PRDI-BF1) Involves Recruitment of Histone Deacetylase. Mol. Cell. Biol. 2000, 20, 2592–2603. [Google Scholar] [CrossRef]
- Kim, K.C.; Geng, L.; Huang, S. Inactivation of a histone methyltransferase by mutations in human cancers. Cancer Res. 2003, 63, 7619–7623. [Google Scholar]
- Ancelin, K.; Lange, U.C.; Hajkova, P.; Schneider, R.; Bannister, A.J.; Kouzarides, T.; Surani, M.A. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol. 2006, 8, 623–630. [Google Scholar] [CrossRef]
- Gyory, I.; Wu, J.; Fejer, G.; Seto, E.; Wright, K.L. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat. Immunol. 2004, 5, 299–308. [Google Scholar]
- Aranda, A.; Pascual, A. Nuclear Hormone Receptors and Gene Expression. Physiol. Rev. 2001, 81, 1269–1304. [Google Scholar]
- Medici, N.; Abbondanza, C.; Nigro, V.; Rossi, V.; Piluso, G.; Belsito, A.; Gallo, L.; Roscigno, A.; Bontempo, P.; Puca, A.A.; et al. Identification of a DNA binding protein cooperating with estrogen receptor as RIZ (retinoblastoma interacting zinc finger protein). Biochem. Biophys. Res. Commun. 1999, 264, 983–989. [Google Scholar] [CrossRef]
- Abbondanza, C.; Medici, N.; Nigro, V.; Rossi, V.; Gallo, L.; Piluso, G.; Belsito, A.; Roscigno, A.; Bontempo, P.; Puca, A.A.; et al. The retinoblastoma-interacting zinc-finger protein RIZ is a downstream effector of estrogen action. Proc. Natl. Acad. Sci. USA 2000, 97, 3130–3135. [Google Scholar]
- Gazzerro, P.; Abbondanza, C.; D’Arcangelo, A.; Rossi, M.; Medici, N.; Moncharmont, B.; Puca, G.A. Modulation of RIZ gene expression is associated to estradiol control of MCF-7 breast cancer cell proliferation. Exp. Cell Res. 2006, 312, 340–349. [Google Scholar]
- Muraosa, Y.; Takahashi, K.; Yoshizawa, M.; Shibahara, S. cDNA cloning of a novel protein containing two zinc-finger domains that may function as a transcription factor for the human heme-oxygenase-1 gene. Eur. J. Biochem. 1996, 235, 471–479. [Google Scholar]
- Xie, M.; Shao, G.; Buyse, I.; Huang, S. Transcriptional repression mediated by the PR domain zinc finger gene RIZ. J. Biol. Chem. 1997, 272, 26360–26366. [Google Scholar]
- Carling, T.; Kim, K.-C.; Yang, X.-H.; Gu, J.; Zhang, X.-K.; Huang, S. A Histone Methyltransferase Is Required for Maximal Response to Female Sex Hormones. Mol. Cell. Biol. 2004, 24, 7032–7042. [Google Scholar] [CrossRef]
- Garcia-Bassets, I.; Kwon, Y.S.; Telese, F.; Prefontaine, G.G.; Hutt, K.R.; Cheng, C.S.; Ju, B.G.; Ohgi, K.A.; Wang, J.; Escoubet-Lozach, L.; et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell 2007, 128, 505–518. [Google Scholar] [CrossRef]
- Metzger, E.; Wissmann, M.; Yin, N.; Muller, J.M.; Schneider, R.; Peters, A.H.F.M.; Gunther, T.; Buettner, R.; Schule, R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 2005, 437, 436–439. [Google Scholar]
- Ju, B.G.; Lunyak, V.V.; Perissi, V.; Garcia-Bassets, I.; Rose, D.W.; Glass, C.K.; Rosenfeld, M.G. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science 2006, 312, 1798–1802. [Google Scholar]
- Perillo, B.; Ombra, M.N.; Bertoni, A.; Cuozzo, C.; Sacchetti, S.; Sasso, A.; Chiariotti, L.; Malorni, A.; Abbondanza, C.; Avvedimento, E.V. DNA Oxidation as Triggered by H3K9me2 Demethylation Drives Estrogen-Induced Gene Expression. Science 2008, 319, 202–206. [Google Scholar] [CrossRef]
- Steele-Perkins, G.; Fang, W.; Yang, X.H.; van Gele, M.; Carling, T.; Gu, J.; Buyse, I.M.; Fletcher, J.A.; Liu, J.; Bronson, R.; et al. Tumor formation and inactivation of RIZ1, an Rb-binding member of a nuclear protein-methyltransferase superfamily. Genes Dev. 2001, 15, 2250–2262. [Google Scholar] [CrossRef]
- Buyse, I.M.; Shao, G.; Huang, S. The retinoblastoma protein binds to RIZ, a zinc-finger protein that shares an epitope with the adenovirus E1A protein. Proc. Natl. Acad. Sci. USA 1995, 92, 4467–4471. [Google Scholar]
- Shapiro, V.S.; Lee, P.; Winoto, A. Identification and cloning of the G3B cDNA encoding a 3' segment of a protein binding to GATA-3. Gene 1995, 163, 329–330. [Google Scholar] [CrossRef]
- Zhu, J.; Yamane, H.; Cote-Sierra, J.; Guo, L.; Paul, W.E. GATA-3 promotes Th2 responses through three different mechanisms: induction of Th2 cytokine production, selective growth of Th2 cells and inhibition of Th1 cell-specific factors. Cell Res. 2006, 16, 3–10. [Google Scholar] [CrossRef]
- Abbondanza, C.; de Rosa, C.; D’Arcangelo, A.; Pacifico, M.; Spizuoco, C.; Piluso, G.; di Zazzo, E.; Gazzerro, P.; Medici, N.; Moncharmont, B.; et al. Identification of a functional estrogen-responsive enhancer element in the promoter 2 of PRDM2 gene in breast cancer cell lines. J. Cell. Physiol. 2012, 227, 964–975. [Google Scholar] [CrossRef]
- O’Sullivan, J.M.; Tan-Wong, S.M.; Morillon, A.; Lee, B.; Coles, J.; Mellor, J.; Proudfoot, N.J. Gene loops juxtapose promoters and terminators in yeast. Nat. Genet. 2004, 36, 1014–1018. [Google Scholar] [CrossRef]
- Ansari, A.; Hampsey, M. A role for the CPF 3'-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 2005, 19, 2969–2978. [Google Scholar] [CrossRef]
- Bentley, D.L. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005, 17, 251–256. [Google Scholar] [CrossRef]
- Abbondanza, C.; de Rosa, C.; Ombra, M.N.; Aceto, F.; Medici, N.; Altucci, L.; Moncharmont, B.; Puca, G.A.; Porcellini, A.; Avvedimento, E.V.; et al. Highlighting chromosome loops in DNA-picked chromatin (DPC). Epigenetics 2011, 6, 979–986. [Google Scholar] [CrossRef]
- Abbondanza, C.; de Nigris, F.; de Rosa, C.; Rossiello, R.; Puca, G.A.; Napoli, C. Silencing of YY1 downregulates RIZ1 promoter in human osteosarcoma. Oncol. Res. 2008, 17, 33–41. [Google Scholar]
- Rossi, V.; Staibano, S.; Pasquali, D.; de Rosa, C.; Mascolo, M.; Bellastella, G.; Visconti, D.; de Bellis, A.; Moncharmont, B.; de Rosa, G.; et al. Expression of RIZ1 protein (Retinoblastoma-interacting zinc-finger protein 1) in prostate cancer epithelial cells changes with cancer grade progression and is modulated in vitro by DHT and E2. J. Cell. Physiol. 2009, 221, 771–777. [Google Scholar] [CrossRef]
- TRANSFAC® 7.0 Public. 2005. Available online: http://www.gene-regulation.com/pub/databases.html/ (accessed on 17 January 2009).
- Gazzerro, P.; Bontempo, P.; Schiavone, E.M.; Abbondanza, C.; Moncharmont, B.; Armetta, I.; Medici, N.; de Simone, M.; Nola, E.; Puca, G.A.; et al. Differentiation of myeloid cell lines correlates with a selective expression of RIZ protein. Mol. Med. 2001, 7, 552–560. [Google Scholar]
- Seale, P.; Bjork, B.; Yang, W.; Kajimura, S.; Chin, S.; Kuang, S.; Scime, A.; Devarakonda, S.; Conroe, H.M.; Erdjument-Bromage, H.; et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature 2008, 454, 961–968. [Google Scholar]
- Eom, G.H.; Kim, K.; Kim, S.-M.; Kee, H.J.; Kim, J.-Y.; Jin, H.M.; Kim, J.-R.; Kim, J.H.; Choe, N.; Kim, K.-B.; et al. Histone methyltransferase PRDM8 regulates mouse testis steroidogenesis. Biochem. Biophys. Res. Commun. 2009, 388, 131–136. [Google Scholar] [CrossRef]
- Pastural, E.; Takahashi, N.; Dong, W.F.; Bainbridge, M.; Hull, A.; Pearson, D.; Huang, S.; Lowsky, R.; DeCoteau, J.F.; Geyer, C.R. RIZ1 repression is associated with insulin-like growth factor-1 signaling activation in chronic myeloid leukemia cell lines. Oncogene 2006, 26, 1586–1594. [Google Scholar]
- Thoenen, H. Neurotrophins and Neuronal Plasticity. Science 1995, 270, 593–598. [Google Scholar]
- Lewin, G.R.; Barde, Y.-A. Physiology of the Neurotrophins. Annu. Rev. Neurosci. 1996, 19, 289–317. [Google Scholar] [CrossRef]
- Chittka, A.; Chao, M.V. Identification of a zinc finger protein whose subcellular distribution is regulated by serum and nerve growth factor. Proc. Natl. Acad. Sci. USA 1999, 96, 10705–10710. [Google Scholar] [CrossRef]
- Chittka, A.; Arevalo, J.C.; Rodriguez-Guzman, M.; Perez, P.; Chao, M.V.; Sendtner, M. The p75NTR-interacting protein SC1 inhibits cell cycle progression by transcriptional repression of cyclin E. J. Cell Biol. 2004, 164, 985–996. [Google Scholar] [CrossRef]
- Chittka, A.; Nitarska, J.; Grazini, U.; Richardson, W.D. Transcription Factor Positive Regulatory Domain 4 (PRDM4) recruits Protein Arginine Methyltransferase 5 (PRMT5) to mediate histone arginine methylation and control neural stem cell proliferation and differentiation. J. Biol. Chem. 2012, 287, 42995–43006. [Google Scholar] [CrossRef]
- Sato, N.; Meijer, L.; Skaltsounis, L.; Greengard, P.; Brivanlou, A.H. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 2004, 10, 55–63. [Google Scholar] [CrossRef]
- Watanabe, S.; Umehara, H.; Murayama, K.; Okabe, M.; Kimura, T.; Nakano, T. Activation of Akt signaling is sufficient to maintain pluripotency in mouse and primate embryonic stem cells. Oncogene 2006, 25, 2697–2707. [Google Scholar] [CrossRef]
- Storm, M.P.; Bone, H.K.; Beck, C.G.; Bourillot, P.-Y.; Schreiber, V.; Damiano, T.; Nelson, A.; Savatier, P.; Welham, M.J. Regulation of Nanog Expression by Phosphoinositide 3-Kinase-dependent Signaling in Murine Embryonic Stem Cells. J. Biol. Chem. 2007, 282, 6265–6273. [Google Scholar]
- Lee, M.Y.; Lim, H.W.; Lee, S.H.; Han, H.J. Smad, PI3K/Akt, and Wnt-Dependent Signaling Pathways Are Involved in BMP-4-Induced ESC Self-Renewal. Stem Cells 2009, 27, 1858–1868. [Google Scholar] [CrossRef]
- Chia, N.-Y.; Chan, Y.-S.; Feng, B.; Lu, X.; Orlov, Y.L.; Moreau, D.; Kumar, P.; Yang, L.; Jiang, J.; Lau, M.-S.; et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature 2010, 468, 316–320. [Google Scholar]
- Bikoff, E.K.; Robertson, E.J. One PRDM is not enough for germ cell development. Nat. Genet. 2008, 40, 934. [Google Scholar] [CrossRef]
- Fujiwara, T.; Dunn, N.R.; Hogan, B.L.M. Bone morphogenetic protein 4 in the extraembryonic mesoderm is required for allantois development and the localization and survival of primordial germ cells in the mouse. Proc. Natl. Acad. Sci. USA 2001, 98, 13739–13744. [Google Scholar]
- Kimura, T.; Nakamura, T.; Murayama, K.; Umehara, H.; Yamano, N.; Watanabe, S.; Taketo, M.M.; Nakano, T. The stabilization of β-catenin leads to impaired primordial germ cell development via aberrant cell cycle progression. Dev. Biol. 2006, 300, 545–553. [Google Scholar] [CrossRef]
- Kemp, C.; Willems, E.; Abdo, S.; Lambiv, L.; Leyns, L. Expression of all Wnt genes and their secreted antagonists during mouse blastocyst and postimplantation development. Dev. Dyn. 2005, 233, 1064–1075. [Google Scholar] [CrossRef]
- Ohinata, Y.; Ohta, H.; Shigeta, M.; Yamanaka, K.; Wakayama, T.; Saitou, M. A Signaling Principle for the Specification of the Germ Cell Lineage in Mice. Cell 2009, 137, 571–584. [Google Scholar] [CrossRef]
- Arnold, S.J.; Maretto, S.; Islam, A.; Bikoff, E.K.; Robertson, E.J. Dose-dependent Smad1, Smad5 and Smad8 signaling in the early mouse embryo. Dev. Biol. 2006, 296, 104–118. [Google Scholar] [CrossRef]
- Ohinata, Y.; Payer, B.; O’Carroll, D.; Ancelin, K.; Ono, Y.; Sano, M.; Barton, S.C.; Obukhanych, T.; Nussenzweig, M.; Tarakhovsky, A.; et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 2005, 436, 207–213. [Google Scholar]
- Surani, M.A.; Hayashi, K.; Hajkova, P. Genetic and Epigenetic Regulators of Pluripotency. Cell 2007, 128, 747–762. [Google Scholar] [CrossRef]
- Bedford, M.T.; Richard, S. Arginine Methylation: An Emerging Regulatorof Protein Function. Mol. Cell 2005, 18, 263–272. [Google Scholar] [CrossRef]
- Tee, W.W.; Pardo, M.; Theunissen, T.W.; Yu, L.; Choudhary, J.S.; Hajkova, P.; Surani, M.A. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 2010, 24, 2772–2777. [Google Scholar] [CrossRef]
- Seki, Y.; Yamaji, M.; Yabuta, Y.; Sano, M.; Shigeta, M.; Matsui, Y.; Saga, Y.; Tachibana, M.; Shinkai, Y.; Saitou, M. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development 2007, 134, 2627–2638. [Google Scholar] [CrossRef]
- Saitou, M.; Barton, S.C.; Surani, M.A. A molecular programme for the specification of germ cell fate in mice. Nature 2002, 418, 293–300. [Google Scholar] [CrossRef]
- Yabuta, Y.; Kurimoto, K.; Ohinata, Y.; Seki, Y.; Saitou, M. Gene Expression Dynamics During Germline Specification in Mice Identified by Quantitative Single-Cell Gene Expression Profiling. Biol. Reprod. 2006, 75, 705–716. [Google Scholar]
- Clark, A.T.; Bodnar, M.S.; Fox, M.; Rodriquez, R.T.; Abeyta, M.J.; Firpo, M.T.; Pera, R.A.R. Spontaneous differentiation of germ cells from human embryonic stem cells in vitro. Hum. Mol. Genet. 2004, 13, 727–739. [Google Scholar] [CrossRef]
- Toyooka, Y.; Tsunekawa, N.; Akasu, R.; Noce, T. Embryonic stem cells can form germ cells in vitro. Proc. Natl. Acad. Sci. USA 2003, 100, 11457–11462. [Google Scholar] [CrossRef]
- Yamaji, M.; Seki, Y.; Kurimoto, K.; Yabuta, Y.; Yuasa, M.; Shigeta, M.; Yamanaka, K.; Ohinata, Y.; Saitou, M. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat. Genet. 2008, 40, 1016–1022. [Google Scholar]
- Kurimoto, K.; Yamaji, M.; Seki, Y.; Saitou, M. Specification of the germ cell lineage in mice: A process orchestrated by the PR-domain proteins, Blimp1 and Prdm14. Cell Cycle 2008, 7, 3514–3518. [Google Scholar] [CrossRef]
- Bao, S.; Leitch, H.G.; Gillich, A.; Nichols, J.; Tang, F.; Kim, S.; Lee, C.; Zwaka, T.; Li, X.; Surani, M.A. The Germ Cell Determinant Blimp1 Is Not Required for Derivation of Pluripotent Stem Cells. Cell Stem Cell 2012, 11, 110–117. [Google Scholar] [CrossRef]
- Ying, Y.; Tao, Q. Epigenetic disruption of the WNT/beta-catenin signaling pathway in human cancers. Epigenetics 2009, 4, 307–312. [Google Scholar] [CrossRef]
- Shu, X.; Geng, H.; Li, L.; Ying, J.; Ma, C.; Wang, Y.; Poon, F.F.; Wang, X.; Ying, Y.; Yeo, W.; et al. The epigenetic modifier PRDM5 functions as a tumor suppressor through modulating WNT/β-catenin signaling and is frequently silenced in multiple tumors. PLoS One 2011, 6, e27346. [Google Scholar]
- Nguyen, L.; Besson, A.; Roberts, J.M.; Guillemot, F. Coupling cell cycle exit, neuronal differentiation and migration in cortical neurogenesis. Cell Cycle 2006, 5, 2314–2318. [Google Scholar] [CrossRef]
- Shirasaki, R.; Pfaff, S.L. Transcriptional codes and the control of neuronal identity. .Annu. Rev. Neurosci. 2002, 25, 251–281. [Google Scholar] [CrossRef]
- Bertrand, N.; Castro, D.S.; Guillemot, F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 2002, 3, 517. [Google Scholar] [CrossRef]
- Ross, S.E.; Greenberg, M.E.; Stiles, C.D. Basic Helix-Loop-Helix Factors in Cortical Development. Neuron 2003, 39, 13–25. [Google Scholar] [CrossRef]
- Bramblett, D.E.; Copeland, N.G.; Jenkins, N.A.; Tsai, M.-J. BHLHB4 Is a bHLH Transcriptional Regulator in Pancreas and Brain That Marks the Dimesencephalic Boundary. Genomics 2002, 79, 402. [Google Scholar] [CrossRef]
- Joshi, P.S.; Molyneaux, B.J.; Feng, L.; Xie, X.; Macklis, J.D.; Gan, L. Bhlhb5 Regulates the Postmitotic Acquisition of Area Identities in Layers II-V of the Developing Neocortex. Neuron 2008, 60, 258. [Google Scholar] [CrossRef]
- Ross, S.E.; Mardinly, A.R.; McCord, A.E.; Zurawski, J.; Cohen, S.; Jung, C.; Hu, L.; Mok, S.I.; Shah, A.; Savner, E.M.; et al. Loss of Inhibitory Interneurons in the Dorsal Spinal Cord and Elevated Itch in Bhlhb5 Mutant Mice. Neuron 2010, 65, 886–898. [Google Scholar] [CrossRef]
- Ohtsuka, T.; Ishibashi, M.; Gradwohl, G.; Nakanishi, S.; Guillemot, F.; Kageyama, R. Hes1 and Hes5 as Notch effectors in mammalian neuronal differentiation. EMBO J. 1999, 18, 2196–2207. [Google Scholar] [CrossRef]
- Nieto, M.; Schuurmans, C.; Britz, O.; Guillemot, F. Neural bHLH Genes Control the Neuronal versus Glial Fate Decision in Cortical Progenitors. Neuron 2001, 29, 401–413. [Google Scholar] [CrossRef]
- Endo, K.; Karim, M.R.; Taniguchi, H.; Krejci, A.; Kinameri, E.; Siebert, M.; Ito, K.; Bray, S.J.; Moore, A.W. Chromatin modification of Notch targets in olfactory receptor neuron diversification. Nat. Neurosci. 2011, 15, 224–233. [Google Scholar] [CrossRef]
- Kinameri, E.; Inoue, T.; Aruga, J.; Imayoshi, I.; Kageyama, R.; Shimogori, T.; Moore, A.W. Prdm Proto-Oncogene Transcription Factor Family Expression and Interaction with the Notch-Hes Pathway in Mouse Neurogenesis. PLoS One 2008, 3, e3859. [Google Scholar]
- Ross, S.E.; McCord, A.E.; Jung, C.; Atan, D.; Mok, S.I.; Hemberg, M.; Kim, T.-K.; Salogiannis, J.; Hu, L.; Cohen, S.; et al. Bhlhb5 and Prdm8 Form a Repressor Complex Involved in Neuronal Circuit Assembly. Neuron 2012, 73, 292–303. [Google Scholar] [CrossRef]
- Ito, Y.; Yeo, J.Y.; Chytil, A.; Han, J.; Bringas, P.; Nakajima, A.; Shuler, C.F.; Moses, H.L.; Chai, Y. Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development 2003, 130, 5269–5280. [Google Scholar]
- Pisano, M.M.; Mukhopadhyay, P.; Greene, R.M. Molecular fingerprinting of TGFß-treated embryonic maxillary mesenchymal cells. Orthodontics Craniofacial Res. 2003, 6, 194. [Google Scholar] [CrossRef]
- D’Angelo, M.; Greene, R.M. Transforming growth factor-beta modulation of glycosaminoglycan production by mesenchymal cells of the developing murine secondary palate. Dev. Biol. 1991, 145, 374–378. [Google Scholar] [CrossRef]
- Mukhopadhyay, P.; Greene, R.M.; Pisano, M.M. Expression profiling of transforming growth factor β superfamily genes in developing orofacial tissue. Birth Defects Res. Part A 2006, 76, 528–543. [Google Scholar] [CrossRef]
- Warner, D.R.; Horn, K.H.; Mudd, L.; Webb, C.L.; Greene, R.M.; Pisano, M.M. PRDM16/MEL1: A novel Smad binding protein expressed in murine embryonic orofacial tissue. Biochim. Biophys. Acta 2007, 1773, 814–820. [Google Scholar]
- Alliston, T.; Ko, T.C.; Cao, Y.; Liang, Y.-Y.; Feng, X.-H.; Chang, C.; Derynck, R. Repression of Bone Morphogenetic Protein and Activin-inducible Transcription by Evi-1. J. Biol. Chem. 2005, 280, 24227–24237. [Google Scholar]
- Bjork, B.C.; Turbe-Doan, A.; Prysak, M.; Herron, B.J.; Beier, D.R. Prdm16 is required for normal palatogenesis in mice. Hum. Mol. Genet. 2010, 19, 774–789. [Google Scholar]
- Sato, T.; Goyama, S.; Nitta, E.; Takeshita, M.; Yoshimi, M.; Nakagawa, M.; Kawazu, M.; Ichikawa, M.; Kurokawa, M. Evi-1 promotes para-aortic splanchnopleural hematopoiesis through up-regulation of GATA-2 and repression of TGF-b signaling. Cancer Sci. 2008, 99, 1407–1413. [Google Scholar]
- Takahata, M.; Inoue, Y.; Tsuda, H.; Imoto, I.; Koinuma, D.; Hayashi, M.; Ichikura, T.; Yamori, T.; Nagasaki, K.; Yoshida, M.; et al. SKI and MEL1 Cooperate to Inhibit Transforming Growth Factor-β Signal in Gastric Cancer Cells. J. Biol. Chem. 2009, 284, 3334–3344. [Google Scholar]
- Jakobsen, L.P.; Borup, R.; Vestergaard, J.; Larsen, L.A.; Lage, K.; Maroun, L.L.; Kjaer, I.; Niemann, C.U.; Andersen, M.; Knudsen, M.A.; et al. Expression analyses of human cleft palate tissue suggest a role for osteopontin and immune related factors in palatal development. Exp. Mol. Med. 2009, 41, 77–85. [Google Scholar] [CrossRef]
- Warner, D.R.; Mukhopadhyay, P.; Webb, C.L.; Greene, R.M.; Pisano, M.M. Chromatin immunoprecipitation-promoter microarray identification of genes regulated by PRDM16 in murine embryonic palate mesenchymal cells. Exp. Biol. Med. (Maywood) 2012, 237, 387–394. [Google Scholar] [CrossRef]
- Ren, B.; Chee, K.J.; Kim, T.H.; Maniatis, T. PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev. 1999, 13, 125–137. [Google Scholar] [CrossRef]
- Diehl, S.A.; Schmidlin, H.; Nagasawa, M.; Blom, B.; Spits, H. IL-6 Triggers IL-21 production by human CD4+ T cells to drive STAT3-dependent plasma cell differentiation in B cells. Immunol. Cell Biol. 2012, 90, 208–211. [Google Scholar]
- Kallies, A.; Hawkins, E.D.; Belz, G.T.; Metcalf, D.; Hommel, M.; Corcoran, L.M.; Hodgkin, P.D.; Nutt, S.L. Transcriptional repressor Blimp-1 is essential for T cell homeostasis and self-tolerance. Nat. Immunol. 2006, 7, 466–474. [Google Scholar]
- Martins, G.A.; Cimmino, L.; Shapiro-Shelef, M.; Szabolcs, M.; Herron, A.; Magnusdottir, E.; Calame, K. Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat. Immunol. 2006, 7, 457–465. [Google Scholar] [CrossRef]
- Gong, D.; Malek, T.R. Cytokine-Dependent Blimp-1 Expression in Activated T Cells Inhibits IL-2 Production. J. Immunol. 2007, 178, 242–252. [Google Scholar]
- Wang, L.; van Panhuys, N.; Hu-Li, J.; Kim, S.; Le Gros, G.; Min, B. Blimp-1 Induced by IL-4 Plays a Critical Role in Suppressing IL-2 Production in Activated CD4 T Cells. J. Immunol. 2008, 181, 5249–5256. [Google Scholar]
- Thiele, S.; Wittmann, J.; Jäck, H.-M.; Pahl, A. miR-9 enhances IL-2 production in activated human CD4+ T cells by repressing Blimp-1. Eur. J. Immunol. 2012, 42, 2100–2108. [Google Scholar] [CrossRef]
- Yu, D.; Rao, S.; Tsai, L.M.; Lee, S.K.; He, Y.; Sutcliffe, E.L.; Srivastava, M.; Linterman, M.; Zheng, L.; Simpson, N.; et al. The Transcriptional Repressor Bcl-6 Directs T Follicular Helper Cell Lineage Commitment. Immunity 2009, 31, 457–468. [Google Scholar] [CrossRef]
- Basso, K.; Dalla-Favera, R. BCL6: Master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv. Immunol. 2010, 105, 193–210. [Google Scholar] [CrossRef]
- Oestreich, K.J.; Huang, A.C.; Weinmann, A.S. The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns. J. Exp. Med. 2011, 208, 1001–1013. [Google Scholar] [CrossRef]
- Oestreich, K.J.; Mohn, S.E.; Weinmann, A.S. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat. Immunol. 2012, 13, 405–411. [Google Scholar]
- Cretney, E.; Xin, A.; Shi, W.; Minnich, M.; Masson, F.; Miasari, M.; Belz, G.T.; Smyth, G.K.; Busslinger, M.; Nutt, S.L.; et al. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat. Immunol. 2011, 12, 304–311. [Google Scholar]
- Zheng, Y.; Josefowicz, S.Z.; Kas, A.; Chu, T.-T.; Gavin, M.A.; Rudensky, A.Y. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 2007, 445, 936–940. [Google Scholar]
- Zheng, Y.; Chaudhry, A.; Kas, A.; deRoos, P.; Kim, J.M.; Chu, T.-T.; Corcoran, L.; Treuting, P.; Klein, U.; Rudensky, A.Y. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control TH2 responses. Nature 2009, 458, 351–356. [Google Scholar]
- Kwon, H.; Thierry-Mieg, D.; Thierry-Mieg, J.; Kim, H.-P.; Oh, J.; Tunyaplin, C.; Carotta, S.; Donovan, C.E.; Goldman, M.L.; Tailor, P.; et al. Analysis of Interleukin-21-Induced Prdm1 Gene Regulation Reveals Functional Cooperation of STAT3 and IRF4 Transcription Factors. Immunity 2009, 31, 941–952. [Google Scholar] [CrossRef]
- Sciammas, R.; Shaffer, A.L.; Schatz, J.H.; Zhao, H.; Staudt, L.M.; Singh, H. Graded Expression of Interferon Regulatory Factor-4 Coordinates Isotype Switching with Plasma Cell Differentiation. Immunity 2006, 25, 225–236. [Google Scholar] [CrossRef]
- Noman, A.S.M.; Koide, N.; Iftakhar-E-Khuda, I.; Dagvadorj, J.; Tumurkhuu, G.; Naiki, Y.; Komatsu, T.; Yoshida, T.; Yokochi, T. Retinoblastoma protein-interacting zinc finger 1, a tumor suppressor, augments lipopolysaccharide-induced proinflammatory cytokine production via enhancing nuclear factor-κB activation. Cell. Immunol. 2010, 264, 114–118. [Google Scholar] [CrossRef]
- Shadat, N.M.A.; Koide, N.; Khuda, I.I.E.; Dagvadorj, J.; Tumurkhuu, G.; Naiki, Y.; Komatsu, T.; Yoshida, T.; Yokochi, T. Retinoblastoma Protein-Interacting Zinc Finger 1 (RIZ1) Regulates the Proliferation of Monocytic Leukemia Cells via Activation of p53. Cancer Invest. 2010, 28, 806–812. [Google Scholar] [CrossRef]
- Duan, Z.; Horwitz, M. Gfi-1 Oncoproteins in Hematopoiesis. Hematology 2003, 8, 339–344. [Google Scholar] [CrossRef]
- Hock, H.; Hamblen, M.J.; Rooke, H.M.; Traver, D.; Bronson, R.T.; Cameron, S.; Orkin, S.H. Intrinsic Requirement for Zinc Finger Transcription Factor Gfi-1 in Neutrophil Differentiation. Immunity 2003, 18, 109–120. [Google Scholar] [CrossRef]
- Karsunky, H.; Zeng, H.; Schmidt, T.; Zevnik, B.; Kluge, R.; Schmid, K.W.; Duhrsen, U.; Moroy, T. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat. Genet. 2002, 30, 295–300. [Google Scholar] [CrossRef]
- Person, R.E.; Li, F.-Q.; Duan, Z.; Benson, K.F.; Wechsler, J.; Papadaki, H.A.; Eliopoulos, G.; Kaufman, C.; Bertolone, S.J.; Nakamoto, B.; et al. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat. Genet. 2003, 34, 308–312. [Google Scholar] [CrossRef]
- Duan, Z.; Person, R.E.; Lee, H.-H.; Huang, S.; Donadieu, J.; Badolato, R.; Grimes, H.L.; Papayannopoulou, T.; Horwitz, M.S. Epigenetic Regulation of Protein-Coding and MicroRNA Genes by the Gfi1-Interacting Tumor Suppressor PRDM5. Mol. Cell. Biol. 2007, 27, 6889–6902. [Google Scholar]
- Ségurel, L.; Leffler, E.M.; Przeworski, M. The Case of the Fickle Fingers: How the PRDM9 Zinc Finger Protein Specifies Meiotic Recombination Hotspots in Humans. PLoS Biol. 2011, 9, e1001211. [Google Scholar] [CrossRef]
- Loyola, A.; Bonaldi, T.; Roche, D.; Imhof, A.; Almouzni, G. PTMs on H3 Variants before Chromatin Assembly Potentiate Their Final Epigenetic State. Mol. Cell 2006, 24, 309–316. [Google Scholar] [CrossRef]
- Pinheiro, I.; Margueron, R.; Shukeir, N.; Eisold, M.; Fritzsch, C.; Richter, F.M.; Mittler, G.; Genoud, C.; Goyama, S.; Kurokawa, M.; et al. Prdm3 and Prdm16 are H3K9me1 Methyltransferases Required for Mammalian Heterochromatin Integrity. Cell 2012, 150, 948–960. [Google Scholar] [CrossRef]
- Olsen, B.R.; Reginato, A.M.; Wang, W. Bone development. Annu. Rev. Cell Dev. Biol. 2000, 16, 191–220. [Google Scholar] [CrossRef]
- Galli, G.G.; Honnens de Lichtenberg, K.; Carrara, M.; Hans, W.; Wuelling, M.; Mentz, B.; Multhaupt, H.A.; Fog, C.K.; Jensen, K.T.; Rappsilber, J.; et al. Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing Bone. PLoS Genet. 2012, 8, e1002711. [Google Scholar] [CrossRef]
- Shing, D.C.; Trubia, M.; Marchesi, F.; Radaelli, E.; Belloni, E.; Tapinassi, C.; Scanziani, E.; Mecucci, C.; Crescenzi, B.; Lahortiga, I.; et al. Overexpression of sPRDM16 coupled with loss of p53 induces myeloid leukemias in mice. J. Clin. Invest. 2007, 117, 3696–3707. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
