Alcohol Modulates the Biogenesis and Composition of Microglia-Derived Exosomes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Alcohol Dosing
2.3. Microglia Morphological Evaluation
2.4. Cell Viability by Trypan Blue Exclusion
2.5. Assessment of Cell Viability via Flow Cytometry
2.6. Assessment of Cell Cycle Progression
2.7. Exosome Isolation and Purification
2.8. NanoSight Analysis
2.9. Dot Blot Analysis
2.10. Enzyme-Linked Immunosorbent Assay (ELISA)
2.11. Statistical Significance
3. Results
3.1. Percent Viability of BV-2 Cells after Alcohol Administration
3.2. Apoptotic Status of BV-2 Cells Treated with EtOH
3.3. Alcohol Exposure Modulates Cell Cycle Progression in BV-2 Microglia Cells
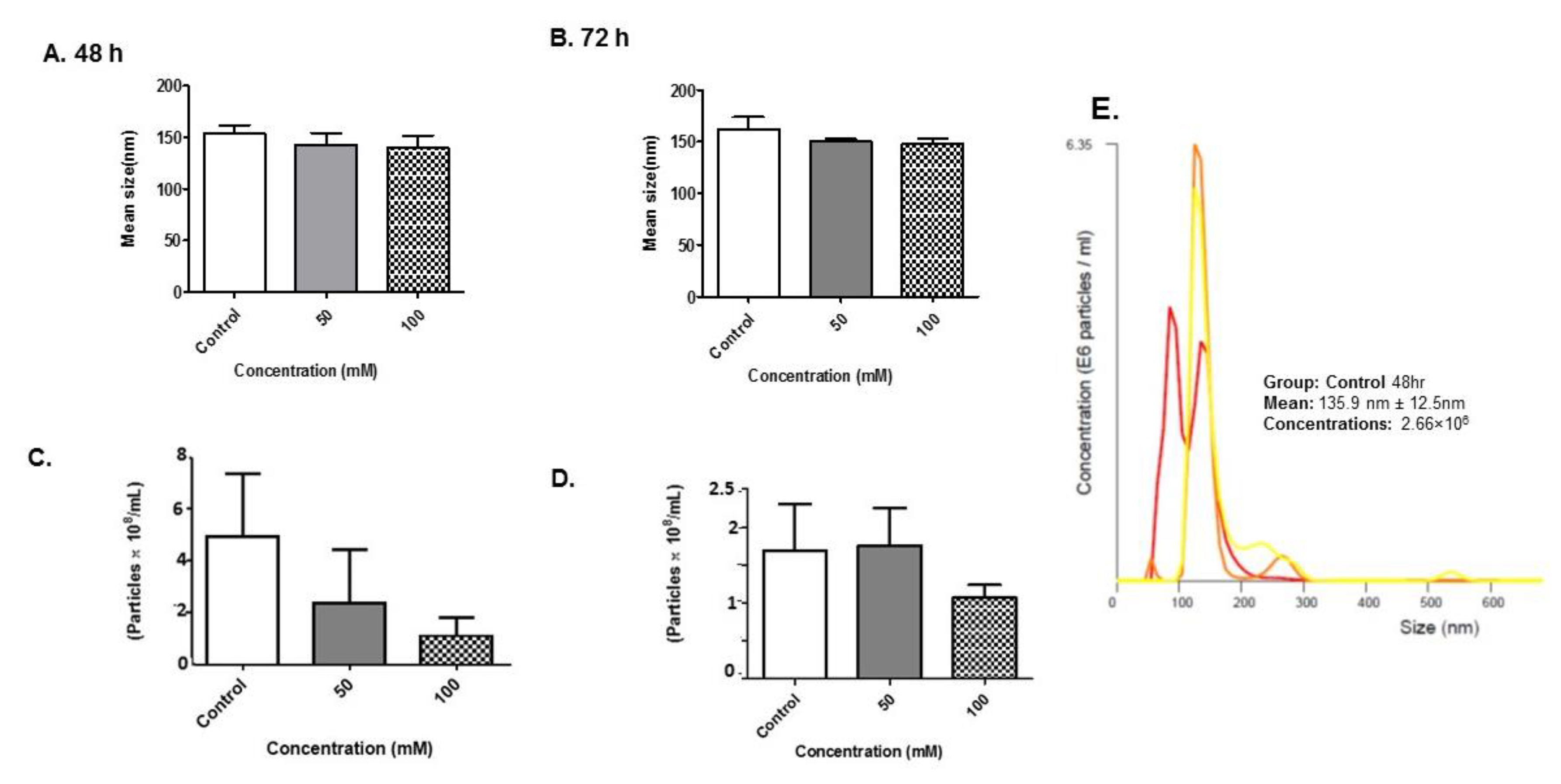
3.4. Size Distribution and Quantity of BV-2-Derived Exosomes after Alcohol Administration
3.5. Expression of Rab Proteins
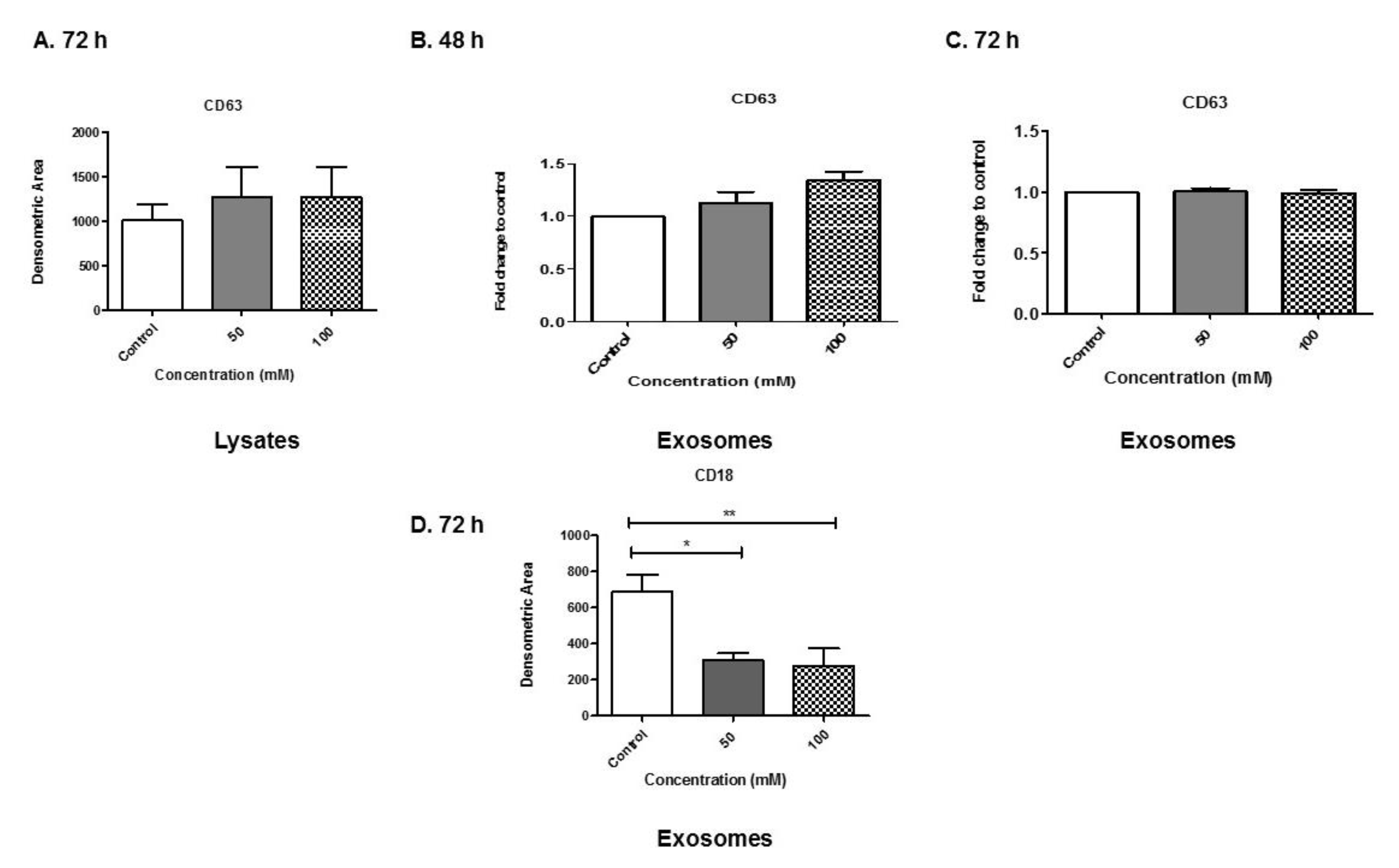
3.6. Analyses of Exosome-Associated Proteins
3.7. Alcohol Dosing Increases Heat Shock Proteins within Exosomes
3.8. Alcohol Dosing Alters Apoptotic Proteins
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Skotland, T.; Sandvig, K.; Llorente, A. Lipids in exosomes: Current knowledge and the way forward. Prog. Lipid. Res. 2017, 66, 30–41. [Google Scholar] [CrossRef]
- Qin, W.; Tsukasaki, Y.; Dasgupta, S.; Mukhopadhyay, N.; Ikebe, M.; Sauter, E.R. Exosomes in Human Breast Milk Promote EMT. Clin. Cancer Res. 2016, 22, 4517–4524. [Google Scholar] [CrossRef]
- Madison, M.N.; Welch, J.L.; Okeoma, C.M. Isolation of Exosomes from Semen for in vitro Uptake and HIV-1 Infection Assays. Bio. Protoc. 2017, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barreiro, K.; Holthofer, H. Urinary extracellular vesicles. A promising shortcut to novel biomarker discoveries. Cell Tissue Res. 2017, 369, 217–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapsogeorgou, E.K.; Abu-Helu, R.F.; Moutsopoulos, H.M.; Manoussakis, M.N. Salivary gland epithelial cell exosomes: A source of autoantigenic ribonucleoproteins. Arthritis Rheum. 2005, 52, 1517–1521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chivet, M.; Hemming, F.; Pernet-Gallay, K.; Fraboulet, S.; Sadoul, R. Emerging role of neuronal exosomes in the central nervous system. Front Physiol. 2012, 3, 145. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Dinkins, M.; He, Q.; Zhu, G.; Poirier, C.; Campbell, A.; Mayer-Proschel, M.; Bieberich, E. Astrocytes secrete exosomes enriched with proapoptotic ceramide and prostate apoptosis response 4 (PAR-4): Potential mechanism of apoptosis induction in Alzheimer disease (AD). J. Biol. Chem. 2012, 287, 21384–21395. [Google Scholar] [CrossRef]
- Fitzner, D.; Schnaars, M.; van Rossum, D.; Krishnamoorthy, G.; Dibaj, P.; Bakhti, M.; Regen, T.; Hanisch, U.K.; Simons, M. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J. Cell Sci. 2011, 124, 447–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glebov, K.; Löchner, M.; Jabs, R.; Lau, T.; Merkel, O.; Schloss, P.; Steinhäuser, C.; Walter, J. Serotonin stimulates secretion of exosomes from microglia cells. Glia 2015, 63, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Thery, C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol. Rep. 2011, 3, 15. [Google Scholar] [CrossRef]
- Sharma, P.; Schiapparelli, L.; Cline, H.T. Exosomes function in cell-cell communication during brain circuit development. Curr. Opin. Neurobiol. 2013, 23, 997–1004. [Google Scholar] [CrossRef] [Green Version]
- Gupta, A.; Pulliam, L. Exosomes as mediators of neuroinflammation. J. Neuroinflammation 2014, 11, 68. [Google Scholar] [CrossRef] [PubMed]
- Bobrie, A.; Colombo, M.; Raposo, G.; Théry, C. Exosome secretion: Molecular mechanisms and roles in immune responses. Traffic 2011, 12, 1659–1668. [Google Scholar] [CrossRef]
- McCarthy, G.M.; Farris, S.P.; Blednov, Y.A.; Harris, R.A.; Mayfield, R.D. Microglial-specific transcriptome changes following chronic alcohol consumption. Neuropharmacology 2018, 128, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S. Alcoholism and its effects on the central nervous system. Curr. Neurovasc. Res. 2013, 10, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Bachtell, R.K.; Jones, J.D.; Heinzerling, K.G.; Beardsley, P.M.; Comer, S.D. Glial and neuroinflammatory targets for treating substance use disorders. Drug Alcohol Depend. 2017, 180, 156–170. [Google Scholar] [CrossRef] [PubMed]
- Moore, E.M.; Migliorini, R.; Infante, M.A.; Riley, E.P. Fetal Alcohol Spectrum Disorders: Recent Neuroimaging Findings. Curr. Dev. Disord. Rep. 2014, 1, 161–172. [Google Scholar] [CrossRef] [Green Version]
- Taylor, P.A.; Jacobson, S.W.; van der Kouwe, A.; Molteno, C.D.; Chen, G.; Wintermark, P.; Alhamud, A.; Jacobson, J.L.; Meintjes, E.M. A DTI-based tractography study of effects on brain structure associated with prenatal alcohol exposure in newborns. Hum. Brain Mapp. 2015, 36, 170–186. [Google Scholar] [CrossRef] [PubMed]
- Uban, K.A.; Herting, M.M.; Wozniak, J.R.; Sowell, E.R.; CIFASD. Sex differences in associations between white matter microstructure and gonadal hormones in children and adolescents with prenatal alcohol exposure. Psychoneuroendocrinology 2017, 83, 111–121. [Google Scholar] [CrossRef]
- Khoury, J.E.; Milligan, K.; Girard, T.A. Executive Functioning in Children and Adolescents Prenatally Exposed to Alcohol: A Meta-Analytic Review. Neuropsychol. Rev. 2015, 25, 149–170. [Google Scholar] [CrossRef]
- Mattson, S.N.; Crocker, N.; Nguyen, T.T. Fetal alcohol spectrum disorders: Neuropsychological and behavioral features. Neuropsychol. Rev. 2011, 21, 81–101. [Google Scholar] [CrossRef]
- Norman, A.L.; O’Brien, J.W.; Spadoni, A.D.; Tapert, S.F.; Jones, K.L.; Riley, E.P.; Mattson, S.N. A functional magnetic resonance imaging study of spatial working memory in children with prenatal alcohol exposure: Contribution of familial history of alcohol use disorders. Alcohol Clin. Exp. Res. 2013, 37, 132–140. [Google Scholar] [CrossRef] [PubMed]
- McClain, J.A.; Morris, S.A.; Deeny, M.A.; Marshall, S.A.; Hayes, D.M.; Kiser, Z.M.; Nixon, K. Adolescent binge alcohol exposure induces long-lasting partial activation of microglia. Brain Behav. Immun. 2011, 25, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Napoli, I.; Kierdorf, K.; Neumann, H. Microglial precursors derived from mouse embryonic stem cells. Glia 2009, 57, 1660–1671. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, Y.S.; Ota, A.; Nakashima, A.; Nagasaki, H.; Kodani, Y.; Mori, K.; Nagatsu, T. Lipopolysaccharide treatment arrests the cell cycle of BV-2 microglial cells in G(1) phase and protects them from UV light-induced apoptosis. J. Neural. Transm. (Vienna) 2015, 122, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Sims, B.; Farrow, A.L.; Williams, S.D.; Bansal, A.; Krendelchtchikov, A.; Gu, L.; Matthews, Q.L. Role of TIM-4 in exosome-dependent entry of HIV-1 into human immune cells. Int. J. Nanomedicine 2017, 12, 4823–4833. [Google Scholar] [CrossRef] [Green Version]
- Sapan, C.V.; Lundblad, R.L.; Price, N.C. Colorimetric protein assay techniques. Biotechnol. Appl. Biochem. 1999, 29, 99–108. [Google Scholar] [PubMed]
- Thery, C.; Boussac, M.; Véron, P.; Ricciardi-Castagnoli, P.; Raposo, G.; Garin, J.; Amigorena, S. Proteomic analysis of dendritic cell-derived exosomes: A secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 2001, 166, 7309–7318. [Google Scholar] [CrossRef] [PubMed]
- Gangalum, R.K.; Bhat, A.M.; Kohan, S.A.; Bhat, S.P. Inhibition of the Expression of the Small Heat Shock Protein alphaB-Crystallin Inhibits Exosome Secretion in Human Retinal Pigment Epithelial Cells in Culture. J. Biol. Chem. 2016, 291, 12930–12942. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Fung, Y.K.; Liu, X.; Pahan, K. Up-regulation of microglial CD11b expression by nitric oxide. J. Biol. Chem. 2006, 281, 14971–14980. [Google Scholar] [CrossRef]
- Moura, C.S.; Lollo, P.C.B.; Morato, P.N.; Amaya-Farfan, J. Dietary Nutrients and Bioactive Substances Modulate Heat Shock Protein (HSP) Expression: A Review. Nutrients 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Santos-Junior, V.A.; Lollo, P.C.B.; Cantero, M.A.; Soares Moura, C.; Amaya-Farfan, J.; Neder Morato, P. Heat Shock Proteins: Protection and Potential Biomarkers for Ischemic Injury of Cardiomyocytes After Surgery. Braz. J. Cardiovasc. Surg. 2018, 33, 291–302. [Google Scholar] [CrossRef]
- Pisitkun, T.; Shen, R.F.; Knepper, M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368–13373. [Google Scholar] [CrossRef] [Green Version]
- Clayton, A.; Turkes, A.; Navabi, H.; Mason, M.D.; Tabi, Z. Induction of heat shock proteins in B-cell exosomes. J. Cell Sci. 2005, 118, 3631–3638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.F.; Fan, J.; Fedesco, M.; Guan, S.; Li, Y.; Bandyopadhyay, B.; Bright, A.M.; Yerushalmi, D.; Liang, M.; Chen, M.; et al. Transforming growth factor alpha (TGFalpha)-stimulated secretion of HSP90alpha: Using the receptor LRP-1/CD91 to promote human skin cell migration against a TGFbeta-rich environment during wound healing. Mol. Cell Biol. 2008, 28, 3344–3358. [Google Scholar] [CrossRef] [PubMed]
- McCready, J.; Sims, J.D.; Chan, D.; Jay, D.G. Secretion of extracellular hsp90alpha via exosomes increases cancer cell motility: A role for plasminogen activation. BMC Cancer 2010, 10, 294. [Google Scholar] [CrossRef] [PubMed]
- Vergara, V.M.; Weiland, B.J.; Hutchison, K.E.; Calhoun, V.D. The Impact of Combinations of Alcohol, Nicotine and Cannabis on Dynamic Brain Connectivity. Neuropsychopharmacology 2018, 43, 877–890. [Google Scholar] [CrossRef]
- Mao, H.; Diehl, A.M.; Li, Y.X. Sonic hedgehog ligand partners with caveolin-1 for intracellular transport. Lab. Invest. 2009, 89, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Fruhbeis, C.; Fröhlich, D.; Kuo, W.P.; Krämer-Albers, E.M. Extracellular vesicles as mediators of neuron-glia communication. Front. Cell Neurosci. 2013, 7, 182. [Google Scholar] [CrossRef]
- Jones, A.W. Evidence-based survey of the elimination rates of ethanol from blood with applications in forensic casework. Forensic Sci. Int. 2010, 200, 1–20. [Google Scholar] [CrossRef]
- Sims, B.; Gu, L.; Krendelchtchikov, A.; Matthews, Q.L. Neural stem cell-derived exosomes mediate viral entry. Int. J. Nanomedicine 2014, 9, 4893–4897. [Google Scholar] [CrossRef] [Green Version]
- Crenshaw, B.J.; Matthews, Q.L.; Alabama State University, Montgomery, AL, USA. Data not shown. 2019.
- Blanc, L.; Vidal, M. New insights into the function of Rab GTPases in the context of exosomal secretion. Small GTPases 2018, 9, 95–106. [Google Scholar] [CrossRef]
- Zhang, M.; Schekman, R. Cell biology. Unconventional secretion, unconventional solutions. Science 2013, 340, 559–561. [Google Scholar] [PubMed]
- Andreu, Z.; Yanez-Mo, M. Tetraspanins in extracellular vesicle formation and function. Front. Immuno. 2014, 5, 442. [Google Scholar] [CrossRef]
- Brites, D.; Fernandes, A. Neuroinflammation and Depression: Microglia Activation, Extracellular Microvesicles and microRNA Dysregulation. Front. Cell Neurosci. 2015, 9, 476. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Knowlton, A.A. HSP60 trafficking in adult cardiac myocytes: Role of the exosomal pathway. Am. J. Physiol. Heart Circ. Physiol. 2007, 292, 3052–3056. [Google Scholar] [CrossRef]
- Yamanaka, K.; Takahashi, N.; Ooie, T.; Kaneda, K.; Yoshimatsu, H.; Saikawa, T. Role of protein kinase C in geranylgeranylacetone-induced expression of heat-shock protein 72 and cardioprotection in the rat heart. J. Mol. Cell Cardiol. 2003, 35, 785–794. [Google Scholar] [CrossRef]
- Ugurlucan, M.; Erer, D.; Karatepe, O.; Ziyade, S.; Haholu, A.; Gungor Ugurlucan, F.; Filizcan, U.; Tireli, E.; Dayioglu, E.; Alpagut, U. Glutamine enhances the heat shock protein 70 expression as a cardioprotective mechanism in left heart tissues in the presence of diabetes mellitus. Expert Opin. Ther. Targets 2010, 14, 1143–1156. [Google Scholar] [CrossRef]
- Malik, Z.A.; Kott, K.S.; Poe, A.J.; Kuo, T.; Chen, L.; Ferrara, K.W.; Knowlton, A.A. Cardiac myocyte exosomes: stability, HSP60, and proteomics. Am. J. Physiol. Heart Circ. Physiol. 2013, 304, 954–965. [Google Scholar] [CrossRef]
- Taylor, A.R.; Robinson, M.B.; Gifondorwa, D.J.; Tytell, M.; Milligan, C.E. Regulation of heat shock protein 70 release in astrocytes: Role of signaling kinases. Dev. Neurobiol. 2007, 67, 1815–1829. [Google Scholar] [CrossRef] [Green Version]
- Stetler, R.A.; Gan, Y.; Zhang, W.; Liou, A.K.; Gao, Y.; Cao, G.; Chen, J. Heat shock proteins: Cellular and molecular mechanisms in the central nervous system. Prog. Neurobiol. 2010, 92, 184–211. [Google Scholar] [CrossRef] [PubMed]
- Yenari, M.A.; Liu, J.; Zheng, Z.; Vexler, Z.S.; Lee, J.E.; Giffard, R.G. Antiapoptotic and anti-inflammatory mechanisms of heat-shock protein protection. Ann. N Y Acad. Sci. 2005, 1053, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Giffard, R.G.; Han, R.Q.; Emery, J.F.; Duan, M.; Pittet, J.F. Regulation of apoptotic and inflammatory cell signaling in cerebral ischemia: The complex roles of heat shock protein 70. Anesthesiology 2008, 109, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Kacimi, R.; Yenari, M.A. Pharmacologic heat shock protein 70 induction confers cytoprotection against inflammation in gliovascular cells. Glia 2015, 63, 1200–1212. [Google Scholar] [CrossRef] [PubMed]
- De Maio, A.; Vazquez, D. Extracellular heat shock proteins: A new location, a new function. Shock 2013, 40, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Tatokoro, M.; Koga, F.; Yoshida, S.; Kihara, K. Heat shock protein 90 targeting therapy: State of the art and future perspective. Excli J. 2015, 14, 48–58. [Google Scholar]
- Buchner, J. Hsp90 & Co.—A holding for folding. Trends. Biochem. Sci. 1999, 24, 136–141. [Google Scholar]
- Welch, W.J.; Feramisco, J.R. Purification of the major mammalian heat shock proteins. J. Biol. Chem. 1982, 257, 14949–14959. [Google Scholar]
- Whitesell, L.; Lin, N.U. HSP90 as a platform for the assembly of more effective cancer chemotherapy. Biochim. Biophys. Acta. 2012, 1823, 756–766. [Google Scholar] [CrossRef] [Green Version]
- Neckers, L.; Kern, A.; Tsutsumi, S. Hsp90 inhibitors disrupt mitochondrial homeostasis in cancer cells. Chem. Biol. 2007, 14, 1204–1206. [Google Scholar] [CrossRef]
- Trepel, J.; Mollapour, M.; Giaccone, G.; Neckers, L. Targeting the dynamic HSP90 complex in cancer. Nat. Rev. Cancer 2010, 10, 537–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitesell, L.; Lindquist, S.L. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer 2005, 5, 761–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.; Piel, W.H.; Gui, L.; Bruford, E.; Monteiro, A. The HSP90 family of genes in the human genome: Insights into their divergence and evolution. Genomics 2005, 86, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Song, H.Y.; Dunbar, J.D.; Zhang, Y.X.; Guo, D.; Donner, D.B. Identification of a protein with homology to hsp90 that binds the type 1 tumor necrosis factor receptor. J. Biol. Chem. 1995, 270, 3574–3581. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.F.; Chen, Y.; Dai, K.; Chen, P.L.; Riley, D.J.; Lee, W.H. A new member of the Hsp90 family of molecular chaperones interacts with the retinoblastoma protein during mitosis and after heat shock. Mol. Cell Biol. 1996, 16, 4691–4699. [Google Scholar] [CrossRef]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crenshaw, B.J.; Kumar, S.; Bell, C.R.; Jones, L.B.; Williams, S.D.; Saldanha, S.N.; Joshi, S.; Sahu, R.; Sims, B.; Matthews, Q.L. Alcohol Modulates the Biogenesis and Composition of Microglia-Derived Exosomes. Biology 2019, 8, 25. https://doi.org/10.3390/biology8020025
Crenshaw BJ, Kumar S, Bell CR, Jones LB, Williams SD, Saldanha SN, Joshi S, Sahu R, Sims B, Matthews QL. Alcohol Modulates the Biogenesis and Composition of Microglia-Derived Exosomes. Biology. 2019; 8(2):25. https://doi.org/10.3390/biology8020025
Chicago/Turabian StyleCrenshaw, Brennetta J., Sanjay Kumar, Courtnee’ R. Bell, Leandra B. Jones, Sparkle D. Williams, Sabita N. Saldanha, Sameer Joshi, Rajnish Sahu, Brian Sims, and Qiana L. Matthews. 2019. "Alcohol Modulates the Biogenesis and Composition of Microglia-Derived Exosomes" Biology 8, no. 2: 25. https://doi.org/10.3390/biology8020025
APA StyleCrenshaw, B. J., Kumar, S., Bell, C. R., Jones, L. B., Williams, S. D., Saldanha, S. N., Joshi, S., Sahu, R., Sims, B., & Matthews, Q. L. (2019). Alcohol Modulates the Biogenesis and Composition of Microglia-Derived Exosomes. Biology, 8(2), 25. https://doi.org/10.3390/biology8020025






