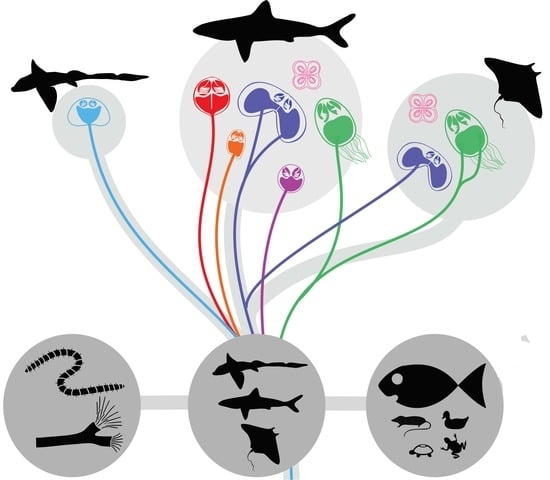
Mechanisms and Drivers for the Establishment of Life Cycle Complexity in Myxozoan Parasites
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Characterization of Myxozoans from Chondrichthyes
2.2. Novel Lineages, Their Phylogeny, and Diversity
2.3. Host-Parasite Cophylogeny and Geographic Character Correlation in Chloromyxum spp.
3. Results
3.1. Preferred Cartilaginous Hosts of Myxozoans
3.2. Origin of Lineages and Comparison with Phylogenetic Position of Teleost Congeners
3.3. Evolution of Myxozoans in Cartilaginous Fish Hosts: Host-Parasite Co-Diversification and Phylogeography
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Moran, N.A. Adaptation and constraint in the complex life cycles of animals. Annu. Rev. Ecol. Syst. 1994, 25, 573–600. [Google Scholar] [CrossRef]
- Poulin, R. Evolutionary Ecology of Parasites, 2nd ed.; Princeton University Press: Princeton, NJ, USA, 2007; pp. 1–342. [Google Scholar]
- Holzer, A.S.; Bartošová-Sojková, P.; Born-Torrijos, A.; Lövy, A.; Hartigan, A.; Fiala, I. The joint evolution of the Myxozoa and their alternate hosts: A cnidarian recipe for success and vast biodiversity. Mol. Ecol. 2018, 27, 1651–1666. [Google Scholar] [CrossRef] [PubMed]
- Licht, M.; Schmuecker, K.; Huelsken, T.; Hanel, R.; Bartsch, P.; Paeckert, M. Contribution to the molecular phylogenetic analysis of extant holocephalan fishes (Holocephali, Chimaeriformes). Org. Divers. Evol. 2012, 12, 421–432. [Google Scholar] [CrossRef]
- Inoue, J.G.; Miya, M.; Lam, K.; Tay, B.H.; Danks, J.A.; Bell, J.; Walker, T.I.; Venka-tesh, B. Evolutionary origin and phylogeny of the modernholocephalans (Chondrichthyes: Chimaeriformes): A mitogenomic perspective. Mol. Biol. Evol. 2010, 27, 2576–2586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holland, J.W.; Okamura, B.; Hartikainen, H.; Secombes, C.J. A novel minicollagen gene links cnidarians and myxozoans. Proc. Biol. Sci. 2011, 278, 546–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shpirer, E.; Diamant, A.; Cartwright, P.; Huchon, D. A genome wide survey reveals multiple nematocyst-specific genes in Myxozoa. BMC Evol. Biol. 2018, 18, 138. [Google Scholar] [CrossRef]
- Chang, E.S.; Neuhof, M.; Rubinstein, N.D.; Diamant, A.; Philippe, H.; Huchon, D.; Cartwright, P. Genomic insights into the evolutionary origin of Myxozoa within Cnidaria. Proc. Natl. Acad. Sci. USA 2015, 112, 14912–14917. [Google Scholar] [CrossRef] [Green Version]
- Hartigan, A.; Wilkinson, M.; Gower, D.J.; Streicher, J.W.; Holzer, A.S.; Okamura, B. Myxozoan infections of caecilians demonstrate broad host specificity and indicate a link with human activity. Int. J. Parasitol. 2016, 46, 375–381. [Google Scholar] [CrossRef]
- Siau, Y.; Gasc, C.; Maillard, C. Premières observations ultrastructurales d‘une myxosporide appartenant au genre Fabespora, parasite de trématode. Protistologica 1981, 17, 131–137. [Google Scholar]
- Overstreet, R.M. Fabespora vermicola sp. n., the first myxosporidan from a platyhelminth. J. Parasitol. 1976, 62, 680–684. [Google Scholar] [CrossRef] [Green Version]
- Yokoyama, H.; Masuda, K. Kudoa sp. (Myxozoa) causing a post-mortem myoliquefaction of North-Pacific giant octopus Paroctopus dofleini (Cephalopoda: Octopodidae). Bull. Eur. Assoc. Fish Pathol. 2001, 21, 266–268. [Google Scholar]
- Freeman, M.A.; Shinn, A.P. Myxosporean hyperparasites of gill monogeneans are basal to the Multivalvulida. Parasit. Vectors 2011, 4, 220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kodádková, A.; Bartošová-Sojková, P.; Holzer, A.S.; Fiala, I. Bipteria vetusta n. sp.—An old parasite in an old host: Tracing the origin of myxosporean parasitism in vertebrates. Int. J. Parasitol. 2015, 45, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, S.D.; Bartholomew, J.L.; Lotan, T. Myxozoans: Ancient metazoan parasites find a home in phylum Cnidaria. Zoology 2018, 129, 66–68. [Google Scholar] [CrossRef]
- Jameson, A.P. Myxosporidia from Californian fishes. J. Parasitol. 1929, 16, 59–86. [Google Scholar] [CrossRef]
- Davis, H.S. The Myxosporidia of the Beaufort region, a systematic and biological study. Fish. Bull. 1917, 35, 199–252. [Google Scholar]
- Gleeson, R.J.; Adlard, R.D. Morphological and genetic analysis of three new species of Ceratomyxa Thélohan, 1892 (Myxozoa: Myxosporea) from carcharhinid sharks off Australia. Syst. Parasitol. 2011, 80, 117–124. [Google Scholar] [CrossRef]
- Gleeson, R.J.; Adlard, R.D. Phylogenetic relationships amongst Chloromyxum Mingazzini, 1890 (Myxozoa: Myxosporea), and the description of six novel species from Australian elasmobranchs. Parasitol. Int. 2012, 61, 267–274. [Google Scholar] [CrossRef]
- Cantatore, D.M.P.; Irigoitia, M.M.; Holzer, A.S.; Bartošová-Sojková, P.; Pecková, H.; Fiala, I.; Timi, J.T. The description of two new species of Chloromyxum from skates in the Argentine Sea reveals that a limited geographic host distribution causes phylogenetic lineage separation of myxozoans in Chondrichthyes. Parasite 2018, 25, 47. [Google Scholar] [CrossRef] [Green Version]
- Weigmann, S. Annotated checklist of the living sharks, batoids and chimaeras (Chondrichthyes) of the world, with a focus on biogeographical diversity. J. Fish Biol. 2016, 88, 837–1037. [Google Scholar] [CrossRef]
- Stein, R.W.; Mull, C.G.; Kuhn, T.S.; Aschliman, N.C.; Davidson, L.N.K.; Joy, J.B.; Smith, G.J.; Dulvy, N.K.; Mooers, A.O. Global priorities for conserving the evolutionary history of sharks, rays and chimaeras. Nat. Ecol. Evol. 2018, 2, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Dulvy, N.K.; Fowler, S.L.; Musick, J.A.; Cavanagh, R.D.; Kyne, P.M.; Harrison, L.R.; Carlson, J.K.; Davidson, L.N.K.; Fordham, S.V.; Francis, M.P.; et al. Extinction risk and conservation of the world’s sharks and rays. eLife 2014, 3, e00590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holzer, A.S.; Sommerville, C.; Wootten, R. Molecular relationships and phylogeny in a community of myxosporeans and actinosporeans based on their 18S rDNA sequences. Int. J. Parasitol. 2004, 34, 1099–1111. [Google Scholar] [CrossRef] [PubMed]
- Holzer, A.S.; Bartošová, P.; Pecková, H.; Tyml, T.; Atkinson, S.; Bartholomew, J.; Sipos, D.; Eszterbauer, E.; Dyková, I. ‘Who’s who’ in renal sphaerosporids (Bivalvulida: Myxozoa) from common carp, Prussian carp and goldfish—Molecular identification of cryptic species, blood stages and new members of Sphaerospora sensu stricto. Parasitology 2012, 140, 46–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4.0b 10. 2001; Sinauer Associates, Inc.: Sunderland, UK, 2002. [Google Scholar]
- Mueller, T.; Fagan, W. Search and navigation in dynamic environments: From individual behaviors to population distributions. Oikos 2008, 117, 654–664. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models using lme4. J. Stat. Softw. 2015, 67. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org/ (accessed on 15 April 2019).
- Merkle, D.; Middendorf, M.; Wieseke, N. A parameter-adaptive dynamic programming approach for inferring cophylogenies. BMC Bioinform. 2010, 11, S60. [Google Scholar] [CrossRef] [Green Version]
- Legendre, P.; Desdevises, Y.; Bazin, E. A statistical test for host–parasite coevolution. Syst. Biol. 2002, 51, 217–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, F. Dispersal-Vicariance Analysis: A New Approach to the Quantification of Historical Biogeography. Syst. Biol. 1997, 46, 195–203. [Google Scholar] [CrossRef]
- Ronquist, F. DIVA Version 1.2. Computer Program for MacOS and Win32. Evolutionary Biology Centre, Uppsala University. 2001. Available online: http://www.ebc.uu.se/systzoo/research/diva/diva.html (accessed on 12 January 2019).
- Yu, Y.; Harris, A.J.; Blair, C.; He, X. RASP (Reconstruct Ancestral State in Phylogenies): A tool for historical biogeography. Mol. Phylogenetics Evol. 2015, 87, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Patra, S.; Bartošová-Sojková, P.; Pecková, H.; Fiala, I.; Eszterbauer, E.; Holzer, A.S. Biodiversity and host-parasite cophylogeny of Sphaerospora (sensu stricto) (Cnidaria: Myxozoa). Parasit. Vectors 2018, 11, 347. [Google Scholar] [CrossRef] [PubMed]
- Fiala, I. The phylogeny of Myxosporea (Myxozoa) based on small subunit ribosomal RNA gene analysis. Int. J. Parasitol. 2006, 36, 1521–1534. [Google Scholar] [CrossRef]
- Okamura, B.; Hartigan, A.; Naldoni, J. Extensive uncharted biodiversity: The parasite dimension. Integr. Comp. Biol. 2018, 58, 1132–1145. [Google Scholar] [CrossRef]
- Hughes, L.C.; Ortí, G.; Huang, Y.; Sun, Y.; Baldwin, C.C.; Thompson, A.W.; Arcila, D.; Betancur, R.R.; Li, C.; Becker, L.; et al. Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomic and genomic data. Proc. Natl. Acad. Sci. USA 2018, 115, 6249–6254. [Google Scholar] [CrossRef] [Green Version]
- Sirin, C.; Santos, M.J.; Rangel, L.F. Morphological and molecular analyses of Bipteria lusitanica n. sp. in wild white seabream, Diplodus sargus (Linnaeus, 1758) in Portugal. Parasitol. Res. 2018, 117, 2035–2041. [Google Scholar] [CrossRef]
- Eiras, J.C.; Lu, Y.S.; Gibson, D.I.; Fiala, I.; Saraiva, A.; Cruz, C.; Santos, M.J. Synopsis of the species of Chloromyxum Mingazinni, 1890 (Myxozoa: Myxosporea: Chloromyxidae). Syst. Parasitol. 2012, 83, 203–225. [Google Scholar] [CrossRef]
- Ikeda, I. Studies on some sporozoan parasites of sipunculoids. In The life history of a new actinomyxidian Tetractinomyxon intermedium g. et sp. nov. Arch. Protistenkd. 1912, 25, 240–272. [Google Scholar]
- Arthur, J.R.; Lom, J. Sphaerospora araii n. sp. (Myxosporea: Sphaerosporidae) from the kidney of a longnose skate (Raja rhina Jordan and Gilbert) from the Pacific Ocean off Canada. Can. J. Zool. 1985, 63, 2902–2906. [Google Scholar] [CrossRef]
- Pariselle, A.; Morand, S.; Deveney, M.R.; Pouyaud, L. Parasite species richness of closely related hosts: Historical scenario and “genetic” hypothesis. In Taxonomy, Ecology and Evolution of Metazoan Parasites; Combes, C., Jourdan, J., Eds.; Presses Universitaires de Perpignan: Perpignan, France, 2003; pp. 147–166. [Google Scholar]
- Gao, C.; Shi, N.N.; Liu, Y.X.; Peay, K.G.; Zheng, Y.; Ding, Q.; Mi, X.C.; Ma, K.P.; Wubet, T.; Buscot, F.; et al. Host plant genus-level diversity is the best predictor of ectomycorrhizal fungal diversity in a Chinese subtropical forest. Mol. Ecol. 2003, 22, 3403–3414. [Google Scholar] [CrossRef] [PubMed]
- Molnár, K.; El-Mansy, A.; Székely, C.; Baska, F. Experimental identification of the actinosporean stage of Sphaerospora renicola Dyková & Lom 1982 (Myxosporea: Sphaerosporidae) in oligochaete alternate hosts. J. Fish Dis. 1999, 22, 143–153. [Google Scholar]
- Bartošová, P.; Fiala, I.; Jirků, M.; Cinková, M.; Caffara, M.; Fioravanti, M.L.; Atkinson, S.D.; Bartholomew, J.L.; Holzer, A.S. Sphaerospora sensu stricto: Taxonomy, diversity and evolution of a unique lineage of myxosporeans (Myxozoa). Mol. Phylogenet. Evol. 2013, 68, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Karlsbakk, E.; Køie, M. Bipteria formosa (Kovaleva et Gaevskaya, 1979) comb. n. (Myxozoa: Myxosporea) in whiting Merlangius merlangus (Teleostei: Gadidae) from Denmark. Folia Parasitol. 2009, 56, 86–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koletić, N.; Novosel, M.; Rajević, N.; Franjević, D. Bryozoans are returning home: Recolonization of freshwater ecosystems inferred from phylogenetic relationships. Ecol. Evol. 2015, 5, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Lucifora, L.O.; de Carvalho, M.R.; Kyne, P.M.; White, W.T. Freshwater sharks and rays. Curr. Biol. 2015, 25, R971–R973. [Google Scholar] [CrossRef] [Green Version]
- Dwyer, G.; Elkinton, J.S. Host dispersal and the spatial spread of insect pathogens. Ecology 1995, 76, 1262–1275. [Google Scholar] [CrossRef]
- Figuerola, J.; Green, A. Haematozoan parasites and migratory behaviour in waterfowl. Evol. Ecol. 2000, 14, 143–153. [Google Scholar] [CrossRef]
- Koprivnikar, J.; Leung, T.L.F. Flying with diverse passengers: Greater richness of parasitic nematodes in migratory birds. Oikos 2015, 124, 399–405. [Google Scholar] [CrossRef]
- Hannon, E.R.; Kinsella, J.M.; Calhoun, D.M.; Joseph, M.B.; Johnson, P.T.J. Endohelminths in bird hosts from northern California and an analysis of the role of life history traits on parasite richness. J. Parasitol. 2016, 102, 199–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nance, R.D.; Worsley, T.R.; Moody, J.B. The supercontinent cycle. Sci. Am. 1988, 259, 72–79. [Google Scholar] [CrossRef]
- Veevers, J.J. Gondwanaland from 650–500 Ma assembly through 320 Ma merger in Pangea to 185–100 Ma breakup: Supercontinental tectonics via stratigraphy and radiometric dating. Earth Sci. Rev. 2004, 68, 1–132. [Google Scholar] [CrossRef]
- Moura, T.; Figueiredo, I.; Bordalo-Machado, P.; Gordo, L.S. Feeding habits of Chimaera monstrosa L. (Chimaeridae) in relation to its ontogenetic development on the southern Portuguese continental slope. Mar. Biol. Res. 2005, 1, 118–126. [Google Scholar] [CrossRef]
- Dunn, M.; Griggs, L.; Forman, J.; Horn, P. Feeding habits and niche separation among the deep-sea chimaeroid fishes Harriotta raleighana, Hydrolagus bemisi and Hydrolagus novaezealandiae. Mar. Ecol. Prog. Ser. 2010, 407, 209–225. [Google Scholar] [CrossRef] [Green Version]
- Sitjà-Bobadilla, A.; Schmidt-Posthaus, H.; Wahli, T.; Holland, J.W.; Secombes, C.J. Fish immune responses to Myxozoa. In Myxozoan Evolution, Ecology and Development; Okamura, B., Gruhl, A., Bartholomew, J., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 253–280. [Google Scholar]
- Agrawal, A. Transposition and evolution of antigen-specific immunity. Science 2000, 290, 1715–1716. [Google Scholar] [CrossRef]
- Trail, D.R.S. Behavioral interactions between parasites and hosts: Host suicide and the evolution of complex life cycles. Am. Nat. 1980, 116, 77–91. [Google Scholar] [CrossRef] [Green Version]
- Brown, S.P.; Renaud, F.; Guegan, J.F.; Thomas, F. Evolution of trophic transmission in parasites: The need to reach a mating place. J. Evol. Biol. 2001, 14, 815–820. [Google Scholar] [CrossRef]
- Parker, G.A.; Chubb, J.C.; Ball, M.A.; Roberts, G.N. Evolution of complex life cycles in helminth parasites. Nature 2003, 425, 480–484. [Google Scholar] [CrossRef]
- Kallert, D.M.; Grabner, D.S.; Yokoyama, H.; El-Matbouli, M.; Eszterbauer, E. Transmission of myxozoans to vertebrate hosts. In Myxozoan Evolution, Ecology and Development; Okamura, B., Gruhl, A., Bartholomew, J., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 235–251. [Google Scholar]
- Liyanage, Y.S.; Yokoyama, H.; Wakabayashi, H. Dynamics of experimental production of Thelohanellus hovorkai (Myxozoa: Myxosporea) in fish and oligochaete alternate hosts. J. Fish Dis. 2003, 26, 575–582. [Google Scholar] [CrossRef]
- Johnson, P.T.J.; Dobson, A.; Lafferty, K.D.; Marcogliese, D.J.; Memmott, J.; Orlofske, S.A.; Poulin, R.; Thieltges, D.W. When parasites become prey: Ecological and epidemiological significance of eating parasites. Trends Ecol. Evol. 2010, 25, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Parker, G.A.; Ball, M.A.; Chubb, J.C. Evolution of complex life cycles in trophically transmitted helminths. I. Host incorporation and trophic ascent. J. Evol. Biol. 2015, 28, 267–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaxter, M.L. Nematoda: Genes, genomes and the evolution of parasitism. Adv. Parasitol. 2003, 54, 101–195. [Google Scholar] [CrossRef] [PubMed]
- Morris, D.J. A new model for myxosporean (Myxozoa) development explains the endogenous budding phenomenon, the nature of cell within cell life stages and evolution of parasitism from a cnidarian ancestor. Int. J. Parasitol. 2012, 42, 829–840. [Google Scholar] [CrossRef] [PubMed]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lisnerová, M.; Fiala, I.; Cantatore, D.; Irigoitia, M.; Timi, J.; Pecková, H.; Bartošová-Sojková, P.; Sandoval, C.M.; Luer, C.; Morris, J.; et al. Mechanisms and Drivers for the Establishment of Life Cycle Complexity in Myxozoan Parasites. Biology 2020, 9, 10. https://doi.org/10.3390/biology9010010
Lisnerová M, Fiala I, Cantatore D, Irigoitia M, Timi J, Pecková H, Bartošová-Sojková P, Sandoval CM, Luer C, Morris J, et al. Mechanisms and Drivers for the Establishment of Life Cycle Complexity in Myxozoan Parasites. Biology. 2020; 9(1):10. https://doi.org/10.3390/biology9010010
Chicago/Turabian StyleLisnerová, Martina, Ivan Fiala, Delfina Cantatore, Manuel Irigoitia, Juan Timi, Hana Pecková, Pavla Bartošová-Sojková, Christian M. Sandoval, Carl Luer, Jack Morris, and et al. 2020. "Mechanisms and Drivers for the Establishment of Life Cycle Complexity in Myxozoan Parasites" Biology 9, no. 1: 10. https://doi.org/10.3390/biology9010010
APA StyleLisnerová, M., Fiala, I., Cantatore, D., Irigoitia, M., Timi, J., Pecková, H., Bartošová-Sojková, P., Sandoval, C. M., Luer, C., Morris, J., & Holzer, A. S. (2020). Mechanisms and Drivers for the Establishment of Life Cycle Complexity in Myxozoan Parasites. Biology, 9(1), 10. https://doi.org/10.3390/biology9010010