Transcriptional Responses of the Heat Shock Protein 20 (Hsp20) and 40 (Hsp40) Genes to Temperature Stress and Alteration of Life Cycle Stages in the Harmful Alga Scrippsiella trochoidea (Dinophyceae)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Scrippsiella trochoidea Culture Maintenance
2.2. Full-Length cDNAs Cloning of StHsp20 and StHsp40
2.3. Sequence Analysis of StHsp20 and StHsp40
2.4. Samples Collection
2.4.1. Temperature Stresses Exposure Treatments
2.4.2. Cells at Different Stages of Life Cycle
2.5. Real-Time Quantitative PCR (qPCR)
3. Results
3.1. cDNA Cloning and Sequencing Characterization of StHsp20 and StHsp40
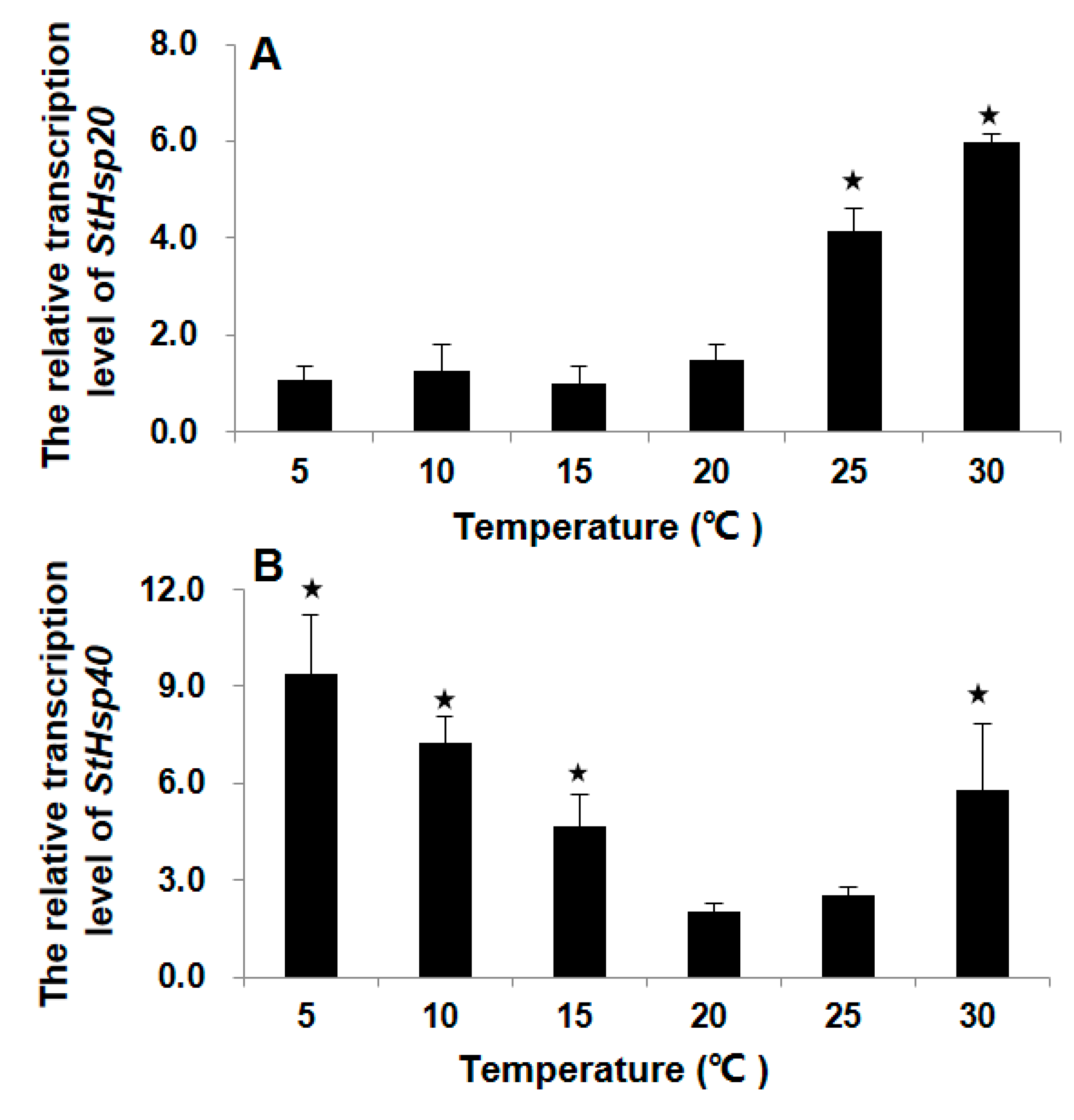
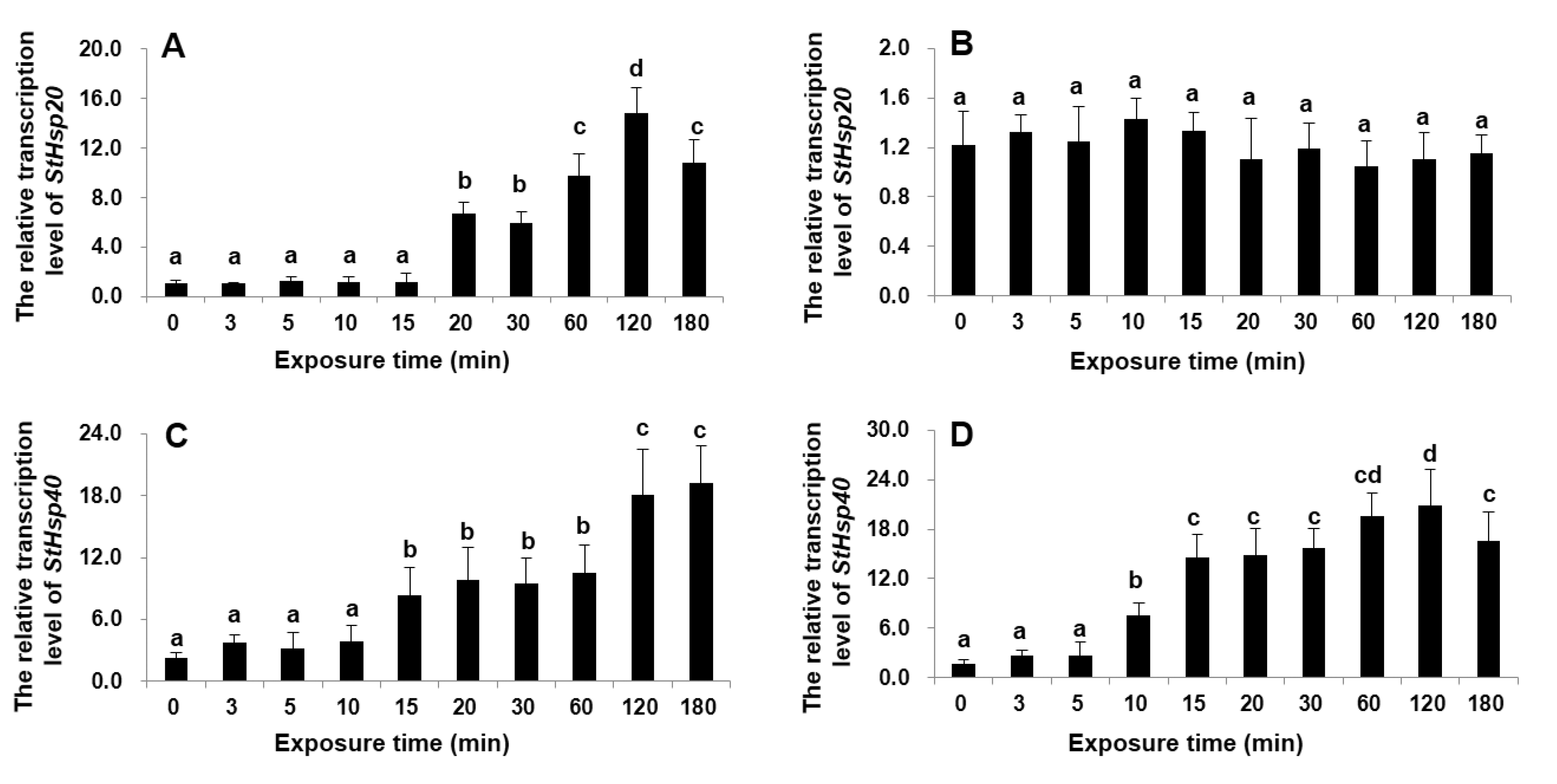
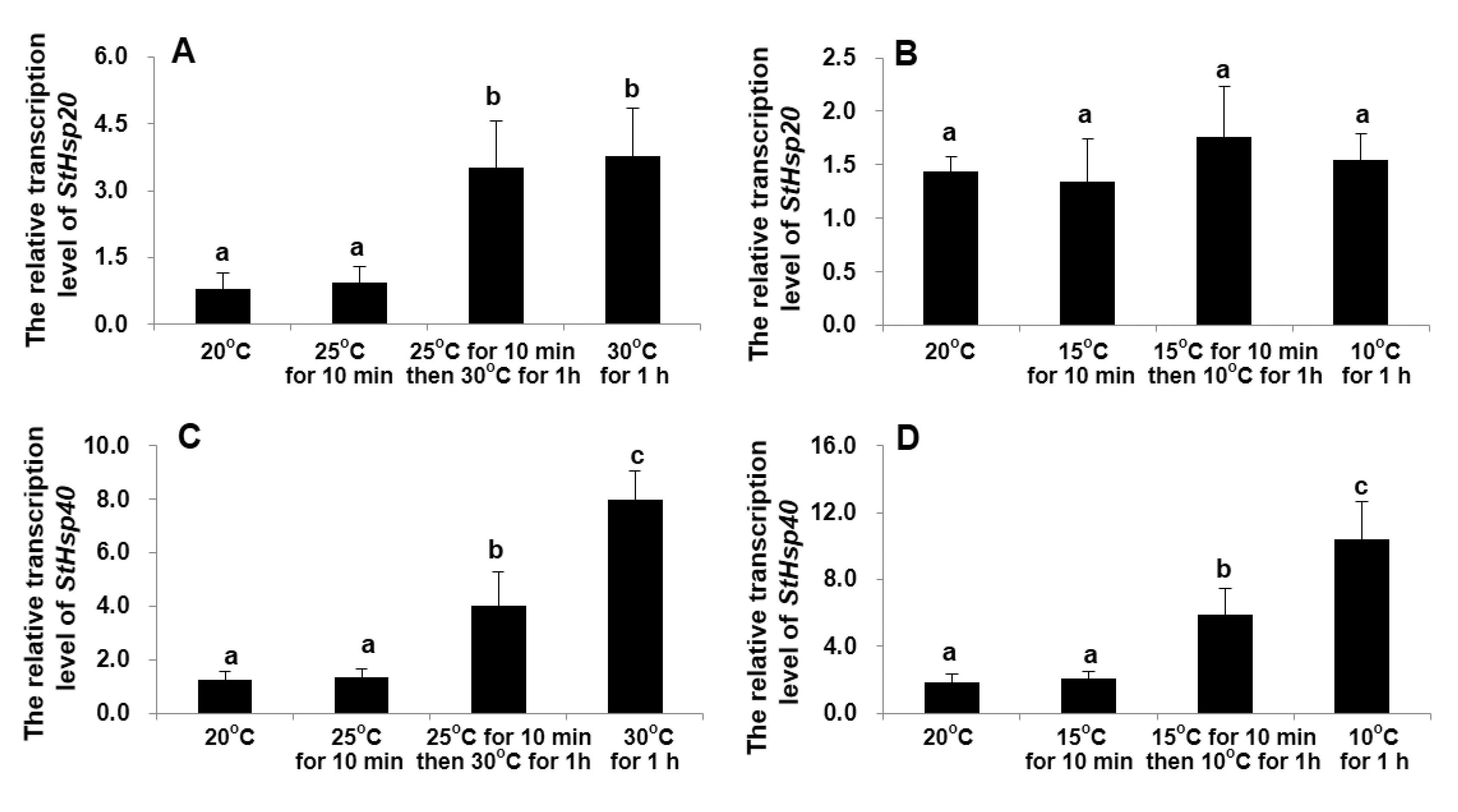
3.2. Transcriptional Responses of StHsp20 and StHsp40 to Temperature Stresses
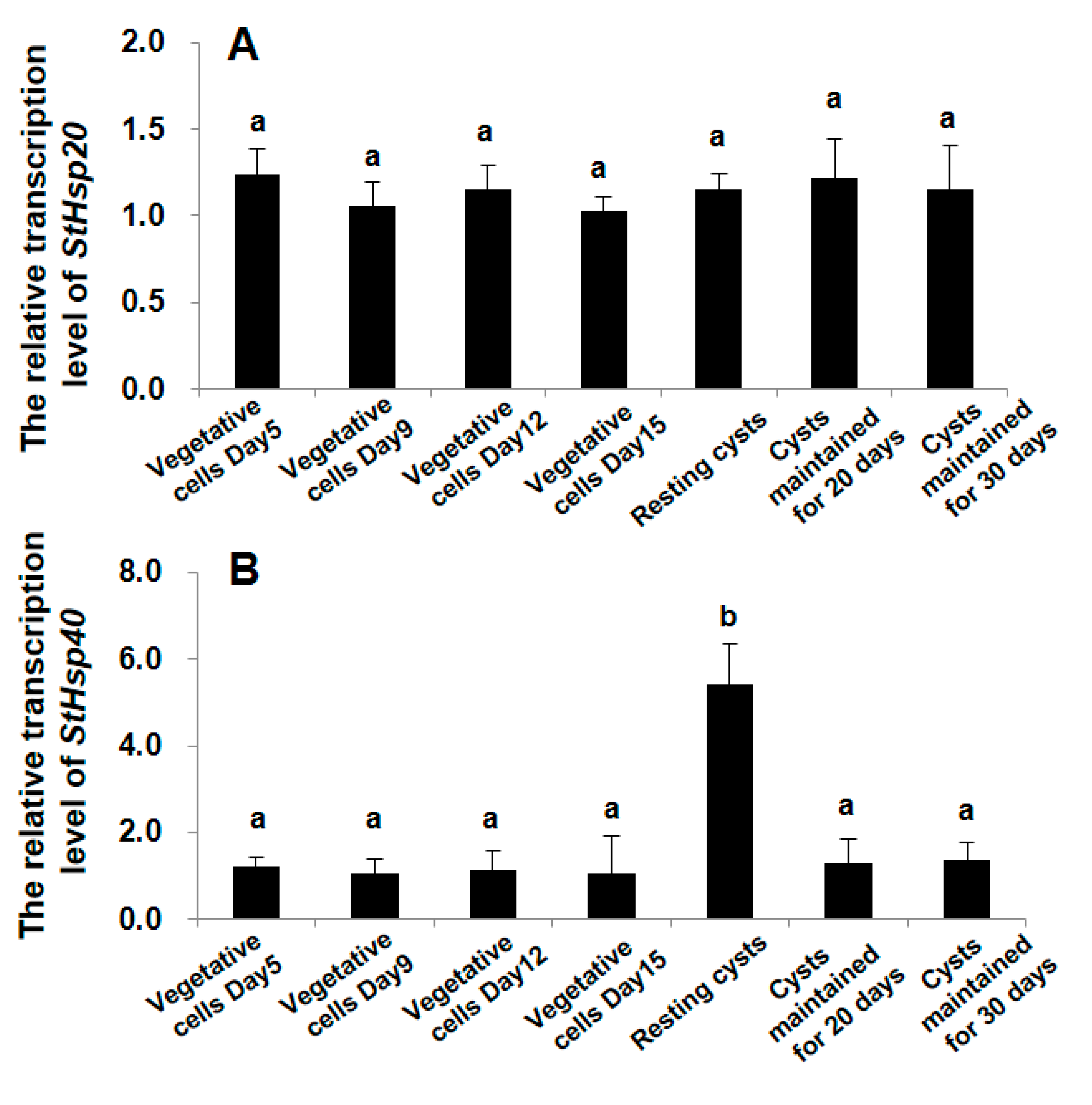
3.3. Transcriptional Responses of StHsp20 and StHsp40 at Different Stages of Growth and Life Cycle
4. Discussion
4.1. Structural Characterization of StHsp20 and StHsp40
4.2. Transcriptions of StHsp20 and StHsp40 in S. trochoidea Cells in Responding to Temperature Shocks
4.3. Differential Transcriptional Responses of StHsp20 and StHsp40 to the Alteration of Life Cycle Stages (Vegetative Cells vs. Resting Cysts)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hackett, J.D.; Anderson, D.M.; Erdner, D.L.; Bhattacharya, D. Dinoflagellates: A remarkable evolutionary experiment. Am. J. Bot. 2004, 91, 1523–1534. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Wang, F.; Zhang, W. Omics analysis for dinoflagellates biology research. Microorganisms 2019, 7, 288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smayda, T.J. Harmful algal blooms: Their ecophysiology and general relevance to blooms in phytoplankton the sea. Limnol. Oceanogr. 1997, 42, 1137–1153. [Google Scholar] [CrossRef]
- Murray, S.A.; Suggett, D.J.; Doblin, M.A.; Kohli, G.S.; Seymour, J.R.; Fabris, M.; Ralph, P.J. Unravelling the functional genetics of dinoflagellates: A review of approaches and opportunities. Perspect. Phycol. 2016, 3, 37–52. [Google Scholar] [CrossRef]
- Tang, Y.Z.; Hu, Z.X.; Deng, Y.Y. Characteristical life history (resting cyst) provides a mechanism for recurrence and geographic expansion of harmful algal blooms of dinoflagellates: A review. Stud. Mar. Sin. 2016, 51, 132–154. [Google Scholar]
- Hallegraeff, G.M.; Bolch, C.J. Transport of diatom and dinoflagellate resting spores in ships’ ballast water: Implications for plankton biogeography and aquaculture. J. Plankton. Res. 1992, 14, 1067–1084. [Google Scholar] [CrossRef]
- Nehring, S. Mechanisms for recurrent nuisance algal blooms in coastal zones: Resting cyst formation as life-strategy of dinoflagellates. In Interdisciplinary Discussion of Coastal Research and Coastal Management Issues and Problems; Lang: Frankfurt, Germany, 1993. [Google Scholar]
- Smayda, T.J. Reflections on the ballast water dispersal-harmful algal bloom paradigm. Harmful Algae 2007, 6, 601–622. [Google Scholar] [CrossRef]
- Bravo, I.; Figueroa, R.I. Towards an ecological understanding of dinoflagellate cyst functions. Microorganisms 2014, 2, 11–32. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.Y.; Hu, Z.X.; Deng, Y.Y.; Tang, Y.Z. Evidence for production of sexual resting cysts by the toxic dinoflagellate Karenia mikimotoi in clonal cultures and marine sediments. J. Phycol. 2020, 56, 121–134. [Google Scholar] [CrossRef]
- Rengefors, K.; Karlsson, I.; Hansson, L.A. Algal cyst dormancy: A temporal escape from herbivory. Proc. R. Soc. Lond. B 1998, 265, 1353–1358. [Google Scholar] [CrossRef]
- Elbrăchter, M. Dinoflagellate reproduction: Progress and conflicts. J. Phycol. 2003, 39, 629–632. [Google Scholar] [CrossRef]
- Xu, N.; Lv, S.H.; Chen, J.F.; He, L.S.; Xie, L.C.; Qi, Y.Z. The influence of water temperature and salinity on the growth of Scrippsiella trochoidea. Mar. Environ. Sci. 2004, 23, 36–38. [Google Scholar]
- Deng, G.; Li, Y.G.; Hu, H.J.; Qi, Y.Z.; Geng, Y.H.; Li, Z.K. Effects of temperature, light and pH on photosynthesis, and of light-dark cycle on growth rate and biomass of Scrippsiella trochoidea and Alexandrium tamarense. J. Wuhan Bot. Res. 2004, 22, 129–135. [Google Scholar]
- Kim, D.I.; Matsuyama, Y.; Nagasoe, S.; Yamaguchi, M.; Yoon, Y.; Oshima, Y.; Imada, N.; Honjo, T. Effects of temperature, salinity and irradiance on the growth of the harmful red tide dinoflagellate Cochlodinium polykrikoides Margalef (Dinophyceae). J. Plankton. Res. 2004, 26, 61–66. [Google Scholar] [CrossRef]
- Matsubara, T.; Nagasoe, S.; Yamasaki, Y.; Shikata, T.; Shimasaki, Y.; Oshima, Y.; Honjo, T. Effects of temperature, salinity, and irradiance on the growth of the dinoflagellate Akashiwo sanguinea. J. Exp. Mar. Biol. Ecol. 2007, 342, 226–230. [Google Scholar] [CrossRef]
- Zhou, M.J.; Yu, R.C. Mechanisms and impacts of harmful algal blooms and the count measures. Chin. J. Nat. 2007, 29, 72–77. [Google Scholar]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, T.; Safdar, W.; Abbasi, B.H.; Naqvi, S.M.S. An overview on the small heat shock proteins. Afr. J. Biotechnol. 2010, 9, 927–949. [Google Scholar]
- Waters, E.R. The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 2013, 64, 391–403. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; MacRae, T.H. Small heat shock proteins: Molecular structure and chaperone function. Cell Mol. Life Sci. 2005, 62, 2460–2476. [Google Scholar] [CrossRef]
- Franck, E.; Madsen, O.; Rheede, T.V.; Ricard, G.; Huynen, M.A.; de Jong, W.W. Evolutionary diversity of vertebrate small heat shock proteins. J. Mol. Evol. 2004, 59, 792–805. [Google Scholar] [CrossRef] [PubMed]
- Vierling, R.; Nguyen, H.T. Heat-shock gene expression in diploid wheat genotypes differing in thermal tolerance. Crop Sci. 1992, 32, 370–377. [Google Scholar] [CrossRef]
- Kim, K.K.; Kim, R.; Kim, S.H. Crystal structure of a small heat shock protein. Nature 1998, 394, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Haslbeck, M.; Kastenmuller, A.; Buchner, J.; Weinkauf, S.; Braun, N. Structural dynamics of archaeal small heat shock proteins. J. Mol. Biol. 2008, 378, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Linder, B.; Jin, Z.; Freedmans, J.H.; Rubin, C.S. Molecular characterization of a novel, developmentally regulated small embryonic chaperone from Caenorhabditis elegans. J. Biol. Chem. 1996, 271, 30158–30166. [Google Scholar] [CrossRef] [Green Version]
- Shirk, P.D.; Broza, R.; Hemphill, M.; Perera, O.P. α-Crystallin protein cognates in eggs of the moth, Plodia interpunctella: Possible chaperones for the follicular epithelium yolk protein. Insect Biochem. Mol. Biol. 1998, 28, 151–161. [Google Scholar] [CrossRef]
- Reineke, A. Identification and expression of a small heat shock protein in two lines of the endoparasitic wasp Venturia canescens. Comp. Biochem. Physiol. Part A 2005, 141, 60–69. [Google Scholar] [CrossRef]
- Huang, L.H.; Wang, C.Z.; Kang, L. Cloning and expression of five heat shock protein genes in relation to cold hardening and development in the leafminer, Liriomyza sativa. J. Insect Physiol. 2009, 55, 279–285. [Google Scholar] [CrossRef]
- Wieske, M.; Benndorf, R.; Behlke, J.; Dolling, R.; Grelle, G.; Bielka, H.; Lutsch, G. Defined sequence segments of the small heat shock proteins HSP25 and α B-crystallin inhibit actin polymerization. Eur. J. Biochem. 2001, 268, 2083–2090. [Google Scholar] [CrossRef] [Green Version]
- Quinlan, R. Cytoskeletal competence requires protein chaperones. Prog. Mol. Sub. Cell Biol. 2002, 28, 219–234. [Google Scholar]
- Wood, K.L.; Voss, O.H.; Huang, Q.; Parihar, A.; Mehta, N.; Batra, S.; Doseff, A.I. The small heat shock protein 27 is a key regulator of CD8+ CD57+ lymphocyte survival. J. Immunol. 2010, 184, 5582–5588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsvetkova, N.M.; Horváth, I.; Török, Z.; Wolkers, W.F.; Balogi, Z.; Shigapova, N.; Crowe, L.M.; Tablin, F.; Vierling, E.; Crowe, J.H.; et al. Small heat-shock proteins regulate membrane lipid polymorphism. Proc. Natl. Acad. Sci. USA 2002, 99, 13504–13509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mackay, D.S.; Andley, U.P.; Shiels, A. Cell death triggered by a novel mutation in the alphaA-crystallin gene underlies autosomal dominant cataract linked to chromosome 21q. Eur. J. Hum. Genet. 2003, 11, 784–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evgrafov, O.V.; Mersiyanova, I.; Irobi, J.; Van Den Bosch, L.; Dierick, I.; Leung, C.L.; Schagina, O.; Verpoorten, N.; Van Impe, K.; Fedotov, V.; et al. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat. Genet. 2004, 36, 602–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selcen, D.; Engel, A.G. Myofibrillar myopathy caused by novel dominant negative a B-crystallin mutations. Ann. Neurol. 2003, 54, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Yochem, J.; Uchida, H.; Sunshine, M.; Saito, H.; Georgopoulos, C.P.; Feiss, M. Genetic analysis of two genes, dnaJ and dnaK, necessary for Escherichia coli and bacteriophage lambda DNA replication. Mol. Gen. Genet. 1978, 164, 9–14. [Google Scholar] [CrossRef]
- Liberek, K.; Georgopoulos, C.; Zylicz, M. Role of the Escherichia coli DnaK and DnaJ heat shock proteins in the initiation of bacteriophage lambda DNA replication. Proc. Natl. Acad. Sci. USA 1988, 85, 6632–6636. [Google Scholar] [CrossRef] [Green Version]
- Qiu, X.B.; Shao, Y.M.; Miao, S.; Wang, L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 2006, 63, 2560–2570. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Craig, E.A. The Hsp70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010, 11, 579–592. [Google Scholar] [CrossRef] [Green Version]
- Pulido, P.; Leister, D. Novel DNA-related proteins in Arabidopsis thaliana. New Phytol. 2018, 217, 480–490. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Zhou, S.; Luo, Y.; Ma, C.; Gong, Y.; Zhou, Y.; Gao, S.; Huang, Z.; Yan, L.; Hu, Y.; et al. The heat shock protein 40 LeDnaJ regulates stress resistance and indole-3-acetic acid biosynthesis in Lentinula edodes. Fungal Genet. Biol. 2018, 118, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Walsh, P.; Bursać, D.; Law, Y.C.; Cyr, D.; Lithgow, T. The J-protein family: Modulating protein assembly, disassembly and translocation. EMBO Rep. 2004, 5, 567–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajan, V.B.; D’Silva, P. Arabidopsis thaliana J-class heat shock proteins: Cellular stress sensors. Funct. Integr. Genom. 2009, 9, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xin, M.; Qin, J.; Peng, H.; Ni, Z.; Yao, Y.; Sun, Q. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Caplan, A.J.; Tsai, J.; Casey, P.J.; Douglas, M.G. Farnesylation of YDJ1p is required for function at elevated growth temperatures in Saccharomyces cerevisiae. J. Biol. Chem. 1992, 267, 18890–18895. [Google Scholar]
- Suetsugu, N.; Kagawa, T.; Wada, M. An auxilin-like J-domain protein, JAC1, regulates phototropin-mediated chloroplast movement in Arabidopsis. Plant Physiol. 2005, 139, 151–162. [Google Scholar] [CrossRef] [Green Version]
- Shen, L.; Yu, H. The J-domain protein J3 mediates the integration of flowering signals in Arabidopsis. Plant Cell 2001, 23, 499–514. [Google Scholar] [CrossRef] [Green Version]
- Levin, R.A.; Beltran, V.H.; Hill, R.; Kjelleberg, S.; Mcdougald, D.; Steinberg, P.D.; Va Oppen, M.J. Sex, scavengers, and chaperones: Transcriptome secrets of divergent Symbiodinium thermal tolerances. Mol. Biol. Evol. 2016, 33, 2201–2215. [Google Scholar] [CrossRef] [Green Version]
- Gierz, S.L.; Foret, S.; Leggat, W. Transcriptomic analysis of thermally stressed Symbiodinium reveals differential expression of stress and metabolism genes. Front Plant Sci. 2017, 8, 271. [Google Scholar] [CrossRef]
- Lei, Q.; Lu, S. Molecular ecological responses of dinoflagellate, Karenia mikimotoi to environmental nitrate stress. Mar. Pollut. Bull. 2011, 62, 2692–2699. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, G.; Wang, Y.; Guo, C.; Zhou, J. Physiological and molecular responses of Prorocentrum donghaiense to dissolved inorganic phosphorus limitation. Mar. Pollut. Bull. 2018, 129, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, Y.; Xie, Z.; Zhang, H.; Lin, L.; Wang, D. iTRAQ-based quantitative proteomic analysis of a toxigenic dinoflagellate Alexandrium catenella and its non-toxic mutant. Proteomics 2015, 15, 4041–4050. [Google Scholar] [CrossRef] [PubMed]
- Steidinger, K.A.; Tangen, K. Dinoflagellates. In Identifying Marine Diatoms and Dinoflagellates; Academic Press: New York, NY, USA, 1996. [Google Scholar]
- Tang, Y.Z.; Gobler, C.J. Lethal effects of Northwest Atlantic Ocean isolates of the dinoflagellate, Scrippsiella trochoidea, on Eastern oyster (Crassostrea virginica) and Northern quahog (Mercenaria mercenaria) larvae. Mar. Biol. 2012, 159, 199–210. [Google Scholar] [CrossRef]
- Wang, Z.; Yu, Z.; Song, X.; Cao, X.; Zhang, Y. Effects of ammonium and nitrate on encystment and growth of Scrippsiella trochoidea. Chin. Sci. Bull. 2014, 59, 4491–4497. [Google Scholar] [CrossRef]
- Deng, Y.Y.; Hu, Z.X.; Shang, L.X.; Peng, Q.C.; Tang, Y.Z. Transcriptomic analyses of Scrippsiella trochoidea reveals processes regulating encystment and dormancy in the life cycle of a dinoflagellate, with a particular attention to the role of abscisic acid. Front. Microbiol. 2017, 8, 2450. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.Y.; Hu, Z.X.; Chai, Z.Y.; Tang, Y.Z. Molecular cloning of heat shock protein 60 (Hsp60) and 10 (Hsp10) genes from the cosmopolitan and harmful dinoflagellate Scrippsiella trochoidea and their differential transcriptions responding to temperature stress and alteration of life cycle. Mar. Biol. 2019, 166, 7. [Google Scholar] [CrossRef]
- Deng, Y.Y.; Hu, Z.X.; Chai, Z.Y.; Tang, Y.Z. Cloning and comparative studies of proliferating cell nuclear antigen (PCNA) genes for nine dinoflagellates. J. Appl. Phycol. 2019, 31, 2969–2979. [Google Scholar] [CrossRef]
- Deng, Y.Y.; Hu, Z.X.; Chai, Z.Y.; Tang, Y.Z. Cloning and partial characterization of a cold shock domain-containing protein gene from the dinoflagellate Scrippsiella trochoidea. J. Eukaryot. Microbiol. 2019, 66, 393–403. [Google Scholar] [CrossRef]
- Guillard, R.R.L. Culture of phytoplankton for feeding marine invertebrates. In Culture of Marine Invertebrate Animals; Springer: Boston, MA, USA, 1975. [Google Scholar]
- Doblin, M.A.; Blackburn, S.I.; Hallegraeff, G.M. Growth and biomass stimulation of the toxic dinoflagellate Gymnodinium catenatum (Graham) by dissolved organic substances. J. Exp. Mar. Biol. Ecol. 1999, 236, 33–47. [Google Scholar] [CrossRef]
- Deng, Y.Y.; Hu, Z.X.; Zhan, Z.F.; Ma, Z.P.; Tang, Y.Z. Differential expressions of an Hsp70 gene in the dinoflagellate Akashiwo sanguinea in response to temperature stress and transition of life cycle and its implications. Harmful Alage 2015, 50, 57–64. [Google Scholar] [CrossRef]
- Zhang, H.; Hou, Y.; Miranda, L.; Campbell, D.A.; Sturm, N.R.; Gaasterland, T.; Lin, S. Spliced leader RNA trans-splicing in dinoflagellates. Proc. Natl. Acad. Sci. USA 2007, 104, 4618–4623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S. Genomic understanding of dinoflagellates. Res. Microbiol. 2011, 162, 551–569. [Google Scholar] [CrossRef] [PubMed]
- Rombel, I.T.; Sykes, K.F.; Rayner, S.; Johnston, S.A. ORF-Finder: A vector for high-throughput gene identification. Gene 2002, 282, 33–41. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.H.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. The Proteomics Protocols Handbook; Humana Press: New Jersey, NJ, USA, 2005. [Google Scholar]
- Geourjon, C.; Deleage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 1995, 11, 681–684. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; Von Heijne, B.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [Green Version]
- Petersen, T.N.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Pfaffl, M. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 2002–2007. [Google Scholar] [CrossRef]
- Radonic, A.; Thulke, S.; Mackay, I.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Bioph. Res. Commun. 2004, 313, 856–862. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Zakrajsek, B.A.; Mills, A.G.; Gorn, V.; Singer, M.J.; Reed, M.W. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: Comparison of endpoint and real-time methods. Anal. Biochem. 2000, 285, 194–204. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basha, E.; Friedrich, K.L.; Vierling, E. The N-terminal arm of small heat shock proteins is important for both chaperone activity and substrate specificity. J. Biol. Chem. 2006, 281, 39943–39952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sørensen, J.G.; Kristensen, T.N.; Loeschcke, V. The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 2003, 6, 1025–1037. [Google Scholar] [CrossRef]
- Feder, M.E.; Hofmann, G.E. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.; Guo, R.; Ki, J. Different transcriptional responses of heat shock protein 20 in the marine diatom Ditylum brightwellii exposed to metals and endocrine-disrupting chemicals. Environ. Toxicol. 2014, 29, 1379–1389. [Google Scholar] [CrossRef] [PubMed]
- Lei, Q.N.; Wu, Y.Y.; Liang, H.Y.; Wang, Z.X.; Zheng, Z.; Deng, Y.W. Molecular cloning and expression analysis of heat shock protein 20 (HSP20) from the pearl oyster Pinctada martensii. Genet. Mol. Res. 2016, 15, gmr.15028799. [Google Scholar] [CrossRef] [PubMed]
- Rhee, J.-S.; Raisuddin, S.; Lee, K.-W.; Seo, J.-S.; Ki, J.-S.; Kim, I.-C.; Park, H.G.; Lee, J.-S. Heat shock protein (Hsp) gene responses of the intertidal copepod Tigriopus japonicus to environmental toxicants. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2009, 149, 104–112. [Google Scholar] [CrossRef]
- Rhee, J.S.; Kim, R.O.; Choi, H.G.; Lee, J.; Lee, Y.M.; Lee, J.S. Molecular and biochemical modulation of heat shock protein 20 (Hsp20) gene by temperature stress and hydrogen peroxide (H2O2) in the monogonont rotifer, Brachionus sp. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2011, 154, 19–27. [Google Scholar] [CrossRef]
- Shen, Y.; Gu, J.; Huang, L.; Zheng, S.; Liu, L.; Xu, W.; Feng, Q.; Kang, L. Cloning and expression analysis of six small heat shock protein genes in the common cutworm, Spodoptera litura. J. Insect Physiol. 2011, 57, 908–914. [Google Scholar] [CrossRef]
- Leadbeater, B.S.; Karpov, S.A. Cyst formation in a freshwater strain of the choanoflagellate Desmarella moniliformis Kent. J. Eukaryot. Microbiol. 2000, 47, 433–439. [Google Scholar] [CrossRef]
- Kristensen, T.N.; Dahlgaard, J.; Loeschcke, V. Inbreeding affects Hsp70 expression in two species of Drosophila even at benign temperatures. Evol. Ecol. Res. 2002, 4, 1209–1216. [Google Scholar]
- Pedersen, K.S.; Kristensen, T.N.; Loeschcke, V. Effects of inbreeding and rate of inbreeding in Drosophila melanogaster-Hsp70 expression and fitness. J. Evol. Biol. 2005, 18, 756–762. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, J.C.; King, V.; Tower, J. Sequence requirements for upregulating expression of Drosophila hsp70 transgenes during aging. Neurobiol. Aging 1999, 20, 545–553. [Google Scholar] [CrossRef]
- Trotter, E.W.; Kao, C.M.F.; Berenfeld, L.; Botstein, D.; Petsko, G.A.; Gray, J.V. Misfolded proteins are competent to mediate a subset of the responses to heat shock in Saccharomyces cerevisiae. J. Biol. Chem. 2002, 277, 44817–44825. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.; Wang, J.; Levichkin, I.V.; Stasinopoulos, S.; Ryan, M.T.; Hoogenraad, N.J. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002, 21, 4411–4419. [Google Scholar] [CrossRef]
- Giebel, L.B.; Dworniczak, B.P.; Bautz, E.K.F. Developmental regulation of a constitutively expressed mouse mRNA encoding a 72-kDa heat shock-like protein. Dev. Boil. 1988, 125, 200–207. [Google Scholar] [CrossRef]
- Gunter, H.M.; Degnan, B.M. Developmental expression of Hsp90, Hsp70 and HSF during morphogenesis in the vetigastropod Haliotis asinina. Dev. Genes Evol. 2007, 217, 603–612. [Google Scholar] [CrossRef]
- Cadman, C.S.; Toorop, P.E.; Hilhorst, H.W.; Finch-Savage, W.E. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J. 2006, 46, 805–822. [Google Scholar] [CrossRef]
- Chibani, K.; Ali-Rachedi, S.; Job, C.; Job, D.; Jullien, M.; Grappin, P. Proteomic analysis of seed dormancy in Arabidopsis. Plant Physiol. 2006, 142, 1493–1510. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.; Zhang, H.; Zhuang, Y.; Tran, B.; Gill, J. Spliced leader-based metatranscriptomic analyses lead to recognition of hidden genomic features in dinoflagellates. Proc. Natl. Acad. Sci. USA 2010, 107, 20033–20038. [Google Scholar] [CrossRef] [Green Version]
- Wisecaver, J.H.; Hackett, J.D. Dinoflagellate genome evolution. Annu. Rev. Microbiol. 2011, 65, 369–387. [Google Scholar] [CrossRef] [PubMed]


| Primer Name | Nucleotide Sequences (5′→3′) | Amplicon Length |
|---|---|---|
| 40-F | GGCGAGGAAGACTTCTATCAGG | 587 bp |
| 40-R | TGCTGCGTCTGCATCACG | |
| 5r-40-outer | CAATCTGCTGGAACTTGTCCTGCG | 496 bp |
| 5r-40-inner | GCGGACTTGATTTCCCTGTCGTTG | 416 bp |
| 3r-40-outer | AGTGGACCTCCCCTTCAAAACCG | 830 bp |
| 3r-40-inner | CCACCCCTCTGGGCGTGATGC | 670 bp |
| 20-F | CCCTTCTTCGCCTTCAACC | 216 bp |
| 20-R | TTCTGTGGAGCGCTTGG | |
| 5r-20-outer | GCTTCAAGGTGCCGTTCTCCA | 417 bp |
| 5r-20-inner | CGTGGCGGCGGGACTTGG | 306 bp |
| 3r-20-outer | CGACATCATTGAAAGAGACGACGCC | 750 bp |
| 3r-20-inner | TTCACTCGAGGTGGAGAACGGCA | 680 bp |
| q40-F | GCATCAAGTGCGAGGGTAGG | 124 bp |
| q40-R | TGCTGCGTCTGCATCACG | |
| q20-F | TGGAGAACGGCACCTTGAA | 183 bp |
| q20-R | TGGGCAGAGTGATGGTGAGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, Y.; Hu, Z.; Shang, L.; Chai, Z.; Tang, Y.Z. Transcriptional Responses of the Heat Shock Protein 20 (Hsp20) and 40 (Hsp40) Genes to Temperature Stress and Alteration of Life Cycle Stages in the Harmful Alga Scrippsiella trochoidea (Dinophyceae). Biology 2020, 9, 408. https://doi.org/10.3390/biology9110408
Deng Y, Hu Z, Shang L, Chai Z, Tang YZ. Transcriptional Responses of the Heat Shock Protein 20 (Hsp20) and 40 (Hsp40) Genes to Temperature Stress and Alteration of Life Cycle Stages in the Harmful Alga Scrippsiella trochoidea (Dinophyceae). Biology. 2020; 9(11):408. https://doi.org/10.3390/biology9110408
Chicago/Turabian StyleDeng, Yunyan, Zhangxi Hu, Lixia Shang, Zhaoyang Chai, and Ying Zhong Tang. 2020. "Transcriptional Responses of the Heat Shock Protein 20 (Hsp20) and 40 (Hsp40) Genes to Temperature Stress and Alteration of Life Cycle Stages in the Harmful Alga Scrippsiella trochoidea (Dinophyceae)" Biology 9, no. 11: 408. https://doi.org/10.3390/biology9110408
APA StyleDeng, Y., Hu, Z., Shang, L., Chai, Z., & Tang, Y. Z. (2020). Transcriptional Responses of the Heat Shock Protein 20 (Hsp20) and 40 (Hsp40) Genes to Temperature Stress and Alteration of Life Cycle Stages in the Harmful Alga Scrippsiella trochoidea (Dinophyceae). Biology, 9(11), 408. https://doi.org/10.3390/biology9110408






