1. Introduction
From the moment of birth, human skin is colonized by numerous microorganisms, which live in community and form the skin microbiota, unique to each individual [
1]. It is now well accepted that the bacterial community on the skin is an integral part of the innate immune system, capable of modulating specific localized inflammatory responses [
2] and contributing to the stability of the skin barrier [
3].
Sequencing-based profiling studies have targeted specific areas of skin, such as the forehead [
4], forearms [
5], or armpits [
6]. The results revealed that the composition of the skin microbiota is primarily determined by the physiology of the skin area, depending on whether it corresponds to a sebaceous, dry, or moist skin microenvironment [
7]. The profiles reported also demonstrated a very high level of inter-individual variability, making it difficult to define what constitutes the core microbiota of healthy skin, in terms of composition, and to what extent that composition can fluctuate.
Beyond this strong inter-individual variability, healthy adults have been shown to stably maintain their skin microbiota for up to two years [
8]. Nevertheless, the skin microbiota is permanently exposed to multiple external factors related to lifestyle, pollution, medication, or hygiene products that may affect the skin microbiota’s equilibrium, and even induce dysbiosis, defined as alteration of the composition and functionality of the skin microbiota [
9].
Skin care products are used daily, and cosmetic formulations usually include several compounds (e.g., preservatives, antibacterial or antiseptic agents, emollient and moisturizing agents, perfumes, surfactants) that could affect the skin microbiota. As suggested by Holland and Bojar [
10], the regular topical use of specific compounds, such as antibacterial agents, may affect the skin’s bacterial ecology. This possibility justifies monitoring of potential changes to the microbiota. In parallel, although cosmetic products are not expected to be sterile, cosmetics manufacturing practices ensure that microorganism numbers remain below the regulatory threshold. Nevertheless, it is quite unlikely that a microorganism present in a product could stably establish and extensively modify the skin microbiota.
In the medical field, the effectiveness of skin disinfection has long been a concern, with the imperative to prevent bacterial infections at surgical sites and the underlying risk of increasing antibiotic resistance. Studies have assessed the efficacy of topical biocides, such as antibiotics [
11], triclosan [
12], benzalkonium chloride or 70% ethanol-based sanitizers [
13], or chlorhexidine gluconate [
14,
15,
16]. In contrast, in the cosmetic field, we lack data on how cosmetic formulas affect the skin microbiota [
17]. Although the impact of deodorants or antiperspirants on axillary flora and of cleansers has been documented [
18,
19], little information is available on the effects of emollients, perfumes, or active ingredients.
The rise of new sequencing technologies and their accessibility have prompted molecular profiling-based assessments of cutaneous bacterial communities to evaluate the impact of cosmetics [
20,
21,
22]. These molecular profiling studies, conducted as part of clinical studies, provide detailed information on compositional changes to the microbiota. However, double-blind, placebo-controlled clinical evaluation is not always possible. First, the launching and processing costs of clinical studies are high, since they require the inclusion of large cohorts. Second, these studies are time-consuming, with topical treatments usually being applied over several weeks. Finally, to facilitate the topical applications and maintain the target cosmetic ingredient intact throughout the duration of the investigation, it must be included in a formulation. The ingredients in this formulation may themselves affect the microbiota and interfere with results.
Therefore, in complement to molecular profiling approaches, faster methods are needed to evaluate the intrinsic impact of a broader spectrum of non-formulated ingredients and to assess how they affect viability of the skin microbiota in the short-term. In this study, we developed a methodology to measure the impact of topical applications on cutaneous bacterial density (i.e., number of bacteria cultivable in a defined medium per unit of skin surface area in a specific zone). Several longitudinal studies with volunteers were undertaken, comparing the effects of application of two reference solutions: physiological saline and chlorhexidine. A scoring method was developed to quantitatively assess the impact of topical applications on skin microbiota viability.
2. Materials and Methods
2.1. Study Design
Four iterative studies were sequentially undertaken from March 2021 to February 2022 at the Givaudan Applied Microbiomics Centre of Excellence (Toulouse, France), and two validation studies were outsourced to BIO-EC (Longjumeau, France) (
Table 1).
A total of 12 healthy volunteers, men and women, ranging in age from 19 to 62 years, were recruited for the in-house studies. A subset of these volunteers participated in several of the reported studies. Two groups of 12 healthy volunteers, men and women, ranging in age from 35 to 65 years, were recruited by BIO-EC in July and December 2022, for the fifth and the sixth studies, respectively.
2.2. Topical Skin Applications and Samplings
For each topical application, a single 5-mL dose of 0.9% sodium chloride (Laboratoires Mercurochrome®, Laboratoires Juva Santé, Paris, France), referred to hereafter as physiological saline, or 0.5% chlorhexidine di-gluconate (Cooper, Melun, France), referred to as chlorhexidine, was used to saturate the 5 × 5-cm folded surface of a 20 × 20-cm sterile gauze pad. The solution was applied by gently dabbing the saturated gauze on a 50-cm² delimited area of skin on the neck, right or left side, with pressure but without rubbing, for 15–20 s. After application, the skin was allowed to air-dry.
For studies involving a single application (studies 1 to 3), the solution was applied just after the initial sample was collected D0 t0 (
Table 1). When six applications were scheduled (studies 4 to 6), the solution was repeatedly applied over 3 days, with two applications per day. In the morning, solutions were applied just after the respective samples were collected (D0 t0, D1 and D2). For the afternoon time points, solutions were applied about 5 h after the morning applications.
After the initial sample was collected (D0 t0), and the solution applied, samples were taken after 3 or 5 h (D0 t3h or D0 t5h), then after 1 day (D1), 2 days (D2), 3 days (D3), or 7 days (D7).
2.3. Sample Collection and Bacterial Counting
Skin bacteria were collected from 50-cm² areas on volunteers’ neck (both right and left) by a non-invasive swabbing method, using sterile cotton-tipped swabs moistened with a sterile solution of 0.9% sodium chloride. Swabs were processed within a maximum of one hour for bacterial counting. The tip of each swab was detached with a sterile surgical blade and transferred to a 1.5-mL tube containing 750 µL of 0.9% sterile sodium chloride solution. The sample biomass was recovered by stirring and pipetting before transferring the biological suspension to a new microtube, taking special care to squeeze and wring out the swab tip. A final volume of 740 μL was reproducibly recovered from each swab sample.
For studies 1 to 4, samples were plated on Tryptic Soy Agar (Becton, Dickinson and Company 7 Loveton Circle P.O. Box 999 Sparks, MD, USA) by an automatic spiral-plater (EasySpiral PRO, Interscience, Saint Nom, France). For each biological suspension, a 1/10 dilution in sterile 0.9% sodium chloride was produced, of which 100 µL was spiral-plated. After the agar had solidified, Petri dishes were inverted and incubated at 37 °C for 24 h. Colony forming units (CFUs) were then counted using an automatic colony counter (Scan 1200, Interscience, Saint Nom, France), the number of CFU·mL−1 was finally deduced taking the dilution factors into account.
For studies 5 and 6, bacteria were counted by a Pour Plate Method. From each bacterial suspension, three serial 10-fold dilutions (1/10, 1/100, 1/1000) were produced with tryptone salt diluent (bioMérieux, Marcy l’Etoile, France). An aliquot (100 µL) of each dilution was placed on the bottom of a 90-mm diameter Petri dish, producing duplicates for each dilution point. Petri dishes were then filled with 15–20 mL of 45 °C tempered TSA medium. The diluted suspensions and TSA were thoroughly mixed by swirling. After incubation, the number of CFUs was counted manually, averaging the values for the two replicates to determine the number of CFU·mL
−1 for each suspension. Only plates with 11–300 CFUs were considered in these calculations. Volunteers for whom an average initial CFU of less than 11 colonies were obtained on the 1/10 dilution, were eliminated from the analysis. The bacterial density of the skin, expressed in CFU per skin surface area (cm
2), referred to hereafter as the skin bacterial density, was calculated as follows:
2.4. Statistical Analysis
The statistical and graphical representations were produced with the R software version 4.2.2 (31 October 2022). As the bacterial density is not a systematically normally distributed variable, and given the size of the volunteer cohorts, statistical analyses were carried out using non-parametric methods. The Wilcoxon two-sample paired signed rank test was used for the comparison of the two paired samples. For the comparison of more than two paired groups, Friedman’s non-parametric tests were performed.
3. Results
3.1. Short-Time Impact of Swabbing
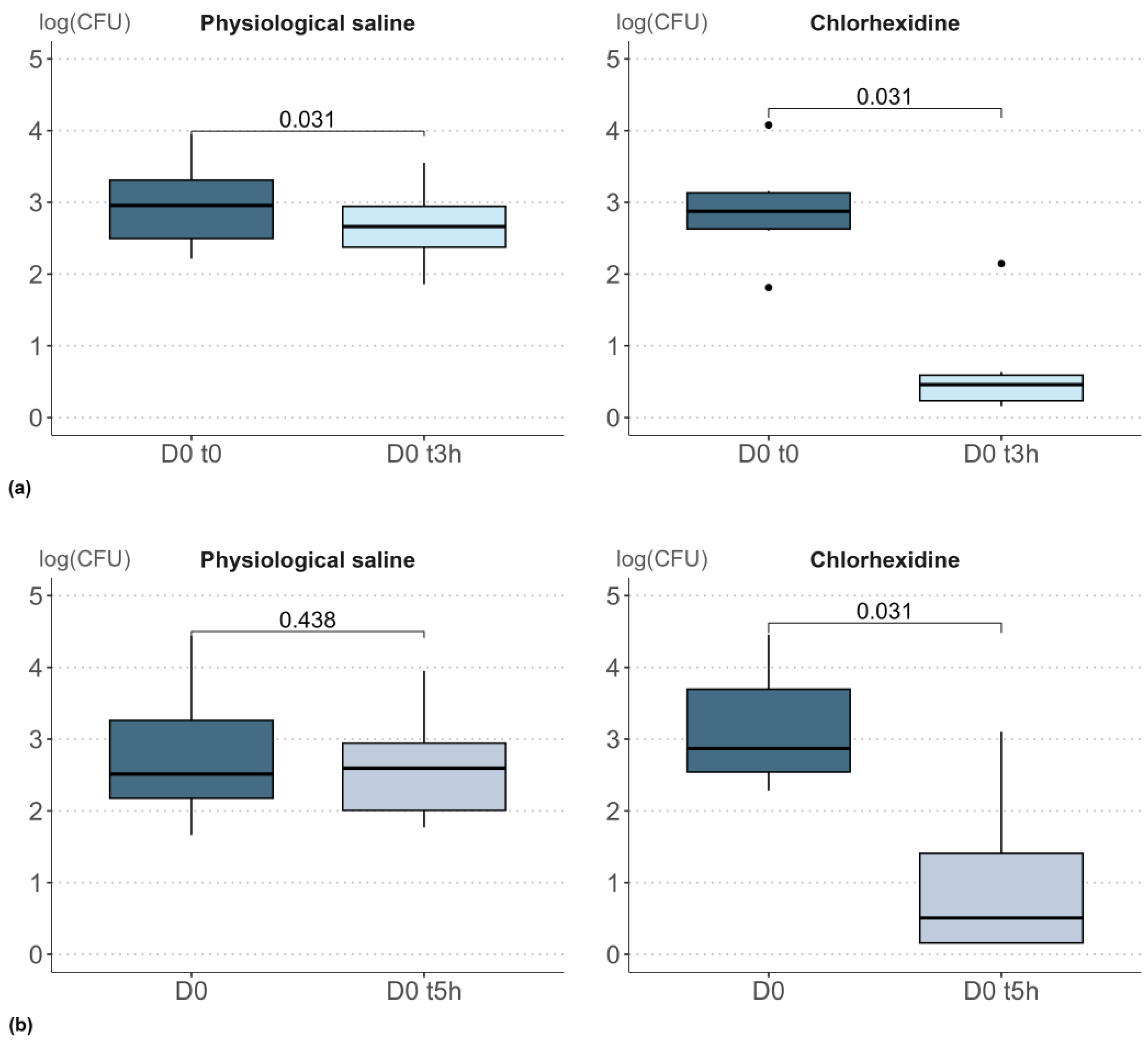
The studies 1 and 2 were carried out to determine how repeat skin sampling in the very short-term, 3 h or 5 h after the initial sampling, affected the bacterial density measured. The results from these studies are presented in
supplementary Table S1. A two-tailed Wilcoxon signed-rank test (α 0.05) was applied to compare the bacterial densities at the initial sampling time to those measured 3 or 5 h later (
Figure 1).
When samples were collected 3 h after a single application of physiological solution, a significant decrease in bacterial density was observed (p = 0.031). The impact of physiological saline, although significant, was very small, with a decrease of just 0.3 log units. A similar decrease between D0 t0 and D0 t3h was observed in all volunteers and reflects the initial removal of biomass. Conversely, when samples were collected 5 h after application of the physiological solution, the decrease of 0.2 log units was not significant (p = 0.438). Therefore, the removal of biomass was no longer detectable in the repeat sample taken 5 h after the initial sample.
Following a single application of chlorhexidine, a stronger impact was observed. Both 3 h and 5 h after the initial sampling, a decrease in bacterial count was observed, with an order of magnitude of −2.2. Based on these outcomes, a 5-h time-lapse was selected for subsequent studies (studies 3 and 4) to prevent any short-term impact of biomass removal.
3.2. Consistency of Skin-Borne Bacterial Density Counts
The pre-application bacterial densities measured on the left and right sides of the neck in studies 1 to 4 are listed in
Supplementary Table S2. The raw data reveal a wide-ranging variation in bacterial density in the neck area, from a minimum value of 46 CFU/cm
2 to a maximum value of 28 × 10
3 CFU/cm
2. The median value was 3 × 10
3 CFU/cm
2. This 3-log magnitude of variation reflects inter-individual variability, and contrasts with the apparent intra-individual consistency between the left and right sides of the neck. A two-tailed Wilcoxon signed-rank test (α 0.05) confirmed that any differences between right and left sides of the neck for the same individual were not statistically significant (
p = 0.310).
Four panelists were enrolled in the March 2021 (study 1), April 2021 (study 2), and February 2022 (study 4) studies. The bacterial densities measured at the initial sampling time (D0 t0) in the three studies were compared by applying a Friedman’s rank sum test (α 0.05). No significant difference was observed over time (p = 0.197). This stability over time of inter-individual differences, and the consistency of densities measured on the left and right sides of the neck, confirmed the measurement of bacterial density to be a reliable metric for longitudinal monitoring of skin bacterial viability.
3.3. Definition of Outlier’s Exclusion Criteria
Considering the wide variability in the bacterial densities measured, a strategic concern was to determine which of the extreme data could be considered acceptable natural variability, and which suggested experimental artifacts and should consequently be excluded.
Analysis of the raw data for the initial sampling time (D0 t0) revealed that some discrepancies between the left and right sides (laterality effect) existed. Although the differences were not significant, a high lateralization deviation (ratio of highest density to lowest density) was observed with three samples (
Table S2). For these samples, left-right deviations of 9.3 (GT055, study 3), 8.3 (GT061, study 3), and 7.1 (GT055, study 4) were recorded. Based on these values, an exclusion threshold of 6.0 was empirically defined for lateralization deviation. The samples from these three volunteers were excluded from further analysis since their lateralization deviation ratio exceeded the exclusion threshold.
The bacterial densities and D0 t5h/D0 t0 ratio for studies 3 and 4 are presented in
Table S3. Application of physiological saline led to a decrease in bacterial density between D0 t0 and D0 t5h, with log values of 0.7 (study 3) and 0.1 (study 4). Application of chlorhexidine produced a stronger decrease, with log values of 2.0 (study 3) and 1.6 (study 4).
Although the [D0 t5h/D0 t0] ratio never exceeded 1.3 after applying physiological saline, an unexpected increase of 2.7 was recorded with samples from volunteer GT055 (study 4). To ensure reliability of results, a second exclusion threshold of 2.5 was empirically defined for the [D0 t5h/D0 t0] ratio. Consequently, a [D0 t5h/D0 t0] ratio exceeding 2.5 should be flagged as an experimental error, since no biological explanation can be found for such a dramatic increase in bacterial density in such a short time-frame, especially when the biomass subtraction effect is considered. Volunteer GT055 was therefore excluded from the dataset for future analyses.
The raw data corresponding to the bacterial densities recorded in longitudinal studies 3 and 4 are presented in
supplementary Tables S4 and S5. Based on our previously-defined exclusion criteria, all samples from volunteers GT055 and GT061 (study 3) and GT055 (study 4) were excluded from the final analysis since their lateralization deviation values and [D0 t5h/D0 t0] ratios exceeded the exclusion thresholds of 6 and 2.5, respectively.
3.4. Impact of a Repeated Topical Applications
The raw data corresponding to the bacterial densities recorded in longitudinal studies 3 and 4 are presented in
Supplementary Tables S4 and S5. Based on our previously-defined exclusion criteria, all samples from volunteers GT055 and GT061 (study 3) and GT055 (study 4) were excluded from the final analysis since their lateralization deviation values and [D0 t5h/D0 t0] ratios exceeded the exclusion thresholds of 6 and 2.5, respectively.
The median bacterial density values (CFU/cm²), expressed in log-scale, for the different sampling time points, are reported in
Table 2, and the distributions of bacterial density values over time are presented in
Figure 2.
As previously observed for study 2, bacterial densities remained relatively stable 5 h after application of physiological saline, with a decrease of just 0.3 log. In contrast, a dramatic decrease in bacterial density was consistently observed 5 h after application of chlorhexidine (study 3), resulting in a maximal drop of 2.6 log. Median bacterial densities recovered slowly on subsequent samples (
Table 2). Analysis of variance based on a Friedman’s rank sum test (α 0.05) confirmed that a single topical application of physiological saline did not significantly alter bacterial density (
p = 0.726). In contrast, the reduction in bacterial density induced by chlorhexidine was significant (
p = 0.001).
When physiological saline was applied twice-daily for three days (study 4), no significant evolution of bacterial density was observed (Friedman’s
p-value = 0.156). The median bacterial density, measured in CFU/cm² and expressed on a log-scale, remained stable and was not affected by the regular sampling (
Table 2). Conversely, repeated applications of chlorhexidine over three days had a very strong and highly significant impact on bacterial density (Friedman’s
p-value = 0.008). A maximal drop of 2.2 log was observed after 5 h, and the recovery of bacterial density was strongly perturbed for the first three days. A median value of 3.1 log (CFU/cm²), close to the median value at D0 t0 (3.4 log), was restored after 7 days (
Figure 2b).
3.5. External Evaluation of the Method
Two studies involving larger groups of volunteers were outsourced to BIO-EC in July 2022 (study 5) and December 2022 (study 6). The raw data for the bacterial densities measured during longitudinal study 5 are presented in
Supplementary Table S6. Based on our previously-defined exclusion criteria, samples from volunteer 12 were excluded from the dataset since the lateralization deviation of 26.2 and [D0 t5h/D0 t0] ratio of 17.2 considerably exceeded the exclusion thresholds of 6 and 2.5, respectively.
The distributions of bacterial density values over time are presented in
Figure 3. As previously observed (study 4,
Figure 2b), the bacterial densities were only mildly affected by repeated applications of physiological saline, with a non-significant Friedman’s rank sum test (α 0.05,
p-value = 0.948). In contrast, the detrimental effect of topical application of the chlorhexidine on bacterial density was significant (
p-value = 0.020).
The raw bacterial density data from longitudinal study 6 are presented in
Supplementary Table S7. Based on our previously-defined exclusion criteria, the samples from volunteer 3 were excluded from the dataset due to a lateralization deviation of 16.3, which considerably exceeded the exclusion threshold of 6. The distributions of bacterial density over time are presented in
Figure 4.
As previously observed, repeated applications of chlorhexidine over three days produced a highly significant decrease of bacterial density (Friedman’s p-value = 3.7 × 10−5).
3.6. Description of the Scoring Process
To qualify and score the intensity of the impact of ingredient application, we selected three reference time points from our longitudinal follow-up:
D0 t5h reflects a very short-term disturbance, after only 5 h, that we refer to as a flash disturbance (FD);
D3 provides information on the cumulative and postponed impact of repeat topical applications over 3 days, we refer to this parameter as cumulative disturbance (CD);
D7 is a measure of the situation after 7 days, i.e., 5 days after discontinuing applications, by which time bacterial density had potentially recovered to the initial value, we refer to this parameter as the resilience pattern (R).
For each reference time point, individual bacterial densities were expressed relative to the initial bacterial density (D0 t0):
FD ratio: bacterial density at D0 t5h/ bacterial density at D0 t0,
CD ratio: bacterial density at D3/ bacterial density at D0 t0),
R ratio: bacterial density at D7/ bacterial density at D0 t0.
As the bacterial density was not systematically normally distributed, the mean metric was considered irrelevant, and the median values of the ratio were retained.
A partial score was attributed to each of the three parameters mentioned above –FD, CD, and R—based on the following principle (
Figure 5):
Comparison of the bacterial densities at D0 t0, D0 t5h, D3, and D7. A Friedman’s rank sum test (α 0.1) was applied to determine whether repeated application had a significant effect.
A maximum of 10 points was awarded for the FD parameter if topical application of the ingredient tested had no significant impact. If a significant impact was observed, between 0 and 10 points were added, based on the median value at 5 h.
Similarly, a maximum of 10 points (between 0 and 10 points) were awarded for the CD parameter, depending on whether the topical application had a significant effect after 3 days.
Finally, a maximum of 5 points (0 to 5) were attributed to the R parameter, depending on whether the topical application produced a significant effect.
A cumulative score was then calculated by adding the points attributed for the three parameters FD, CD, and R. The final score was thus within a range from 0 to 25, which we split into five viability score values: A (20 to 25 points), B (16 to 20 points), C (11 to 15 points), D (6 to 10 points), and E (0 to 5 points).
3.7. Establishing the Chlorhexidine and Physiological Saline Viability Score
A viability score was first estimated for the results from study 5, conducted in July 2022. The distributions of bacterial densities at D0 t0, D0 t5h, D3, and D7, after repeated applications of physiological saline, were compared. According to Friedman’s rank sum test (α 0.1), the variations observed were not significant (p = 0.948). The maximum number of points—10 (FD), 10 (CD), and 5 (R)—were therefore assigned by default, since no significant effect was detected. This maximum value of 25 corresponds to a viability score of A for physiological saline.
In contrast, the bacterial density was significantly affected by the repeated application of chlorhexidine (
p = 0.020). The median bacterial density ratios relative to the density measured for D0 t0 for the three selected sampling time points are reported in
Table 3.
A strong effect was observed after 5 h and after 3 days, with median values of 0.129 and 0.293 for D0 t5h/D0 t0 and D3/D0 t0, respectively. Based on these median values, the FD, CD, and R scores attributed were 1, 2, and 2, respectively. The cumulative score was therefore 5, which corresponds to the lowest viability score—E.
The viability score for chlorhexidine was estimated once again, based on the results from study 6, conducted in December 2022. Repeated application of chlorhexidine led to a strong and significant decrease in bacterial density, according to the Friedman’s rank sum test analysis (p = 3.7 × 10−5). Based on median bacterial densities of 0.044, 0.066, and 0.586, the FD, CD, and R parameters were all awarded null scores. The cumulative score was therefore 0, which was lower than for study 5, but once again corresponded to a viability score of E.
Applying the scoring process to data from studies 3 and 4, corresponding to either a single (study 3) or repeated (study 4) chlorhexidine applications, also returned a viability score of E, based on cumulative point values of 2 and 1, respectively.
4. Discussion
The main objective of this study was to establish the reliability of a swabbing and bacterial counting approach to assess how topical application of an ingredient affected viability of the skin microbiota.
Although the facial area, particularly the cheeks, is frequently targeted in clinical skin studies, we chose to study skin on the neck area for several reasons. Firstly, preliminary in-house studies indicated that the neck area shares similarities with the cheek area, both in terms of bacterial density and bacterial diversity (data not shown). Secondly, the neck area is substantially less exposed to confounding factors (UV exposure, skin care creams, make-up, uncontrolled hand touches). Thirdly, as it is situated below the beard, sampling from the neck area also facilitates swabbing for male volunteers. Finally, given the proximity between the cheek area and the eyes, application of chlorhexidine to the neck considerably limits the risk of ocular projections during application.
Since swabbing the skin intrinsically leads to removal of biological material from its surface, the bacterial count could be strongly perturbed, or even skewed, by repeated swabbing in a short time-frame. Our preliminary experiments established that re-swabbing within 5 h did not significantly hinder the estimation of bacterial density, despite the prior removal of biomass. This result suggests that the initial swabbing does not exhaust the skin’s bacterial reservoir. Rather, it only removes some of the bacterial cells. However, when swabbing was repeated in the shorter 3-h time-frame, a significant decrease in bacterial density was recorded. It can therefore be assumed that bacterial repopulation processes are engaged rapidly after the initial sampling. A longer lapse, of 5 h, appears to be preferable, limiting the short-term impact of biomass removal.
Given the many perturbations to which the skin is exposed on a daily basis, a second concern related to the robustness of skin bacterial density as a biological metric. The order of magnitude of the median bacterial density at D0 t0 (3 × 10
3 CFU/cm
2) was consistent with reported values [
23]. However, a very broad inter-individual variation was observed, spanning 3 log. The extent of this inter-individual variability was in line with differences in the composition of skin bacterial communities reported elsewhere [
24]. In addition, both the consistency of the left and right bacterial densities and the stability of the bacterial densities over time suggested that this inter-individual variability was not artifactual, being maintained over time. Indeed, the intra-individual stability of the bacterial density over several months was remarkable, reflecting the stability of the skin microbiota [
8]. The skin bacterial density should therefore be considered a reliable cutaneous feature.
As the swabbing-counting approach was validated as a reliable measure of bacterial density on the skin, the subsequent step was to establish how responsive this metric was to topical skin treatments. To this end, results following application of two compounds were compared. Physiological saline was used as a negative control; it was expected not to have any impact on skin bacterial density. The local antiseptic, chlorhexidine, was used as a positive control; it was expected to reduce bacterial density.
Across all the studies described in this paper, physiological saline was confirmed to be a completely innocuous topical ingredient, having no impact on the skin’s bacterial density. Conversely, also as expected, topical application of chlorhexidine systematically led to a reduction in bacterial density. This effect was observed even after a single application of chlorhexidine. This result is fully in line with previous findings [
15], where an impact of chlorhexidine on bacterial numbers was recorded after 3 h. In contrast, using an alternative approach based on 16S rRNA sequencing analysis of skin bacterial communities, Wiemken et al. [
25] reported that a single topical application of chlorhexidine to the calf area had no detectable influence on the composition of the skin’s microbiome composition. Similarly, Mougeot et al. [
26] concluded that a single topical application of chlorhexidine to the dorsal surface of the forearm did not significantly alter the composition of the bacterial community. Several hypotheses can be put forward to explain this apparent discrepancy. First, the impact of chlorhexidine on the relative abundances of bacterial taxa could go unnoticed in sequencing studies because chlorhexidine is a broad-spectrum antimicrobial agent potentially exerting an equal effect on all members of the bacterial community. Second, a single application of chlorhexidine should not be disruptive enough to alter the structure of a healthy, stable skin microbiota. Third, since the sequencing approach relies on extracted nucleic acids, it cannot distinguish between live and dead bacteria or free DNA, and consequently it may not be suitable for detecting short-term alterations of the skin microbiota. As a method that exclusively focuses on active bacteria, and their ability to grow in culture, the colony-counting approach appears to be particularly suitable when studying how topical applications affect bacterial viability in the short-term.
In our study, the impact of multiple topical applications was assessed by examining three time points in the longitudinal follow-up. These observations were used to establish the viability score.
The very short-term impact, 5 h after the first topical application, was referred to as Flash Disturbance (FD) and reflects an immediate deleterious effect that could be assimilated to a bactericidal effect. Topical application of chlorhexidine clearly produced a 2-log drop in the bacterial density measured, fully in line with its broad-spectrum antimicrobial properties. The strong impact confirmed the prior observation that topical application of a 0.5% chlorhexidine solution induced a strong and significant decrease in bacterial numbers. The effect was similar at 0 h (just after application and drying) or at 3 h [
15]. It should be emphasized that Macias et al. employed a sampling strategy that involved separately evaluating bacterial densities after 0, 3, or 24 h on three distinct cohorts of volunteers; our longitudinal evaluation involved a single cohort.
A second time point was then examined, 3 days after the first topical application. The effect observed was referred to as cumulative disturbance (CD). This time point reflects both instantaneous toxicity of the applied ingredient and/or any perturbation of bacterial recolonization following the topical application(s). After a single topical application of chlorhexidine (study 3), the initial bacterial density was progressively restored, with a return to bacterial counts close to those recorded at D0 t0 (
Figure 2). A similar recolonization process after a single application of chlorhexidine was reported elsewhere [
16], where it was termed post-antisepsis bacterial regrowth. This bacterial regrowth supports the two assumptions that a single chlorhexidine treatment does not fully eradicate the skin microbiota, and that swabbing does not exhaustively remove the whole bacterial population. When chlorhexidine was applied twice-daily for three days (study 4), bacterial regrowth was still observed, but the return to the initial bacterial density was clearly slowed by the repeated topical applications. As a reflection of the recolonization process after 3 days, the CD should detect the impact of bactericidal ingredients, in complement to the information provided by the FD.
A third and last time point, 7 days after the first topical application, was considered and referred to as “resilience”. Resilience has been defined as the capacity of an ecosystem to maintain its state and recover from perturbations [
27]. Overall resilience can be considered as a combination of the capacity of the system to persist during an impact (resistance) and to return to baseline after a perturbation (recovery) [
27]. Here, resilience was determined with respect to the initial bacterial density, taking two situations into account, no effect of the ingredient applied on bacterial density (resistance), and effect of the ingredient but recovery of initial bacterial density after discontinuing applications (recovery).
Our scoring process to quantify the impact of topical application of an ingredient on skin bacterial density is based on analysis and rating of three parameters related to: short-term impact, cumulative impact, and the ability of the skin microbiota to maintain or recover its bacterial density. A partial score is first obtained by awarding 0 to 10 points to the FD and 0 to 10 points to the CD. As resilience encompasses both resistance to perturbation, which is already addressed through FD and CD, and the recovery capacity, this parameter was only awarded 0 to 5 points. The sum of the points awarded for the three parameters gives a cumulative score, between 0 (highly perturbing ingredient that strongly alters the skin’s bacterial density and affects resilience of the skin’s microbiota) and 25 (neutral ingredient that has no adverse effects on the skin’s bacterial density). To make it easily understood, this cumulative score was equally distributed across five viability score categories, associated with five letters: A (21 to 25 points), B (16 to 20 points), C (11 to 15 points), D (6 to 10 points), and E (0 to 5 points).
The viability score is not intended to indicate whether the ingredient is intrinsically good or bad, but to establish, under given conditions of concentration and topical application, how it affects skin bacterial density in the short-term. Several certification approaches have been proposed over the past few years within the cosmetic industry to claim that cosmetic ingredients are safe for skin microbiota or can even improve or restore a healthy skin microbiome. A “Microbiome Friendly” certification was recently proposed by the German company MyMicrobiome. This certification relies on in vitro tests, including mono-culture and co-culture of targeted bacterial strains, with cultures treated or not with the ingredients to be evaluated. This type of approach can provide valuable information on the interaction between an ingredient and specific target bacteria, representative of skin environments. However, the growth conditions in planktonic bacterial cultures are characterized by high nutrient availability, high growth rates, high bacterial densities, and low complexity of artificial bacterial co-cultures. These in vitro growth conditions markedly contrast with the harsher conditions prevailing on the skin’s surface. Under these conditions, any attempt to extrapolate in vitro test results rapidly reaches its limits, and any related claims may be regarded as weakly supported.
As an alternative to these in vitro strategies, sequencing technologies have emerged. Their rise has considerably expanded the scientific community’s knowledge of skin microbial communities [
1]. Whether based on the 16S rRNA gene (small subunit of ribosomal RNA) or the whole genome (WGS), sequencing can be used to describe the bacterial composition of the microbiota, and any changes it undergoes. The differences lie in the level of taxonomic precision—genus level for 16S sequencing or species level for WGS. Private contracting companies now propose sequencing services to objectively classify cosmetic ingredients as “Microbiome Friendly”. However, longitudinal studies require sufficiently large cohorts of volunteers to absorb the high inter-individual variability, and the target ingredient or vehicle formula must be applied to a specific area of skin for several weeks, making these studies both time-consuming and expensive. Although this very powerful approach can detect how a topically applied ingredient affects the composition of the skin microbiota, it cannot distinguish between live, dead, or quiescent bacteria and free DNA, and does not seem suitable for use to detect short-term alterations to the skin microbiota’s composition.