Accurate Transmission-Less Attenuation Correction Method for Amyloid-β Brain PET Using Deep Neural Network
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dataset
2.2. Network Architecture
2.3. Training and Validation
2.4. Image Analysis
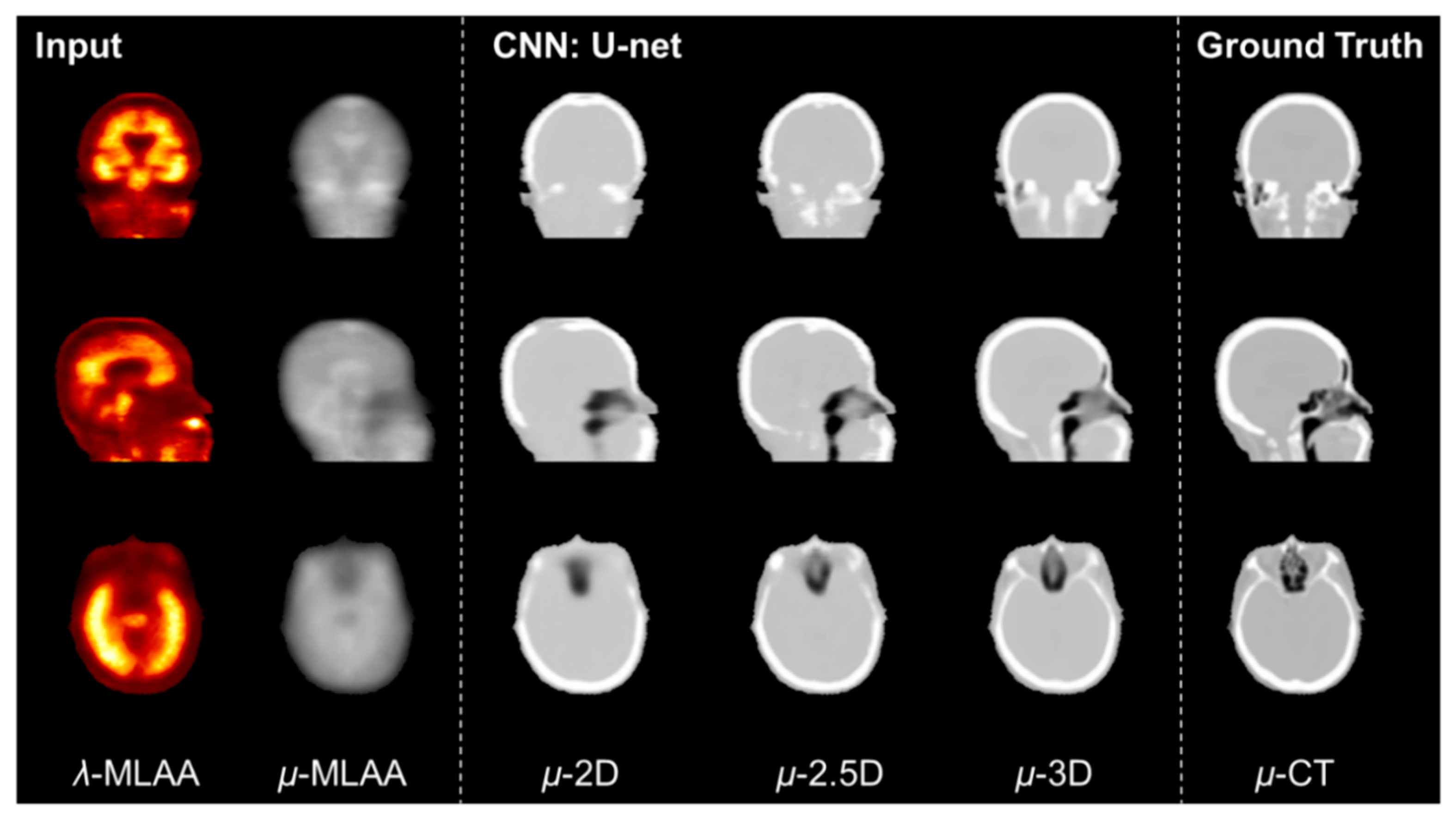
2.5. Comparison between 2D, 2.5D, and 3D U-Net
2.6. Comparison between Single-Image Input and Multi-Input
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PET | Positron emission tomography |
| CT | Computed tomography |
| MRI | Magnetic resonance imaging |
| AC | Attenuation correction |
| μ-map | Attenuation map |
| Aβ | Amyloid-β |
| OSEM | Ordered-subset expectation maximisation |
| μ-CT | CT-derived attenuation map |
| MLAA | Maximum likelihood reconstruction of activity and attenuation |
| λ-MLAA | Activity maps obtained from the MLAA |
| μ-MLAA | Attenuation maps obtained from the MLAA |
| ReLU | Rectified linear units |
| μ-CNN | U-net output |
| NRMSE | Normalized root mean square error |
| PSNR | Peak signal-to-noise ratio |
| SUV | Standard uptake value |
| SUVr | SUV ratio relative to the reference region |
| ROI | Region of interest |
| GAN | Generative adversarial network |
References
- Cherry, S.R.; Dahlbom, M.; Phelps, M.E. PET: Physics, Instrumentation, and Scanners. In PET; Springer: New York, NY, USA, 2004; pp. 1–124. [Google Scholar]
- Bailey, D.L. Transmission scanning in emission tomography. Eur. J. Nucl. Med. Mol. Imaging 1998, 25, 774–787. [Google Scholar] [CrossRef]
- Zaidi, H.; Hasegawa, B. Determination of the attenuation map in emission tomography. J. Nucl. Med. 2003, 44, 291–315. [Google Scholar]
- Kinahan, P.; Hasegawa, B.H.; Beyer, T. X-ray-based attenuation correction for positron emission tomography/computed tomography scanners. Semin. Nucl. Med. 2003, 33, 166–179. [Google Scholar] [CrossRef]
- Townsend, D.W. Dual-Modality Imaging: Combining Anatomy and Function. J. Nucl. Med. 2008, 49, 938–955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, Y.Y.; Lee, J.S.; Yang, S.-O. Musculoskeletal Lesions: Nuclear Medicine Imaging Pitfalls. In Pitfalls in Musculoskeletal Radiology; Springer Science and Business Media LLC: New York, NY, USA, 2017; pp. 951–976. [Google Scholar]
- Goerres, G.W.; Ziegler, S.I.; Burger, C.; Berthold, T.; Von Schulthess, G.K.; Buck, A. Artifacts at PET and PET/CT Caused by Metallic Hip Prosthetic Material. Radiology 2003, 226, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Kamel, E.M.; Burger, C.; Buck, A.; Von Schulthess, G.K.; Goerres, G.W. Impact of metallic dental implants on CT-based attenuation correction in a combined PET/CT scanner. Eur. Radiol. 2003, 13, 724–728. [Google Scholar] [CrossRef]
- Lodge, M.A.; Mhlanga, J.C.; Cho, S.Y.; Wahl, R.L. Effect of Patient Arm Motion in Whole-Body PET/CT. J. Nucl. Med. 2011, 52, 1891–1897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mawlawi, O.; Erasmus, J.J.; Pan, T.; Cody, D.D.; Campbell, R.; Lonn, A.H.; Kohlmyer, S.; Macapinlac, H.A.; Podoloff, D.A. Truncation Artifact on PET/CT: Impact on Measurements of Activity Concentration and Assessment of a Correction Algorithm. Am. J. Roentgenol. 2006, 186, 1458–1467. [Google Scholar] [CrossRef]
- Keereman, V.; Mollet, P.; Berker, Y.; Schulz, V.; Vandenberghe, S. Challenges and current methods for attenuation correction in PET/MR. Magma Magn. Reson. Mater. Phys. Biol. Med. 2012, 26, 81–98. [Google Scholar] [CrossRef]
- Vandenberghe, S.; Marsden, P.K. PET-MRI: A review of challenges and solutions in the development of integrated multimodality imaging. Phys. Med. Biol. 2015, 60, R115–R154. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.J.; Lee, J.S.; Lee, J.M. Integrated whole body MR/PET: Where are we? Korean J. Radiol. 2015, 16, 32–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; An, H. Attenuation Correction of PET/MR Imaging. Magn. Reson. Imaging Clin. N. Am. 2017, 25, 245–255. [Google Scholar] [CrossRef] [Green Version]
- An, H.J.; Seo, S.; Kang, H.; Choi, H.; Cheon, G.J.; Kim, H.-J.; Lee, D.S.; Song, I.-C.; Kim, Y.K.; Lee, J.S. MRI-Based Attenuation Correction for PET/MRI Using Multiphase Level-Set Method. J. Nucl. Med. 2016, 57, 587–593. [Google Scholar] [CrossRef] [Green Version]
- Catana, C.; Van Der Kouwe, A.; Benner, T.; Michel, C.J.; Hamm, M.; Fenchel, M.; Fischl, B.; Rosen, B.; Schmand, M.; Sorensen, A.G. Toward Implementing an MRI-Based PET Attenuation-Correction Method for Neurologic Studies on the MR-PET Brain Prototype. J. Nucl. Med. 2010, 51, 1431–1438. [Google Scholar] [CrossRef] [Green Version]
- Delso, G.; Wiesinger, F.; Sacolick, L.I.; Kaushik, S.S.; Shanbhag, D.D.; Hüllner, M.; Veit-Haibach, P. Clinical Evaluation of Zero-Echo-Time MR Imaging for the Segmentation of the Skull. J. Nucl. Med. 2015, 56, 417–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keereman, V.; Fierens, Y.; Broux, T.; De Deene, Y.; Lonneux, M.; Vandenberghe, S. MRI-Based Attenuation Correction for PET/MRI Using Ultrashort Echo Time Sequences. J. Nucl. Med. 2010, 51, 812–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montandon, M.-L.; Zaidi, H. Atlas-guided non-uniform attenuation correction in cerebral 3D PET imaging. NeuroImage 2005, 25, 278–286. [Google Scholar] [CrossRef]
- Yang, J.; Wiesinger, F.; Kaushik, S.; Shanbhag, D.; Hope, T.A.; Larson, P.E.Z.; Seo, Y. Evaluation of Sinus/Edge-Corrected Zero-Echo-Time-Based Attenuation Correction in Brain PET/MRI. J. Nucl. Med. 2017, 58, 1873–1879. [Google Scholar] [CrossRef] [Green Version]
- Hofmann, M.; Bezrukov, I.; Mantlik, F.; Aschoff, P.; Steinke, F.; Beyer, T.; Pichler, B.J.; Schölkopf, B. MRI-Based Attenuation Correction for Whole-Body PET/MRI: Quantitative Evaluation of Segmentation- and Atlas-Based Methods. J. Nucl. Med. 2011, 52, 1392–1399. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H.; Lee, J.S.; Song, I.-C.; Lee, D.S. Comparison of Segmentation-Based Attenuation Correction Methods for PET/MRI: Evaluation of Bone and Liver Standardized Uptake Value with Oncologic PET/CT Data. J. Nucl. Med. 2012, 53, 1878–1882. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Möller, A.; Souvatzoglou, M.; Delso, G.; Bundschuh, R.A.; Chefd’Hotel, C.; Ziegler, S.I.; Navab, N.; Schwaiger, M.; Nekolla, S.G. Tissue Classification as a Potential Approach for Attenuation Correction in Whole-Body PET/MRI: Evaluation with PET/CT Data. J. Nucl. Med. 2009, 50, 520–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.S. A Review of Deep-Learning-Based Approaches for Attenuation Correction in Positron Emission Tomography. IEEE Trans. Radiat. Plasma Med. Sci. 2021, 5, 160–184. [Google Scholar] [CrossRef]
- Jang, H.; Liu, F.; Zhao, G.; Bradshaw, T.; McMillan, A.B. Technical Note: Deep learning based MRAC using rapid ultrashort echo time imaging. Med. Phys. 2018, 45, 3697–3704. [Google Scholar] [CrossRef] [PubMed]
- Ladefoged, C.N.; Marner, L.; Hindsholm, A.; Law, I.; Højgaard, L.; Andersen, F.L. Deep Learning Based Attenuation Correction of PET/MRI in Pediatric Brain Tumor Patients: Evaluation in a Clinical Setting. Front. Neurosci. 2019, 12, 1005. [Google Scholar] [CrossRef] [PubMed]
- Leynes, A.P.; Yang, J.; Wiesinger, F.; Kaushik, S.S.; Shanbhag, D.D.; Seo, Y.; Hope, T.A.; Larson, P.E.Z. Zero-Echo-Time and Dixon Deep Pseudo-CT (ZeDD CT): Direct Generation of Pseudo-CT Images for Pelvic PET/MRI Attenuation Correction Using Deep Convolutional Neural Networks with Multiparametric MRI. J. Nucl. Med. 2017, 59, 852–858. [Google Scholar] [CrossRef] [PubMed]
- Torrado-Carvajal, A.; Vera-Olmos, J.; Izquierdo-Garcia, D.; Catalano, O.A.; Morales, M.A.; Margolin, J.; Soricelli, A.; Salvatore, M.; Malpica, N.; Catana, C. Dixon-VIBE Deep Learning (DIVIDE) Pseudo-CT Synthesis for Pelvis PET/MR Attenuation Correction. J. Nucl. Med. 2019, 60, 429–435. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, A.M.; Tashima, H.; Yoshida, E.; Nishikido, F.; Yamaya, T. Simulation study comparing the helmet-chin PET with a cylindrical PET of the same number of detectors. Phys. Med. Biol. 2017, 62, 4541–4550. [Google Scholar] [CrossRef]
- Gonzalez, A.J.; Sanchez, F.; Benlloch, J.M. Organ-Dedicated Molecular Imaging Systems. IEEE Trans. Radiat. Plasma Med. Sci. 2018, 2, 388–403. [Google Scholar] [CrossRef]
- Majewski, S.R.; Proffitt, J.; Brefczynski-Lewis, J.; Stolin, A.; Weisenberger, A.; Xi, W.; Wojcik, R. HelmetPET: A silicon photomultiplier based wearable brain imager. In Proceedings of the 2011 IEEE Nuclear Science Symposium Conference Record, Valencia, Spain, 23–29 October 2012; pp. 4030–4034. [Google Scholar] [CrossRef]
- Yamamoto, S.; Honda, M.; Oohashi, T.; Shimizu, K.; Senda, M. Development of a Brain PET System, PET-Hat: A Wearable PET System for Brain Research. IEEE Trans. Nucl. Sci. 2011, 58, 668–673. [Google Scholar] [CrossRef]
- Bergström, M.; Litton, J.; Eriksson, L.; Bohm, C.; Blomqvist, G. Determination of Object Contour from Projections for Attenuation Correction in Cranial Positron Emission Tomography. J. Comput. Assist. Tomogr. 1982, 6, 365–372. [Google Scholar] [CrossRef]
- Kops, E.R.; Herzog, H. Alternative methods for attenuation correction for PET images in MR-PET scanners. In Proceedings of the 2007 IEEE Nuclear Science Symposium Conference Record, Honolulu, HI, USA, 26 October–3 November 2007; Volume 6, pp. 4327–4330. [Google Scholar] [CrossRef]
- Sekine, T.; Buck, A.; Delso, G.; ter Voert, E.; Huellner, M.; Veit-Haibach, P.; Warnock, G. Evaluation of Atlas-Based Attenuation Correction for Integrated PET/MR in Human Brain: Application of a Head Atlas and Comparison to True CT-Based Attenuation Correction. J. Nucl. Med. 2016, 57, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Hooper, P.K.; Meikle, S.; Eberl, S.; Fulham, M.J. Validation of postinjection transmission measurements for attenuation correction in neurological FDG-PET studies. J. Nucl. Med. 1996, 37, 128–136. [Google Scholar]
- Kaneko, K.; Kuwabara, Y.; Sasaki, M.; Koga, H.; Abe, K.; Baba, S.; Hayashi, K.; Honda, H. Validation of quantitative accuracy of the post-injection transmission-based and transmissionless attenuation correction techniques in neurological FDG-PET. Nucl. Med. Commun. 2004, 25, 1095–1102. [Google Scholar] [CrossRef]
- Dong, X.; Lei, Y.; Wang, T.; Higgins, K.; Liu, T.; Curran, W.J.; Mao, H.; Nye, J.A.; Yang, X. Deep learning-based attenuation correction in the absence of structural information for whole-body positron emission tomography imaging. Phys. Med. Biol. 2020, 65, 055011. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Wang, T.; Lei, Y.; Higgins, K.; Liu, T.; Curran, W.J.; Mao, H.; A Nye, J.; Yang, X. Synthetic CT generation from non-attenuation corrected PET images for whole-body PET imaging. Phys. Med. Biol. 2019, 64, 215016. [Google Scholar] [CrossRef] [PubMed]
- Hwang, D.; Kang, S.K.; Kim, K.Y.; Seo, S.; Paeng, J.C.; Lee, D.S.; Lee, J.S. Generation of PET Attenuation Map for Whole-Body Time-of-Flight 18F-FDG PET/MRI Using a Deep Neural Network Trained with Simultaneously Reconstructed Activity and Attenuation Maps. J. Nucl. Med. 2019, 60, 1183–1189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, D.; Kim, K.Y.; Kang, S.K.; Seo, S.; Paeng, J.C.; Lee, D.S.; Lee, J.S. Improving the Accuracy of Simultaneously Reconstructed Activity and Attenuation Maps Using Deep Learning. J. Nucl. Med. 2018, 59, 1624–1629. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Jang, H.; Kijowski, R.; Zhao, G.; Bradshaw, T.; McMillan, A.B. A deep learning approach for 18F-FDG PET attenuation correction. EJNMMI Phys. 2018, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Onofrey, J.A.; Revilla, E.M.; Toyonaga, T.; Menard, D.; Ankrah, J.; Carson, R.E.; Liu, C.; Lu, Y. A Novel Loss Function Incorporating Imaging Acquisition Physics for PET Attenuation Map Generation Using Deep Learning. In Transactions on Petri Nets and Other Models of Concurrency XV; Springer Science and Business Media LLC: New York, NY, USA, 2019; pp. 723–731. [Google Scholar]
- Shiri, I.; Ghafarian, P.; Geramifar, P.; Leung, K.H.-Y.; Oghli, M.G.; Oveisi, M.; Rahmim, A.; Ay, M.R. Direct attenuation correction of brain PET images using only emission data via a deep convolutional encoder-decoder (Deep-DAC). Eur. Radiol. 2019, 29, 6867–6879. [Google Scholar] [CrossRef]
- Su, Y.; Rubin, B.B.; McConathy, J.; Laforest, R.; Qi, J.; Sharma, A.; Priatna, A.; Benzinger, T.L. Impact of MR-Based Attenuation Correction on Neurologic PET Studies. J. Nucl. Med. 2016, 57, 913–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, K.; Han, P.K.; Johnson, K.A.; El Fakhri, G.; Ma, C.; Li, Q. Attenuation correction using deep Learning and integrated UTE/multi-echo Dixon sequence: Evaluation in amyloid and tau PET imaging. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 1351–1361. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Han, Y.; Ye, J.C. Framing U-Net via Deep Convolutional Framelets: Application to Sparse-View CT. IEEE Trans. Med Imaging 2018, 37, 1418–1429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hegazy, M.A.A.; Cho, M.H.; Cho, M.H.; Lee, S.Y. U-net based metal segmentation on projection domain for metal artifact reduction in dental CT. Biomed. Eng. Lett. 2019, 9, 375–385. [Google Scholar] [CrossRef]
- Lee, M.S.; Hwang, D.; Kim, J.H.; Lee, J.S. Deep-dose: A voxel dose estimation method using deep convolutional neural network for personalized internal dosimetry. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Bae, S.; Seo, S.; Park, S.; Bang, J.-I.; Han, J.H.; Lee, W.W.; Lee, J.S. Measurement of Glomerular Filtration Rate using Quantitative SPECT/CT and Deep-learning-based Kidney Segmentation. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.; Hwang, D.; Kim, K.Y.; Kang, S.K.; Kim, Y.K.; Lee, J.S. Computed tomography super-resolution using deep convolutional neural network. Phys. Med. Biol. 2018, 63, 145011. [Google Scholar] [CrossRef]
- Sevastopolsky, A. Optic disc and cup segmentation methods for glaucoma detection with modification of U-Net convolutional neural network. Pattern Recognit. Image Anal. 2017, 27, 618–624. [Google Scholar] [CrossRef] [Green Version]
- Yie, S.Y.; Kang, S.K.; Hwang, D.; Lee, J.S. Self-supervised PET Denoising. Nucl. Med. Mol. Imaging 2020, 54, 299–304. [Google Scholar] [CrossRef]
- Kang, S.K.; Shin, S.A.; Seo, S.; Byun, M.S.; Lee, D.Y.; Kim, Y.K.; Lee, D.S.; Lee, J.S. Deep learning-Based 3D inpainting of brain MR images. Sci. Rep. 2021, 11, 1–11. [Google Scholar] [CrossRef]
- Aasheim, L.B.; Karlberg, A.; Goa, P.E.; Håberg, A.; Sørhaug, S.; Fagerli, U.-M.; Eikenes, L. PET/MR brain imaging: Evaluation of clinical UTE-based attenuation correction. Eur. J. Nucl. Med. Mol. Imaging 2015, 42, 1439–1446. [Google Scholar] [CrossRef] [PubMed]
- Defrise, M.; Rezaei, A.; Nuyts, J. Time-of-flight PET data determine the attenuation sinogram up to a constant. Phys. Med. Biol. 2012, 57, 885–899. [Google Scholar] [CrossRef]
- Rezaei, A.; Defrise, M.; Bal, G.; Michel, C.; Conti, M.; Watson, C.; Nuyts, J. Simultaneous Reconstruction of Activity and Attenuation in Time-of-Flight PET. IEEE Trans. Med. Imaging 2012, 31, 2224–2233. [Google Scholar] [CrossRef]
- Salomon, A.; Goedicke, A.; Schweizer, B.; Aach, T.; Schulz, V. Simultaneous Reconstruction of Activity and Attenuation for PET/MR. IEEE Trans. Med. Imaging 2011, 30, 804–813. [Google Scholar] [CrossRef]
- Glorot, X.; Bengio, Y. Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the International Conference on Artificial Intelligence and Statistics, Sardinia, Italy, 13 May 2010; pp. 249–256. [Google Scholar]
- Kang, K.W.; Lee, D.S.; Cho, J.H.; Lee, J.S.; Yeo, J.S.; Lee, S.K.; Chung, J.-K.; Lee, M.C. Quantification of F-18 FDG PET images in temporal lobe epilepsy patients using probabilistic brain atlas. NeuroImage 2001, 14, 1–6. [Google Scholar] [CrossRef]
- Lee, J.S.; Lee, D.S. Analysis of functional brain images using population-based probabilistic atlas. Curr. Med. Imaging 2005, 1, 81–87. [Google Scholar] [CrossRef]
- Van Sluis, J.J.; De Jong, J.; Schaar, J.; Noordzij, W.; Van Snick, P.; Dierckx, R.; Borra, R.; Willemsen, A.; Boellaard, R. Performance Characteristics of the Digital Biograph Vision PET/CT System. J. Nucl. Med. 2019, 60, 1031–1036. [Google Scholar] [CrossRef]
- Levin, C.S.; Maramraju, S.H.; Khalighi, M.M.; Deller, T.W.; Delso, G.; Jansen, F. Design Features and Mutual Compatibility Studies of the Time-of-Flight PET Capable GE SIGNA PET/MR System. IEEE Trans. Med. Imaging 2016, 35, 1907–1914. [Google Scholar] [CrossRef] [PubMed]
- Son, J.-W.; Kim, K.Y.; Yoon, H.S.; Won, J.Y.; Ko, G.B.; Lee, M.S.; Lee, J.S. Proof-of-concept prototype time-of-flight PET system based on high-quantum-efficiency multianode PMTs. Med. Phys. 2017, 44, 5314–5324. [Google Scholar] [CrossRef] [PubMed]
- Mehranian, A.; Zaidi, H. Joint Estimation of Activity and Attenuation in Whole-Body TOF PET/MRI Using Constrained Gaussian Mixture Models. IEEE Trans. Med. Imaging 2015, 34, 1808–1821. [Google Scholar] [CrossRef] [PubMed]
- Arabi, H.; Zeng, G.; Zheng, G.; Zaidi, H. Novel adversarial semantic structure deep learning for MRI-guided attenuation correction in brain PET/MRI. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2746–2759. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.; Lee, D. Generation of Structural MR Images from Amyloid PET: Application to MR-Less Quantification. J. Nucl. Med. 2017, 59, 1111–1117. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.K.; Seo, S.; Shin, S.A.; Byun, M.S.; Lee, D.Y.; Kim, Y.K.; Lee, N.S.; Lee, J.S. Adaptive template generation for amyloid PET using a deep learning approach. Hum. Brain Mapp. 2018, 39, 3769–3778. [Google Scholar] [CrossRef] [PubMed]
- Hwang, D.; Kim, K.Y.; Kang, S.K.; Choi, H.; Seo, S.; Paeng, J.C.; Lee, D.S.; Lee, J.S. Accurate attenuation correction for whole-body Ga-68-DOTATOC PET studies using deep learning. J. Nucl. Med. 2019, 60, 568. [Google Scholar]
- Chen, X.; Nguyen, B.P.; Chui, C.-K.; Ong, S.-H. Reworking Multilabel Brain Tumor Segmentation: An Automated Framework Using Structured Kernel Sparse Representation. IEEE Syst. Man Cybern. Mag. 2017, 3, 18–22. [Google Scholar] [CrossRef]
- Bentley, P.; Ganesalingam, J.; Jones, A.L.C.; Mahady, K.; Epton, S.; Rinne, P.; Sharma, P.; Halse, O.; Mehta, A.; Rueckert, D. Prediction of stroke thrombolysis outcome using CT brain machine learning. NeuroImage Clin. 2014, 4, 635–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.W.; Hui, R.; Tian, Z. Classification of CT brain images based on deep learning networks. Comput. Methods Programs Biomed. 2017, 138, 49–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.; Lu, Z.; Zhang, Y.-D. Pathological brain detection based on AlexNet and transfer learning. J. Comput. Sci. 2019, 30, 41–47. [Google Scholar] [CrossRef]






| Method | Bone | Air |
|---|---|---|
| MLAA | 0.073 ± 0.081 | 0.055 ± 0.015 |
| 2D U-net | 0.718 ± 0.048 | 0.400 ± 0.074 |
| 2.5D U-net | 0.702 ± 0.047 | 0.424 ± 0.062 |
| 3D U-net | 0.826 ± 0.032 | 0.674 ± 0.057 |
| Method | Regression Line | R2 |
|---|---|---|
| MLAA | y = 0.30x + 0.035 | 0.05 |
| 2D U-net | y = 0.71x + 0.029 | 0.68 |
| 2.5D U-net | y = 0.69x + 0.030 | 0.67 |
| 3D U-net | y = 0.83x + 0.017 | 0.80 |
| Method | Attenuation Map | Activity Map | ||
|---|---|---|---|---|
| NRMSE | PSNR (dB) | NRMSE | PSNR (dB) | |
| MLAA | 0.197 | 8.06 | 0.132 | 17.78 |
| 2D U-net | 0.049 | 26.21 | 0.017 | 35.73 |
| 2.5D U-net | 0.053 | 25.95 | 0.017 | 35.44 |
| 3D U-net | 0.039 | 28.31 | 0.011 | 38.77 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, B.-H.; Hwang, D.; Kang, S.-K.; Kim, K.-Y.; Choi, H.; Seo, S.; Lee, J.-S. Accurate Transmission-Less Attenuation Correction Method for Amyloid-β Brain PET Using Deep Neural Network. Electronics 2021, 10, 1836. https://doi.org/10.3390/electronics10151836
Choi B-H, Hwang D, Kang S-K, Kim K-Y, Choi H, Seo S, Lee J-S. Accurate Transmission-Less Attenuation Correction Method for Amyloid-β Brain PET Using Deep Neural Network. Electronics. 2021; 10(15):1836. https://doi.org/10.3390/electronics10151836
Chicago/Turabian StyleChoi, Bo-Hye, Donghwi Hwang, Seung-Kwan Kang, Kyeong-Yun Kim, Hongyoon Choi, Seongho Seo, and Jae-Sung Lee. 2021. "Accurate Transmission-Less Attenuation Correction Method for Amyloid-β Brain PET Using Deep Neural Network" Electronics 10, no. 15: 1836. https://doi.org/10.3390/electronics10151836







