Detection of Diseases in Tomato Leaves by Color Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Image Acquisition
2.2. Color Models
2.3. Image Histogram
2.4. Otsu Method
2.5. Related Components
2.6. Support Vector Machines
2.7. K-Nearest Neighbor (K-NN)
2.8. Multilayer Perceptron (MLP)
2.9. GLCM Gray Level Co-Occurrence Matrix and Color Moments
- Medium:
- Standard deviation:
- Correlation:
- Entropy:
- Dissimilarity:
- Contrast:
- Homogeneity:
- Energy:
- Maximum probability:
3. Proposed Model
3.1. Segmentation
- The grayscale image is obtained using the average of the three channels of the RGB model, that is,;
- The negative operator is applied;
- A median filter with a 3 × 3 mask is applied;
- The threshold is set using the Otsu method (Equations (4)–(10)).
3.2. Feature Extraction
3.3. Classification
4. Experiments and Results
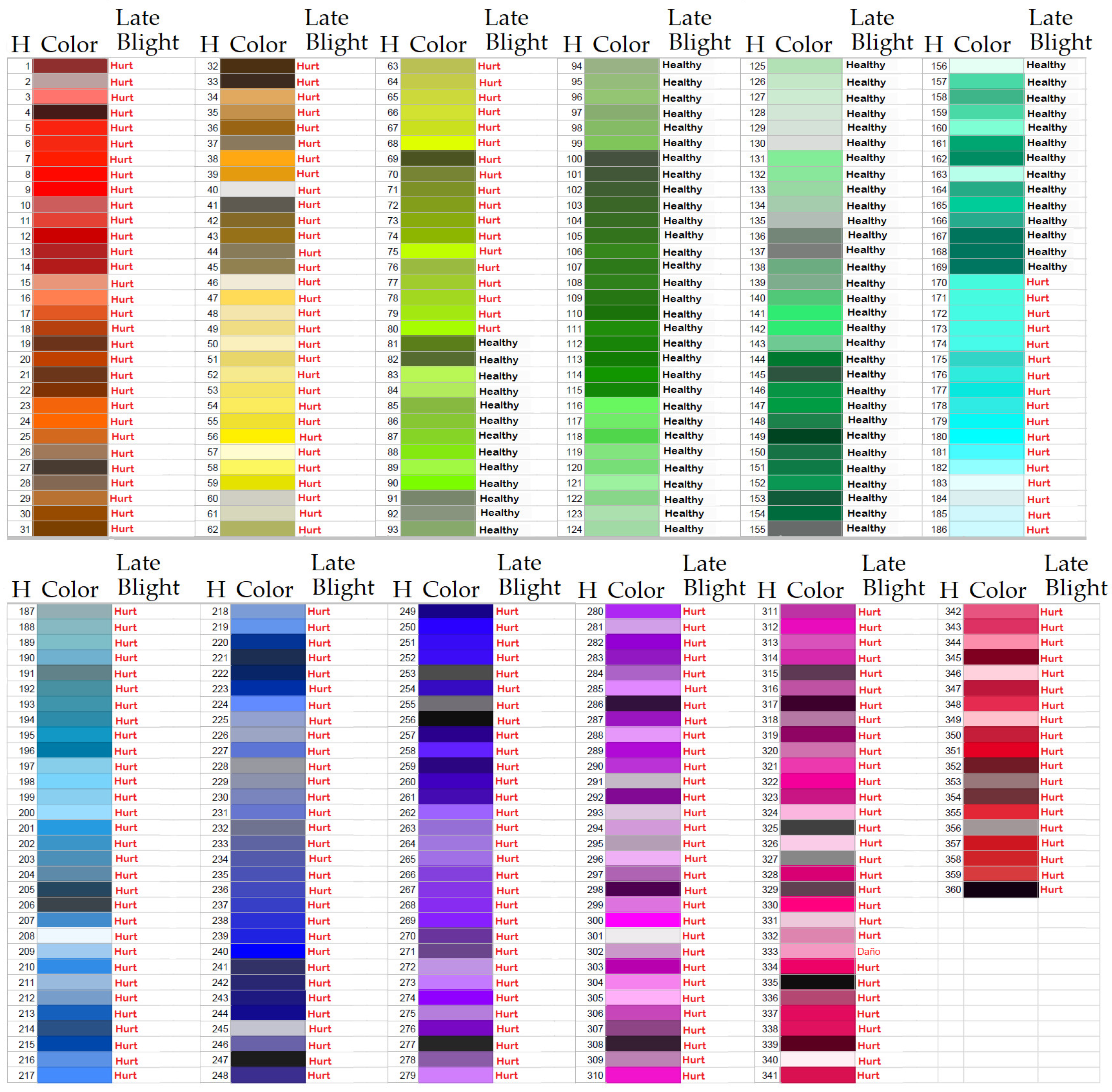
- Segmentation: First, the RGB model was used, and by means of the average of the three channels R, G and B, it was converted to gray scale, then the negative of the image was obtained and on it, immediately, a median filter was applied of a 3×3 mask. Then, it was thresholded by the Otsu method to obtain the optimal binarization threshold, the image was binarized and the related components of the negative of the threshold image by means of Otsu were calculated. The related components of greater area were eliminated and those related components that were not delimited by an area of black pixels where obtained by the binarization by means of Otsu. In this way, the area of the segmented leaf was obtained. Once the color leaf was segmented, the HSV color model was considered, and on the HSV model it was segmented by threshold on the H component, considering the color thresholds in which a plant is damaged by any of the diseases considered, or is healthy. These threshold values were validated by experts in the area of phytopathology. For the case of late blight, Figure 4 shows the color threshold.
- Features extraction: Four color moments for each RGB component and nine statistical measurements were used for texture analysis by GLCM, obtaining a total of 21 features.
- Classification: MLP, K-NN and SVM classifiers were used, using the architecture proposed in Figure 6, i.e., all three classifiers were considered to decide on the output of the classification. Table 1 shows the arguments considered for each of the classifiers that were used for the proposed methodology.
5. Discussion
6. Conclusions and Future Work
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Organización de las Naciones Unidas para la Alimentación y la Agricultura. Food and Agriculture Organization of the United Nations. Available online: http://www.fao.org/home/es/ (accessed on 24 April 2021).
- Fideicomisos Instituidos en Relación con la Agricultura. Available online: https://www.fira.gob.mx/ (accessed on 24 April 2021).
- Li, Y.; Wang, H.; Dang, L.M.; Sadeghi-Niaraki, A.; Moon, H. Crop pest recognition in natural scenes using convolutional neural networks. Comput. Electron. Agric. 2020, 169, 105174. [Google Scholar] [CrossRef]
- Arsenovic, M.; Karanovic, M.; Sladojevic, S.; Anderla, A.; Stefanovic, D. Solving Current Limitations of Deep Learning Based Approaches for Plant Disease Detection. Symmetry 2019, 11, 939. [Google Scholar] [CrossRef] [Green Version]
- Atila, Ü.; Uçar, M.; Akyol, K.; Uçar, E. Plant leaf disease classification using EfficientNet deep learning model. Ecol. Informatics 2021, 61, 101182. [Google Scholar] [CrossRef]
- Nazki, H.; Yoon, S.; Fuentes, A.; Park, D.S. Unsupervised image translation using adversarial networks for improved plant disease recognition. Comput. Electron. Agric. 2020, 168, 105117. [Google Scholar] [CrossRef]
- Kumbar, B.; Mahmood, R.; Nagesha, S.; Nagaraja, M.; Prashant, D.; Kerima, O.Z.; Karosiya, A.; Chavan, M. Field application of Bacillus subtilis isolates for controlling late blight disease of potato caused by Phytophthora infestans. Biocatal. Agric. Biotechnol. 2019, 22, 101366. [Google Scholar] [CrossRef]
- Charalampopoulos, I.; Droulia, F. The Agro-Meteorological Caused Famines as an Evolutionary Factor in the Formation of Civilisation and History: Representative Cases in Europe. Climate 2020, 9, 5. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, S.; Shen, J.; Wu, Z.; Du, Z.; Gao, F. The phylogeographic history of tomato mosaic virus in Eurasia. Virology 2021, 554, 42–47. [Google Scholar] [CrossRef]
- Klap, C.; Luria, N.; Smith, E.; Bakelman, E.; Belausov, E.; Laskar, O.; Lachman, O.; Gal-On, A.; Dombrovsky, A. The Potential Risk of Plant-Virus Disease Initiation by Infected Tomatoes. Plants 2020, 9, 623. [Google Scholar] [CrossRef]
- Ávila, M.C.R.; Lourenço, V.; Quezado-Duval, A.M.; Becker, W.F.; De Abreu-Tarazi, M.F.; Borges, L.C.; Nascimento, A.D.R. Field validation of TOMCAST modified to manage Septoria leaf spot on tomato in the central-west region of Brazil. Crop. Prot. 2020, 138, 105333. [Google Scholar] [CrossRef]
- Mulugeta, T.; Miuhinyuza, J.B.; Gouws-Meyer, R.; Matsaunyane, L.; Andreasson, E.; Alexandersson, E. Botanicals and plant stregtheners for potato and tomato cultivation in Africa. J. Integr. Agric. 2020, 19, 406–427. [Google Scholar] [CrossRef]
- Arnal, J.G. Plant disease identification from individual lesions and spots using deep learning. Biosyst. Eng. 2019, 180, 96–107. [Google Scholar] [CrossRef]
- Saleem, M.H.; Potgieter, J.; Arif, K.M. Plant Disease Detection and Classification by Deep Learning. Plants 2019, 8, 468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, V.; Misra, A. Detection of plant leaf diseases using image segmentation and soft computing techniques. Inf. Process. Agric. 2017, 4, 41–49. [Google Scholar] [CrossRef] [Green Version]
- Vianna, G.K.; Cunha, G.V.; Oliveira, G.S. A Neural Network Classifier for Estimation of the Degree of Infestation by Late Blight on Tomato Leaves. Int. J. Comput. Inf. Eng. 2017, 11, 18–24. [Google Scholar]
- Sabrol, H.; Kumar, S. Recognition of Tomato Late Blight by using DWT and Component Analysis. Int. J. Electr. Comput. Eng. 2017, 7, 194–199. [Google Scholar] [CrossRef] [Green Version]
- Mattos, A.P.; Tolentino, J.B.; Itako, A.T. Determination of the severity of Septoria leaf spot in tomato by using digital im-ages. Australas. Plant Pathol. 2020, 49, 329–356. [Google Scholar] [CrossRef]
- Zhang, K.; Wu, Q.; Liu, A.; Meng, X. Can Deep Learning Identify Tomato Leaf Disease? Adv. Multimedia 2018, 2018, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Vetal, S.; Khule, R.S. Tomato plant disease detection using image processing. Int. J. Adv. Res. Comput. Commun. Eng. 2017, 6, 293–297. [Google Scholar] [CrossRef]
- Sabrol, H.; Satish, K. Tomato plant disease classification in digital images using classification tree. In Proceedings of the IEEE 2016 International Conference on Communication and Signal Processing (ICCSP), Melmaruvathur, India, 6–8 April 2016; pp. 1242–1246. [Google Scholar]
- Saleem, G.; Akhtar, M.; Ahmed, N.; Qureshi, W. Automated analysis of visual leaf shape features for plant classification. Comput. Electron. Agric. 2019, 157, 270–280. [Google Scholar] [CrossRef]
- Din, M.Z.; Adnan, S.M.; Ahmad, W.; Aziz, S.; Rashid, J.; Ismail, W.; Iqba, M.J. Classification of Disease in Tomato Plants’ Leaf Using Image Segmentation and SVM. Tech. J. Univ. Eng. Technol. 2018, 23, 81–88. [Google Scholar]
- Khan, S.; Saboo, M.H.; Narvekar, M.; Sanghvi, D.J. Novel fusion of color balancing and Superpixel based approach for detection of Tomato plant diseases in natural complex environment. J. King Saud Univ. Comput. Inf. Sci. 2020, 1319–1578. [Google Scholar] [CrossRef]
- Saeed, F.; Khan, M.A.; Sharif, M.; Mittal, M.; Goyal, L.M.; Roy, S. Deep neural network features fusion and selection based on PLS regression with an application for crops diseases classification. Appl. Soft Comput. 2021, 103, 107164. [Google Scholar] [CrossRef]
- Kumar, S.D.; Esakkirajan, S.; Bama, S.; Keerthiveena, B. A microcontroller based machine vision approach for tomato grading and sorting using SVM classifier. Microprocess. Microsyst. 2020, 76, 103090. [Google Scholar] [CrossRef]
- Tian, K.; Li, J.; Zeng, J.; Evans, A.; Zhang, L. Segmentation of tomato leaf images based on adaptive clustering number of K-means algorithm. Comput. Electron. Agric. 2019, 165, 104962. [Google Scholar] [CrossRef]
- Sabrol, H.; Kumar, S. Fuzzy and Neural Network based Tomato Plant Disease Classification using Natural Outdoor Images. Indian J. Sci. Technol. 2016, 9, 1–8. [Google Scholar] [CrossRef]
- Petrellis, N. Plant Disease Diagnosis for Smart Phone Applications with Extensible Set of Diseases. Appl. Sci. 2019, 9, 1952. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Sun, C.; Zou, T.; Li, L.; Wang, L.; Liu, H. SVM-based image partitioning for vision recognition of AVG guide paths under complex illumination conditions. Robot. Comput. Integr. Manuf. 2020, 61, 101856. [Google Scholar] [CrossRef]
- Hosseini, S.; Zade, B.M.H. New hybrid method for attack detection using combination of evolutionary algorithms, SVM, and ANN. Comput. Netw. 2020, 173, 107168. [Google Scholar] [CrossRef]
- Saadatfar, H.; Khosravi, S.; Joloudari, J.H.; Mosavi, A.; Shamshirband, S. A New K-Nearest Neighbors Classifier for Big Data Based on Efficient Data Pruning. Mathematics 2020, 8, 286. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.-C. Multilayer Perceptron for Robust Nonlinear Interval Regression Analysis Using Genetic Algorithms. Sci. World J. 2014, 2014, 1–8. [Google Scholar] [CrossRef]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using Deep Learning for Image-Based Plant Disease Detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, D.P.; Salathé, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics through machine learning and crowdsourcing. arXiv 2015, arXiv:1511.08060. [Google Scholar]
- Xie, C.; Shao, Y.; Li, X.; He, Y. Detection of early blight and late blight diseases on tomato leaves using hyperspectral imaging. Sci. Rep. 2015, 5, 16564. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Lu, Z.; Qi, W.; Chen, Y. Plant Disease Classification Using Deep Learning Methods. In Proceedings of the 4th International Conference on Machine Learning and Soft Computing, ACM, Haiphong City, Vietnam, 17–19 January 2020; pp. 5–9. [Google Scholar]
- Sharma, P.; Berwal, Y.P.S.; Ghai, W. KrishiMitr (Farmer’s Friend): Using Machine Learning to Identify Diseases in Plants. In Proceedings of the IEEE International Conference on Internet of Things and Intelligence System (IOTAIS), Bali, Indonesia, 1–3 November 2018; pp. 29–34. [Google Scholar]
- Durmus, H.; Gunes, E.O.; Kirci, M. Disease detection on the leaves of the tomato plants by using deep learning. In Proceedings of the IEEE 2017 6th International Conference on Agro-Geoinformatics, Fairfax, VA, USA, 7–10 August 2017; pp. 1–5. [Google Scholar]
- Fuentes, A.; Yoon, S.; Kim, S.C.; Park, D.S. A Robust Deep-Learning-Based Detector for Real-Time Tomato Plant Diseases and Pests Recognition. Sensors 2017, 17, 2022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Sun, Y.; Wang, J. Automatic Image-Based Plant Disease Severity Estimation Using Deep Learning. Comput. Intell. Neurosci. 2017, 2017, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Prabakaran, G.; Vaithiyanathan, D.; Ganesan, M. FPGA based effective agriculture productivity prediction system using fuzzy support vector machine. Math. Comput. Simul. 2021, 185, 1–16. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, Y. Village precision poverty alleviation and smart agriculture based on FPGA and machine learning. Microprocess. Microsyst. 2020, 103469. [Google Scholar] [CrossRef]
- Huang, C.-H.; Chen, P.-J.; Lin, Y.-J.; Chen, B.-W.; Zheng, J.-X. A robot-based intelligent management design for agricultural cyber-physical systems. Comput. Electron. Agric. 2021, 181, 105967. [Google Scholar] [CrossRef]
- Rodríguez-Robles, J.; Martin, Á.; Martin, S.; Ruipérez-Valiente, J.; Castro, M. Autonomous Sensor Network for Rural Agriculture Environments, Low Cost, and Energy Self-Charge. Sustainability 2020, 12, 5913. [Google Scholar] [CrossRef]
- Zervopoulos, A.; Tsipis, A.; Alvanou, A.G.; Bezas, K.; Papamichail, A.; Vergis, S.; Stylidou, A.; Tsoumanis, G.; Komianos, V.; Koufoudakis, G.; et al. Wireless Sensor Network Synchronization for Precision Agriculture Applications. Agriculture 2020, 10, 89. [Google Scholar] [CrossRef] [Green Version]
- Karar, M.E.; Alsunaydi, F.; Albusaymi, S.; Alotaibi, S. A new mobile application of agricultural pests recognition using deep learning in cloud computing system. Alex. Eng. J. 2021, 60, 4423–4432. [Google Scholar] [CrossRef]
- Hyun, S.; Yang, S.M.; Kim, J.; Kim, K.S.; Shin, J.H.; Lee, S.M.; Lee, B.-W.; Beresford, R.M.; Fleisher, D.H. Development of a mobile computing framework to aid decision-making on organic fertilizer management using a crop growth model. Comput. Electron. Agric. 2021, 181, 105936. [Google Scholar] [CrossRef]
- Cicioğlu, M.; Çalhan, A. Smart agriculture with internet of things in cornfields. Comput. Electr. Eng. 2021, 90, 106982. [Google Scholar] [CrossRef]
- Merwe, D.; Burchfield, D.R.; Witt, T.D.; Price, K.P.; Sharda, A. Chapter One—Drones in agriculture. Adv. Agron. 2020, 162, 1–30. [Google Scholar]






| Sorter | Accuracy | Arguments |
|---|---|---|
| MLP | healthy-sick | 1 hidden layer |
| SVM | healthy-sick | kernel linear |
| K-NN | healthy-sick | k = 9 |
| MLP | type-disease | 2 hidden layers |
| SVM | type-disease | kernel rbf |
| K-NN | type-disease | k = 3 |
| Sorter | Accuracy | — |
|---|---|---|
| J48 | 77.70% | - |
| KStar | 77.29% | - |
| Random Tree | 74.79% | - |
| LWL | 61.45% | - |
| Decision Stump | 61.04% | - |
| Naive Bayes Updateable | 61.16% | - |
| PART | 76.45% | - |
| Decision Table | 74.37% | RMSE |
| MLP | 89.02% | 2 hidden layers |
| SVM | 89.03% | Kernel linear |
| K-NN | 85.39% | k = 3 |
| K-NN | 83.01% | k = 5 |
| K-NN | 70.84% | k = 9 |
| Proposed | 86.45% | - |
| Sorter | Accuracy | — |
|---|---|---|
| J48 | 92.91% | - |
| KStar | 92.91% | - |
| Random Tree | 90.93% | - |
| LWL | 78.75% | - |
| Decision Stump | 77.5% | - |
| Naive Bayes Updateable | 78.33% | - |
| PART | 91.45% | - |
| Decision Table | 90.72% | RMSE |
| MLP | 98.05% | 1 hidden layer |
| SVM | 97.52% | Kernel linear |
| SVM | 97.27% | Kernel rbf |
| K-NN | 94.92% | k = 1 |
| K-NN | 93.09% | k = 3 |
| K-NN | 95.05% | k = 9 |
| Proposed | 97.39% | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luna-Benoso, B.; Martínez-Perales, J.C.; Cortés-Galicia, J.; Flores-Carapia, R.; Silva-García, V.M. Detection of Diseases in Tomato Leaves by Color Analysis. Electronics 2021, 10, 1055. https://doi.org/10.3390/electronics10091055
Luna-Benoso B, Martínez-Perales JC, Cortés-Galicia J, Flores-Carapia R, Silva-García VM. Detection of Diseases in Tomato Leaves by Color Analysis. Electronics. 2021; 10(9):1055. https://doi.org/10.3390/electronics10091055
Chicago/Turabian StyleLuna-Benoso, Benjamín, José Cruz Martínez-Perales, Jorge Cortés-Galicia, Rolando Flores-Carapia, and Víctor Manuel Silva-García. 2021. "Detection of Diseases in Tomato Leaves by Color Analysis" Electronics 10, no. 9: 1055. https://doi.org/10.3390/electronics10091055








