Abstract
Diabetic retinopathy (DR) is the primary cause of blindness in developing and developed countries. Early-stage DR detection reduces the risk of blindness in Diabetes Mellitus (DM) patients. There has been a sharp rise in the prevalence of DM in recent years, especially in low- and middle-income countries. In this context, automated artificial intelligence-based DM screening is a crucial tool to help classify the considerable amount of Retinal Fundus Images (RFI). However, retinal image quality assessment has shown to be fundamental in real-world DR screening processes to avoid out-of-distribution data, drift, and images lacking relevant anatomical information. This work analyzes the spatial domain features and image quality assessment metrics for carrying out Deep Learning (DL) classification and detecting notable features in RFI. In addition, a novel lightweight convolutional neural network is proposed specifically for binary classification at a low computational cost. The training results are comparable to state-of-the-art neural networks, which are widely used in DL applications. The implemented architecture achieves 98.6% area under the curve, and 97.66%, and 98.33% sensitivity and specificity, respectively. Moreover, the object detection model trained achieves 94.5% mean average precision. Furthermore, the proposed approach can be integrated into any automated RFI analysis system.
1. Introduction
The global population with Diabetes Mellitus (DM) was 463 million in 2019, and it is estimated that it will reach 700 million by 2045 [1]. Diabetic retinopathy (DR) is due to a complication of DM [2] and is the primary cause of blindness in developing and developed countries [3]. The estimated DR prevalence rate is 35.90% in Africa, 33.30% in North America and the Caribbean, and 13.37% [1] in South and Central America. According to the International Council of Ophthalmology (ICO) [2], early-stage DR screening reduces the risk of vision loss and blindness in DM patients.
Traditional screening is performed by a well-trained clinical ophthalmologist who analyzes and assesses Retinal Fundus Images (RFI) for the severity of DR. Nevertheless, the increase in DR prevalence is not proportional to the number of experts available, hindering patients from receiving timely diagnosis and treatment, which eventually leads to irreversible vision loss [4]. Therefore, there is a need for an automated diagnostic system that helps ophthalmologists increase the screening process efficiency [5].
In recent years, Deep Learning (DL) has become a powerful tool for medical image analysis using RFI. These DR detection tools initially focused on using traditional machine learning methods to detect macula and the Optic Disc (OD) in RFI to perform retinal vessel segmentation and classify the presence/absence of DR. In this context, Convolutional Neural Networks (CNN) for DR detection have predominantly focused on identifying referable DR patients, achieving outstanding results [6,7,8]. In addition, these DL systems have been implemented in primary care clinical programs designed to follow clinical DR guidelines. Therefore, such algorithms have shown outstanding lab classification performance, although the accuracy of these DL algorithms significantly decreases when using real-world data [9,10,11]. One possible reason is that CNN performance is susceptible to quality distortions, particularly blur and noise [12]. Even though DL systems are generally trained and tested on high-quality image datasets, the input images cannot be assumed to be of a high-quality level in practical applications. Thus, some DL methods provide image quality assessment, which is one of the most necessary additions to primary DR screening [8].
The quality of an RFI is a characteristic of its amount of visual degradation, such as noise, artifacts, flashlights, or a lack of focus. Consequently, Image Quality Assessment (IQA) has been introduced in various computer vision system applications. This ensures and improves the quality of visual contents at the receiving end [13,14,15,16]. Objective IQA can be achieved in three ways: full-reference image quality assessment, which requires both distorted and reference images; reduced-reference image quality assessment by only using metrics from partial information from the reference image; and No-Reference Image Quality Assessment (NR-IQA), which predicts the perceptual quality of images without accessing a reference image [16].
There are several state-of-the-art works based on different methodologies. For example, SalStructlQA [17] evaluates RFI based on extensive and small-sized protruding structures. On the other hand, Dias et al. [18] and Yao et al. [19] assess the quality of RFI in the spatial domain with indicators such as color, focus, contrast, and illumination. In addition, feed-forward neural networks or support vector machines were trained with subjective human labeling, which resulted in an estimate measure to determine if the image is ungradable or gradable. Moreover, Yu et al. [20] and Zago et al. [21] proposed using CNNs trained with manual labeling RFI. Nevertheless, one constraint of these approaches is that they are limited to the perception of human labeling to perform the classification. For example, if a model is fitted into a dataset that could be labeled as gradable with a non-visible high noise due to JPEG compression, it could affect another stage in the DR diagnosis.
This work presents a new semi-automated workflow to pre-process, label, train, and classify the suitability of RFI associated with DR, which is defined as images with acceptable quality and anatomically gradable. The pre-processing of the RFI starts with image resizing, preserving their aspect ratio. This is achieved by removing the background from the RFI using automated semantic segmentation based on CNN.
The contributions of this work are summarized as follows:
- A lightweight CNN model was proposed for quality binary classification (low and high), which was trained with a dataset generated from public databases in two steps. First, a pre-selection labeling considers quality metrics such as noise, blur, and contrast, as well as the relationship between these metrics. Additionally, an image quality score evaluator assigns a numerical quality level value to each RFI. The second step for labeling is carried out by human judgment based on their visual perception of anatomical features, artifacts, sharpness, and overall visual assessment. Human labeling cannot be solely relied upon due to the possibility of distortions not visible to the human eye. For this, quality metrics, numerical evaluation, pre-selection labeling, and human criteria selection ensure that images correspond to each class.
- A small CNN model (YOLOv5 nano) is trained to detect the most relevant regions for DR screening, such as the macula and the OD, in order to ensure the presence of anatomical regions. These two models ensure that making the suitable decision is more transparent because the anatomical features are present.
In each workflow stage, the proposed CNN model has been compared to some state-of-the-art models, including the new vision transformers, obtaining similar performance. Finally, a gradient visualization technique explains the proposed model’s behavior. It is important to note that the models integrated for pre-processing, classification, and object detection are those with the fewest parameters to keep computational costs low and can be integrated into any RFI analysis system for DR.
2. Methods and Data
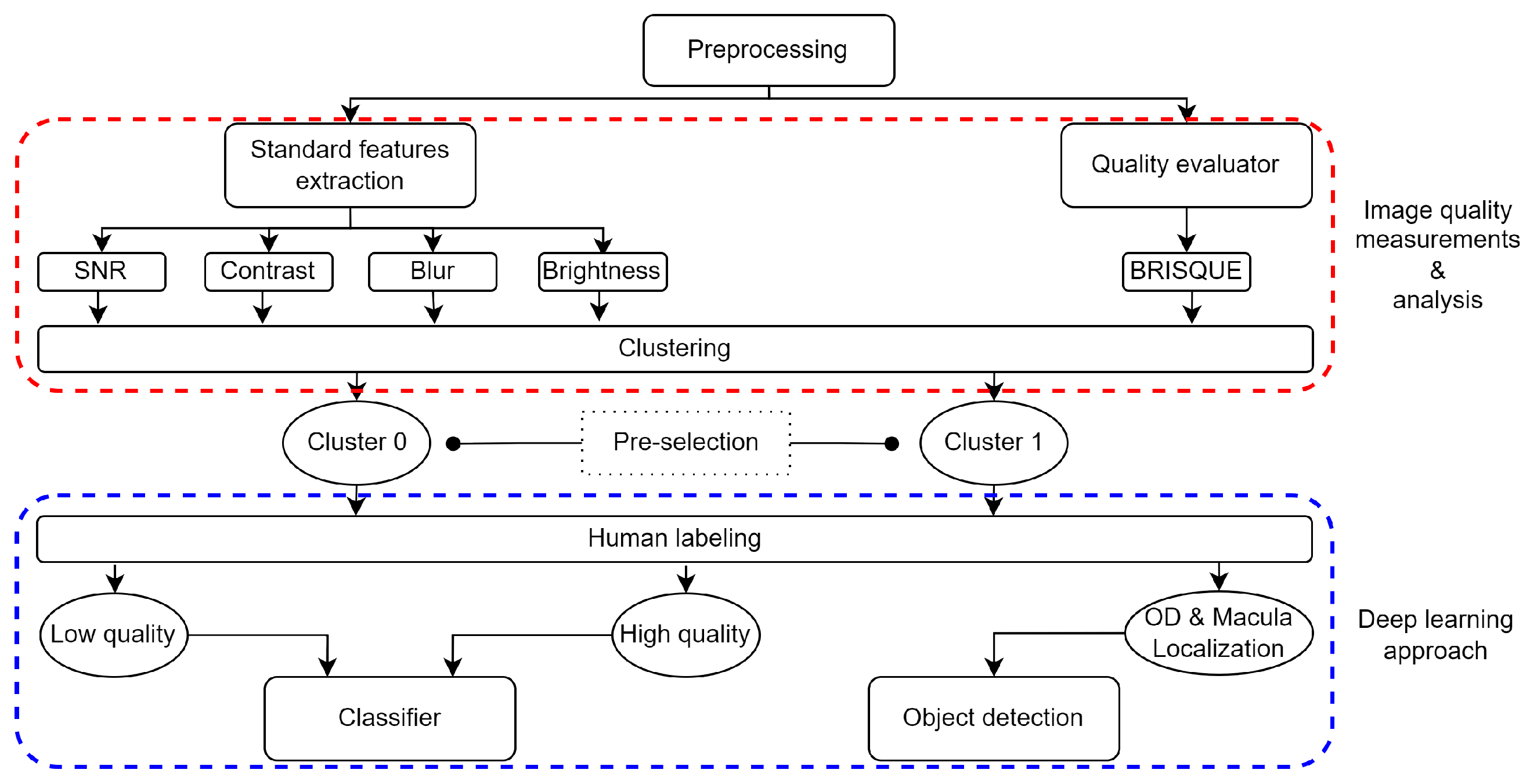
Figure 1 shows the workflow used in this work. It is divided into a pre-processing stage, IQA measurements, and analysis as stage one obtains a pre-labeled dataset. In stage two, human labeling following pre-selected images for DL automated systems are trained and tested.
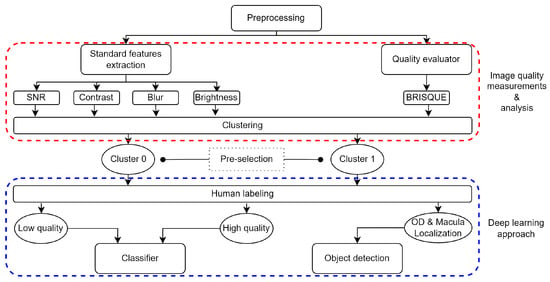
Figure 1.
Workflow diagram: pre-processing, analysis, labeling, classification, and detection. The suitable decision is based on quality classification (by a CNN) and RFI anatomical feature detection.
The first stage is based on NR-IQA by extracting standard features (SF), which include signal-to-noise Ratio (SNR), contrast value (C), blur index (B), and brightness (L), each one represented by scalar values. In addition, this analysis is combined with the evaluation method based on the Image Quality Evaluator (IQE), which is guided by a correlation analysis between the SF and the score obtained by the IQE function. Correlation analysis is not limited to statistical methods; hence, an unsupervised machine learning model is proposed that clusters items based on their measured SF and IQE value.
In the second stage, a human rater performs the medical suitability analysis, performing a two-class labeling method. It defines label-1 for images with valid features for high-quality classification and label-0 for low-quality classification. Once the labeling is completed, the validated dataset is used to train supervised learning classification models. Additionally, DL detection models are implemented to ensure the presence of anatomical elements.
2.1. Images Dataset
The public dataset repositories used in this work are (i) Kaggle Diabetic Retinopathy Detection provided by EyePACS [22], (ii) the Indian Diabetic Retinopathy Image Dataset (IDRiD) [23], and (iii) the Methods to Evaluate Segmentation and Indexing Techniques in the field of Retinal Ophthalmology (MESSIDOR II) [24] proposed for segmentation and DR grade, (iv) Asia Pacific Tele-Ophthalmology Society (APTOS) SymposiumAPTOS2019 [25] in the competition “Detect diabetic retinopathy to stop blindness before it’s too late”, (v) the Digital Retinal Images for Vessel Extraction (DRIVE) [26] and (vi) the Diabetic Retinopathy Images Database for Quality Testing of Retinal Images (DRIMDB) [27].
2.2. Pre-Processing
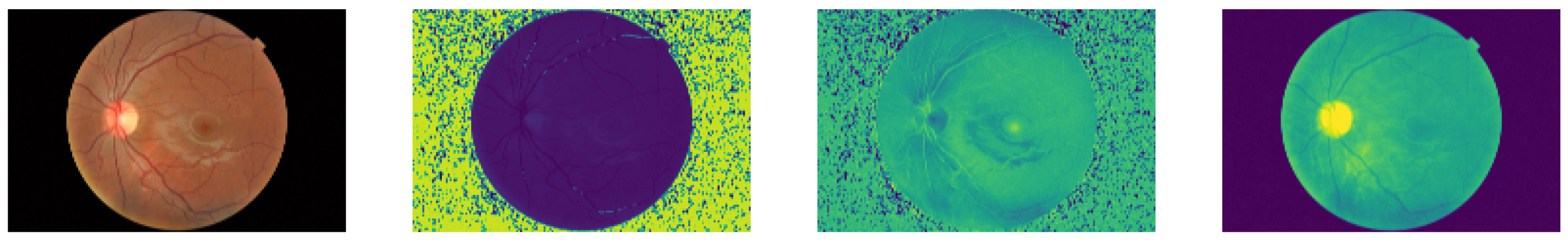
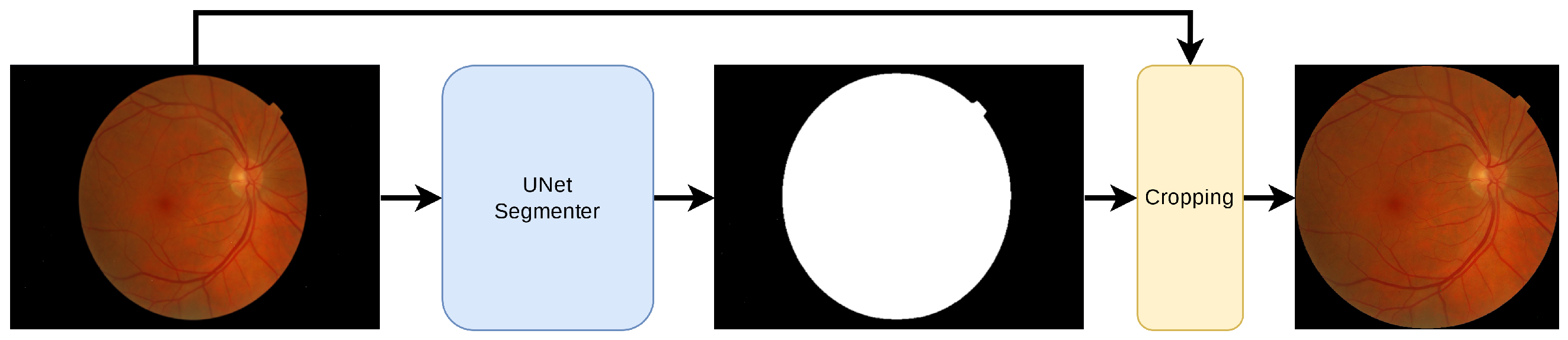
The public RFI databases were generated with different resolutions and compression formats, generating undesirable noise in the image background. For instance, see the HSV representation of an RFI in Figure 2, where speckle noise is present in the saturation and hue channels. As a result, a pre-processing technique based on Briseno et al. [28] is used for background segmentation, automated cropping, and resizing to 224 × 224, preserving their aspect ratios. Figure 3 shows the pre-processing flow diagram in which, given an RFI, the forward pass is performed through the segmentation CNN model (UNet [29]) to obtain a binary mask. After that, generating a bounding box representing the region of interest is possible, which means that everything outside this reference is cropped. Finally, a zero-fill stage is added to maintain a 1:1 aspect ratio.

Figure 2.
Apparently good quality RFI and its three channels in HSV color space. The background noise can be seen, which is mostly associated with image compression.
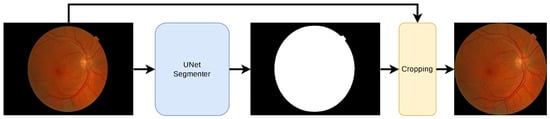
Figure 3.
Flowchart for the pre-processing of an RFI. The input image is of size H × W. An automatic segmentation model detects the retinal circle. The max and min coordinates are obtained in X and Y used as a reference to remove the background. Zero padding is then added if necessary. The output is an image of H × H with an aspect ratio of 1:1.
The dataset used to train the pre-processing segmentation model comprises 420 images. Twenty of these images correspond to a binary mask from DRIVE [26], and the rest are manually annotated from the EyePACS [22] dataset. Initial training is carried out with traditional UNet [29] architecture, which is one of the most popular CNNs for semantic segmentation in medical applications. In addition, to compare the performance, several segmentation models (CNN and one visual transformer Swin-Unet [30]) were trained and tested under the same conditions.
2.3. Image Quality Standard Features
Standard features (SF) numerically measure quality degradations (apparent or non-apparent). First, the signal-to-noise ratio (SNR) is a function that measures the mean value of an image divided by its standard deviation as noise (see Equation (1)). For SNR values greater than 1, the presence of a signal is greater than the noise level, and for values less than 1, the presence of noise is more significant.
where , , are the red, green, and blue components, while and are the mean and standard deviation, respectively.
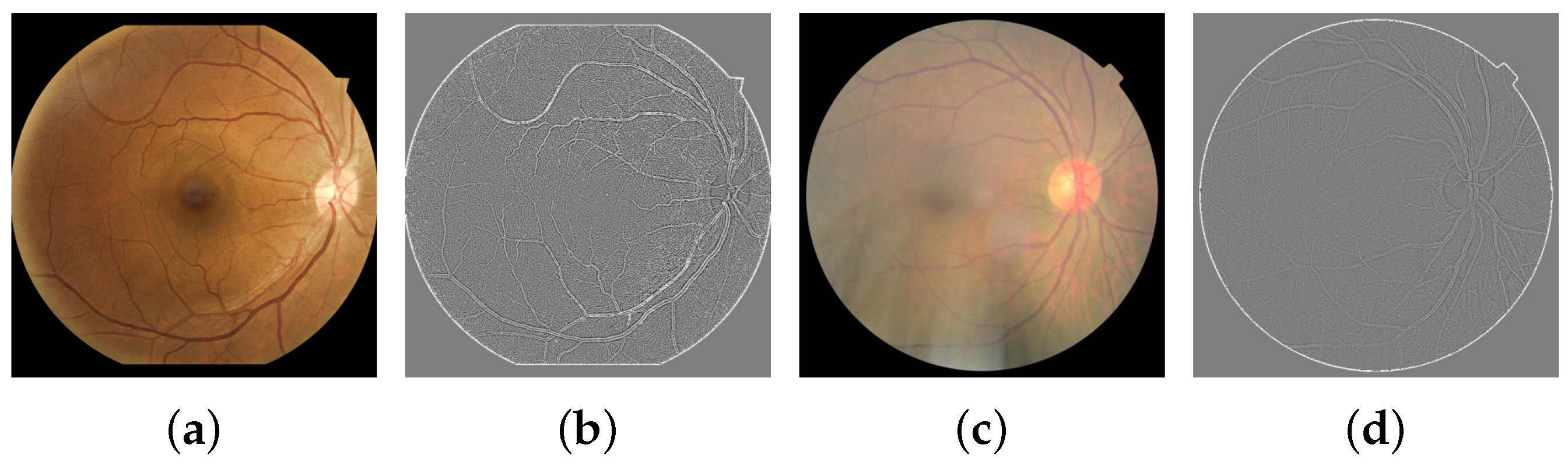
The second SF is the blur index obtained from its variance value of visible edges. The assumption is that if an image contains a high variance, there is a wide spread of responses, representing an in-focus image. On the other hand, an image with low edge variance is blurry. One option for edge detection is the Laplacian operator Δ, which is a finite approximation of a two-dimensional second derivative expressed in Equation (2). The blur coefficient is obtained by applying convolution from Equation (3) and then computing its variance with Equation (4). Typical and blurred RFI examples for this feature extraction are shown in Figure 4.
where is a grayscale image, is the Laplacian operator, and is the image variance.
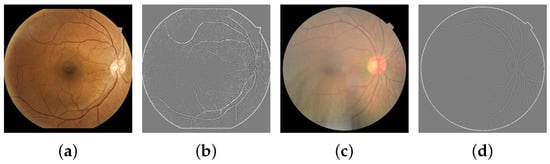
Figure 4.
Original images and their Laplacian representations. (a) Normal RFI. (b) Laplacian, B = 19.75. (c) Blurred RFI. (d) Laplacian, B = 10.57.
The third feature is the contrast level. This SF is computed according to Michelson contrast (peak-to-peak contrast), which is the relation between the spread and the sum of the two luminances in the histogram shown in Equation (5).
where and are the maximum and minimum values of the contrast histogram, respectively.
Finally, the brightness is the proposed fourth SF and is obtained with Equation (6), which is the level of the Luma channel (Y). This feature is calculated by taking the average from each channel RGB and converting it to YUV.
where , , and are the mean value of the red, green, and blue image channels, respectively.
2.4. Image Quality Scoring Evaluator
The image quality evaluator is performed based on the score given by the method of Mittal et al. [31] also known as BRISQUE, which is a statistical analysis of the natural scene using features extraction and defined as a method for IQA. This method uses a generic reference-less model based on natural scene statistics operating within the spatial domain.
Extraction starts with natural scene statistics, which are then passed through a mean subtracted contrast normalization process to predict the type of distortion affecting the image. This process is also repeated to capture neighborhood relations. Furthermore, the generated images are used to calculate a 36 × 1 feature vector. Moreover, an asymmetric generalized Gaussian distribution fits into each of the four pairwise product images. Finally, a support vector machine is used to fit a regression function. BRISQUE [31] is used in this work as the RFI IQE and can be supported by the SF measurements and their correlations.
2.5. Clustering
Clustering is a machine learning technique that involves grouping data points. Data points in the same group should have similar unique properties, while data points in different groups should have very different properties. Clustering is an unsupervised learning method featuring a joint statistical data analysis technique used in various fields.
The KMeans algorithm [32] is based on partitioning an N-dimensional population into k sets based on a sample. The process groups in the sense of variation within the class. If p is the probability mass function for the population, is a partition of , and , for is the conditional mean of p over the set . The mathematical expression for KMeans is given by:
where J represents the quadratic sum of the inaccuracy of samples of class n and its mean value, which is also called the sum of sample distances and their mean value. The input data vector is represented by , and is the central group mean for each .
The clustering algorithm is employed in this work to find a pattern in the SF and the IQE for all available data. The aim is to obtain two separated groups by assuming that the elements of Cluster 0 belong to low-quality features and the elements of Cluster 1 belong to high-quality features. It is essential to highlight that this stage only considers the numerical relationship of the dataset, and a visual validation will be necessary after this task.
2.6. Human Visual Opinion
Although labeling the dataset has been obtained by clustering, it has been a numerical analysis. However, RFI has definite features, mainly visualizing anatomical elements and visible details. Therefore, a human specialist must be validated to determine whether an image meets the selection criteria of low (class 0) or high quality (class 1).
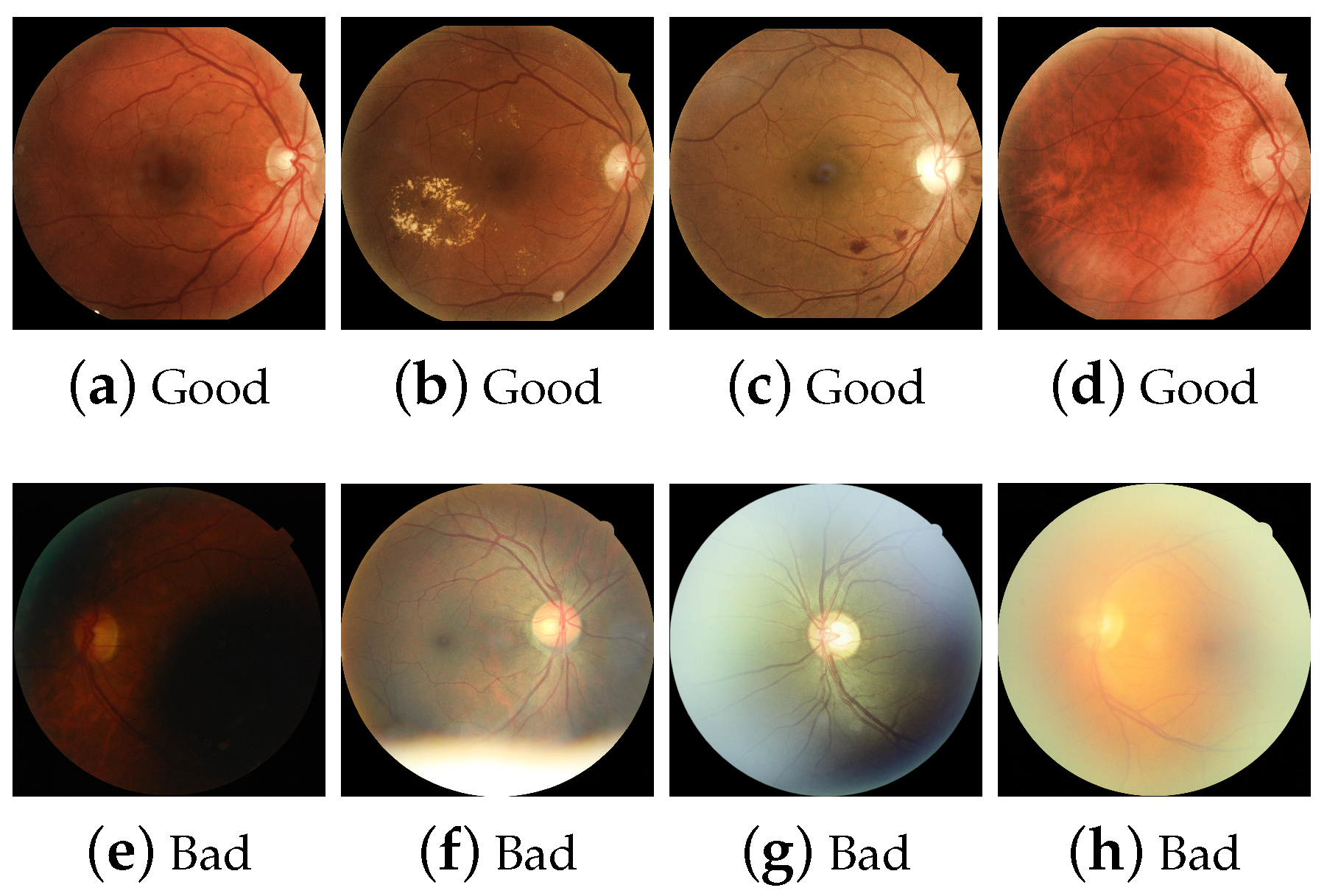
The human observer has an evaluation criterion shown in Table 1. Some aspects are considered that may alter the capture of an RFI, for example, if artifacts or elements inherent to the camera are encountered due to patient movement. The sharpness criterion is the distinction of significant separable elements. For example, microaneurysms, neovessels, and capillaries may be small compared to hemorrhages, exudates, or more significant elements such as venous dilatation, macula, and OD. The field of view is the presence of OD and macula. Some examples of these assessments are shown in Figure 5.

Table 1.
Human visual perception criteria.
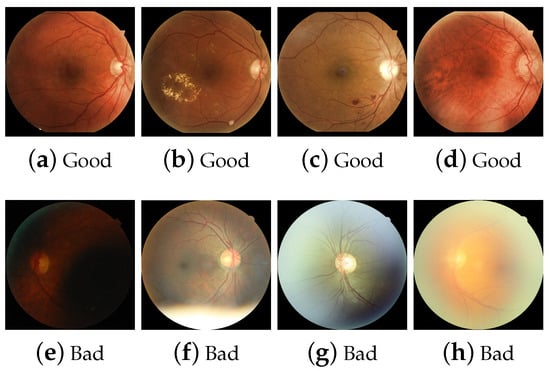
Figure 5.
Examples of human labeling based on visual content considerations, (a) visible OD, macula, and vessels, (b) visible exudates, (c) visible hemorrhages, (d) visible neo-vessels and capillaries, (e) dark image, (f) flash artifact, (g) macula not visible and flash artifact, (h) blurred image.
2.7. Classification
In this work, CNN-based and Transformers-based models are performed to compare the classification scores and the number of parameters for the application. Furthermore, one of its most remarkable contributions is its low computational cost.
2.7.1. CNN-Based Models
In computer vision, architecture models based on CNNs have become relevant to solving object classification, segmentation, and detection problems. In different applications and areas of knowledge, these models have solved automation and optimization problems, comparing their performance in the ImageNet competition [33].
Some of the most popular networks are implemented for classifying images into low and high quality, including Inceptionv3 [34], Resnet [35], and VGG [36]. In addition, transfer learning is carried out on these models with the initialization of ImageNet weights.
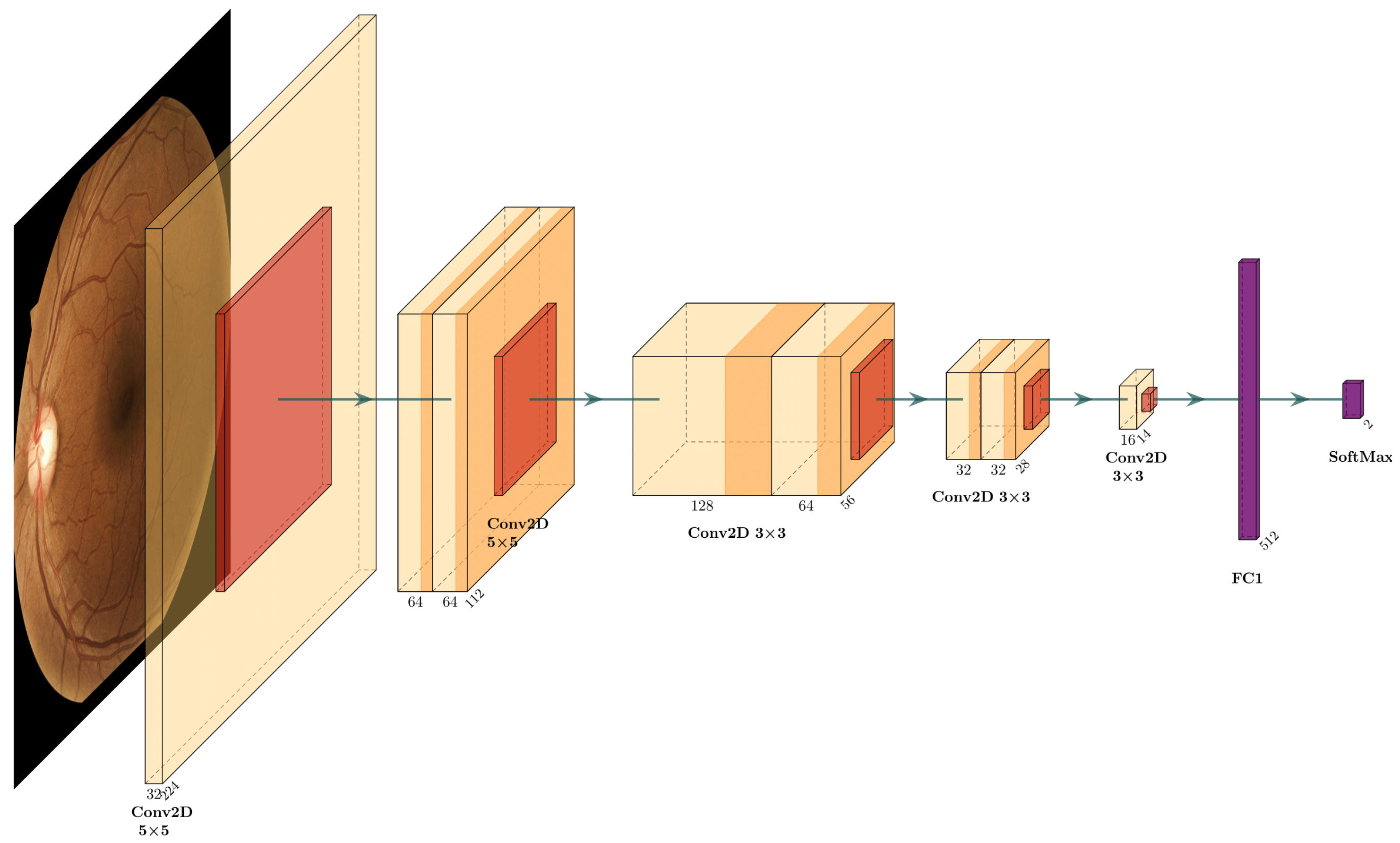
The proposed DRNet-Q architecture is shown in Figure 6, which is separated into three convolutional stages and two fully connected layers. The first stage is the input layer with 325 × 5 filters, with a 2 × 2 zero-padding to maintain the aspect ratio at the output of the convolution and the max-pooling operation. In this layer, it is possible to extract the most general features of the image. The intermediate layers are called double convolution downsampling. These double convolution blocks contain the ReLU activation function for each output, batch normalization, and downsampling with max-pooling. The first block has 64 filters with 5 × 5 kernels and 2 × 2 padding, the second stage of hidden convolutional layers comprises two double convolution blocks of 128, 64, and 32 filters, with a 3 × 3 kernel and a 1 × 1 padding, respectively. The output convolutional layer has 16 filters with a 16 × 7 × 7 shape obtaining 784 features, which are the input of the fully connected layer with 512 perceptron units and ReLU activation function.
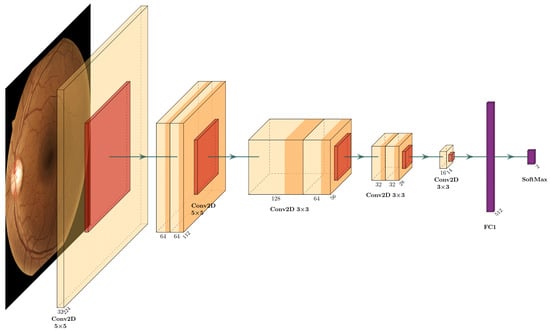
Figure 6.
DRNet-Q proposed CNN architecture.
Finally, the output layer has two perceptron units with a SoftMax probability activation function to classify low-quality (Q0) and high-quality (Q1). As shown in the summary in Table 2, the proposed CNN has the minimum number of trainable parameters with 739,000, facilitating low computational and memory cost application on any DR system.

Table 2.
Detailed DRNet-Q summary.
2.7.2. Self-Attention Transformers
Transformers are a novel paradigm of neural network architectures in natural language processing and computer vision. They are based on Multi-Head Self Attention (MSA) proposed by Vaswani et al. [37]. In this context, Dosovitskiy et al. [38] proposed a Transformer-based image classifier based on translating an image to 16 × 16 words, called tokens, and an MSA encoder. This model surpassed state-of-the-art CNNs on many milestones at ImageNet [33]. However, the model could only effectively solve simple classification tasks and did not perform well in complex vision tasks, such as semantic segmentation and object detection. Liu et al. [39] introduced the Swin Transformer (an acronym for Shifted WINdow transformer) by generating tokens from the window’s MSA operation with a pixel shift before the window partitioning stage. The Swin Transformer architectures are presented in different sizes by the number of parameters, heads, and embedded dimensions. For example, the Swin Tiny version has an embedded dimension of 96 channels, with a layered architecture [2, 2, 6, 2] corresponding to two and six Swin blocks, respectively. On the other hand, the number of MSA modules for each layer is [3, 6, 12, 24]. For comparative purposes, a custom version of Swin is implemented with a 24-channel embedded dimension, [2, 2, 2, 2] architecture layers, and [2, 2, 2, 2] MSA modules.
A new proposal has been introduced by Hassani et al. [40], Neighborhood Attention Transformer (NAT). NAT utilizes a multi-level hierarchical design, similar to Swin [39], meaning that feature maps are downsampled between levels instead of all at once. The Neighborhood Attention Transformer model has different setups with different depths, MSA modules, and embedded channels. For example, the NAT Mini version has a 64-channel embedded dimension with a layered architecture [3, 4, 6, 5]. For comparative purposes, a custom version of NATCustom is trained, which has an embedded dimension of 24 channels, a depth of [2, 2, 2, 2], and [2, 2, 2, 2] MSA modules.
2.8. Object Detection
The detection phase identifies the eye macula and OD in an RFI. Detection is achieved with the YOLOv5 model [41]. YOLOv5 is a compound-scaled model which has fast, compact object detection features. Object detection works by extracting features from input images and then feeding these into a prediction system. It generates boxes around these objects to define what they essentially are. These boxes are drawn around the identified feature and subsequently classified and named. This implementation uses a fine-tuned and trained YOLOv5 specifically for RFI anatomical macula and OD object detection.
The YOLOv5 network has three main segments: the backbone, bottleneck, and head. The backbone is a CNN that collects and generates the image features. The bottleneck phase consists of layers that mix and combine these features into a single output, which is passed onto the prediction stage for further analysis. The head then takes the information to predict the box position and object type.
In the YOLOv5 detector model, these three main segments are trained and configured for RFI, as shown in Figure 7. The CNN modules are based on DenseNET [42], improving performance and the number of parameters required. PANet is the neck of the YOLOv5 detector, which generates feature aggregation.
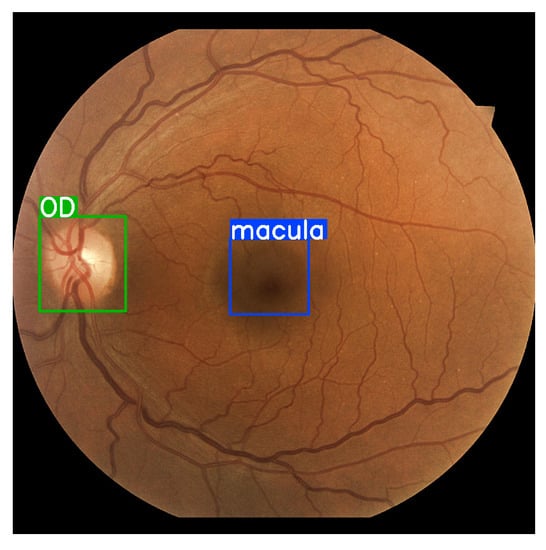
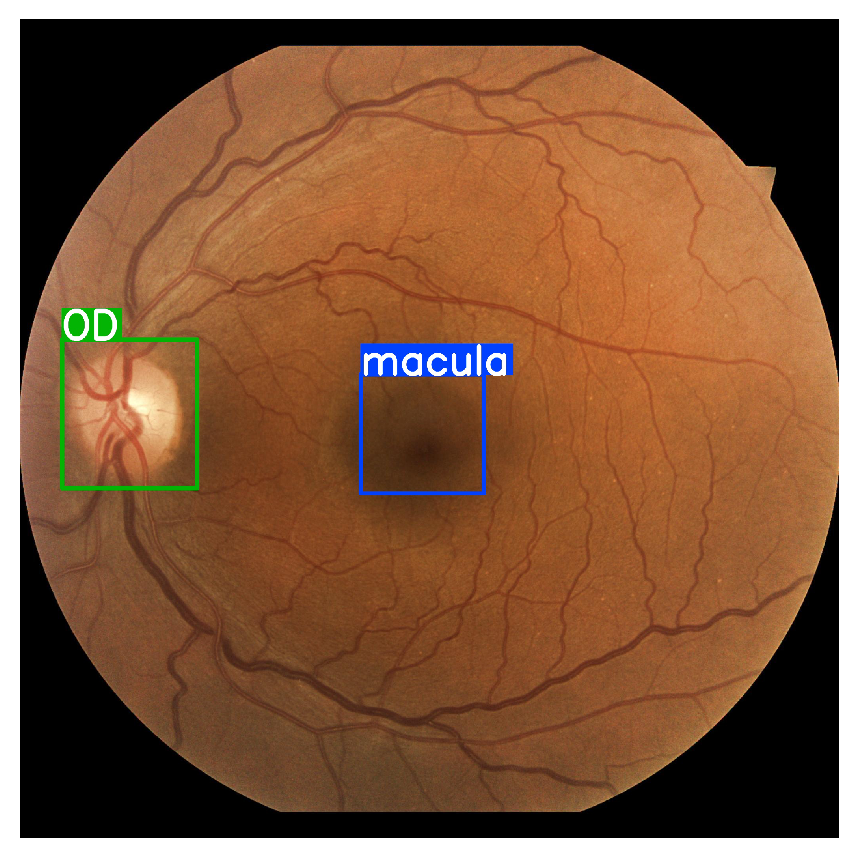
Figure 7.
Anatomic interest areas for object detection macula and optic disc (OD).
3. Results
3.1. Hardware and Resources
All models were trained on the PyTorch [43] framework in python scripting language, on the Ubuntu 20.04 operating system, AMD EPYC 7543 32-Core Processor, 480 GB RAM, 2× A40 NVIDIA Data Center GPU 48 GB each. The Tensorboard library is used to track and manage training. This setup will be valid for the experiments under the “trained under PyTorch” category.
3.2. Pre-Processing
The pre-processing stage consists of removing redundant data. Several neural network models are trained, some based on CNNs and others on Transformers. The amount of data is 420 images separated into a ratio of 80/20, with 336 for training and 84 for validation. The hyper-parameter configuration includes Adam optimizer, learning rate 0.001, 0.9, 0.999, weight decay of 1 × 10, batch size 64, 50 epochs, and a step learning scheduler with a gamma of 0.8 every ten epochs. The loss function used is the binary-cross-entropy with logistics.
Table 3 shows the neural network model training results. In models based on CNN as UNet [29] with 32 filters in the first layer and four layers of depth, the training results obtained are 0.9967 and 0.0167 of accuracy and loss function, respectively, with 467,000 parameters. This result is comparable to the SwinUnet [30] Transformer-based model with 1.7 million parameters, where performance is reduced to 0.9929 and 0.0226 accuracy and loss, respectively. On the other hand, compared with backbone ImageNet CNN models, UNet-Resnet18 [44], UNet-Resnet50 [44], and UNet-VGG16 [44] show slight significant performance improvement compared to the considerable difference in the number of parameters. For example, the UNet-Resnet50 [44] model has 32 million parameters and has a difference in loss function of 0.0016 to its neat reduced version UNet pure.

Table 3.
Training results for semantic segmentation models for pre-processing.
With this, it is clear that a CNN-based UNet model or a Transformer-based SwinUnet with few parameters is sufficient to solve the problem of cropping the region of interest for the RFI at a lower computational cost.
3.3. Standard Features and Quality Evaluator
After the redundant data are removed in the pre-processing stage, the measurement of the standard features is performed on every RFI available. In addition, the images are evaluated by BRISQUE [31] IQE. It is relevant to analyze the relationship between the SF and the IQE.
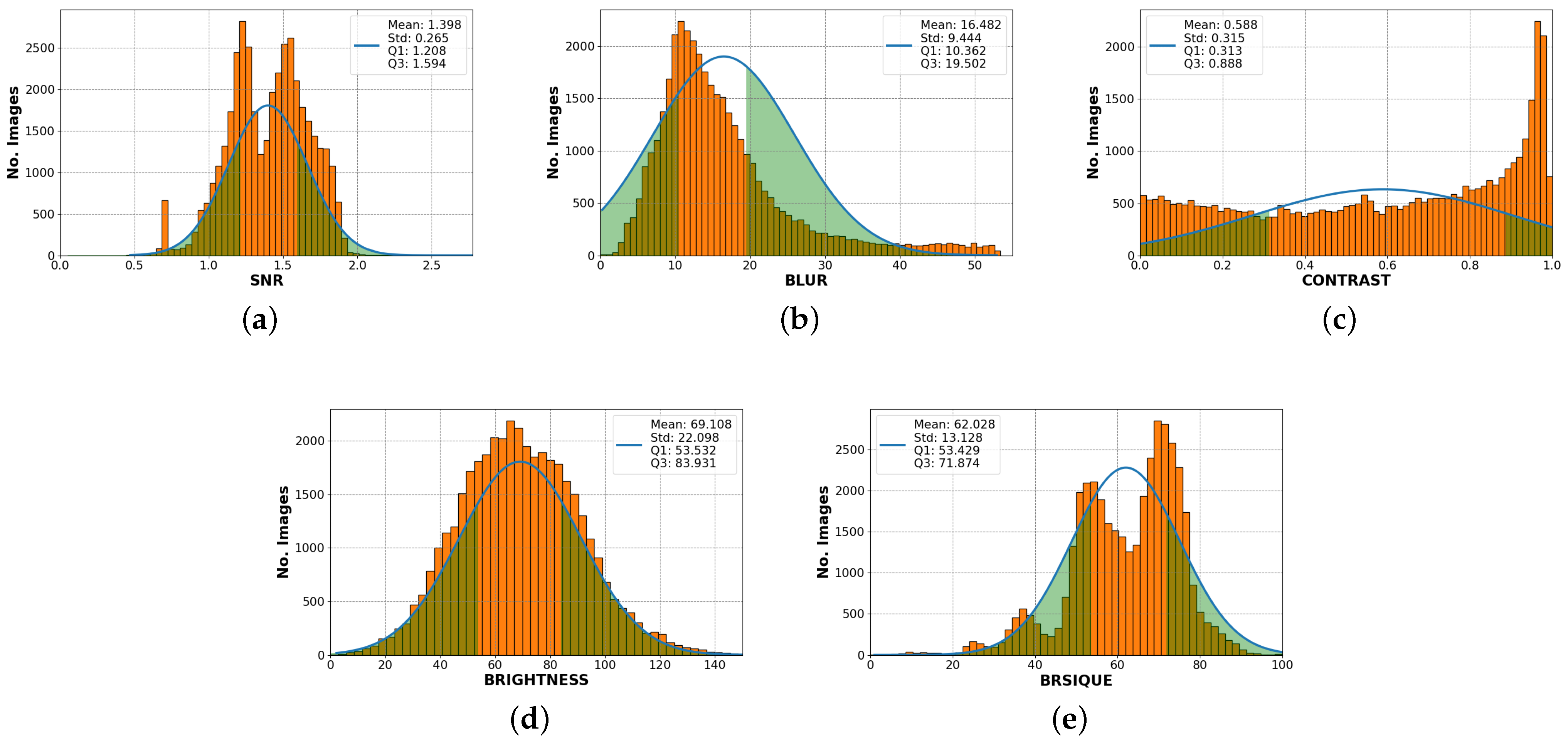
First, it is necessary to analyze data distribution measurements. Figure 8 shows the histogram for each SF and the BRISQUE evaluation score distribution. A comparison is made with a Gaussian fit, with the respective values mean, standard deviation, quartile 1 (Q1), and quartile 3 (Q3) values. On the other hand, the signal-to-noise ratio distribution is greater than 1. There is a greater presence in Q1 at 1.208 and another essential set is in Q3 at 1.594. The mean is 1.398, and the standard deviation is 0.265. In the case of the blur coefficient, the peak of the histogram is close to the mean of 16.482, the standard deviation is 9.444, and the result of this feature indicates that a significant amount of data has a value between and . In addition, the brightness level histogram distribution has a Gaussian fit, which means that it is probable to find images with balanced brightness levels. Additionally, image distribution by contrast level is uniform with a standard deviation value of 0.315 and a very high peak near 1, which corresponds to low-contrast images. This also means that the available dataset has a wide range of image contrasts. Finally, in the BRISQUE evaluation score distribution, two peaks are found in Q1 53.429 and Q3 71.874, with a mean of 64.805 and a standard deviation of 13.128, indicating a significant number of better quality images in Q1 and a significant number of lower quality images score grouped in Q2.
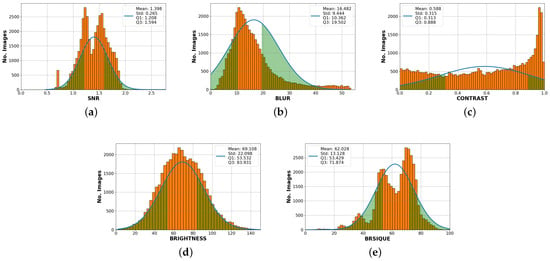
Figure 8.
Histogram display of each standard feature fitted to Gaussian curve. (a) The signal-to-noise ratio has two peaks in quartile 1 (Q1) and quartile 3 (Q3), with a mean of 1.398. (b) Blur has a high element concentration in Q1 with 10,362. (c) The contrast has a uniform distribution. (d) The brightness fits the Gaussian distribution. (e) BRISQUE has two peaks in Q1 and Q3. However, the highest concentration of elements is below the mean.
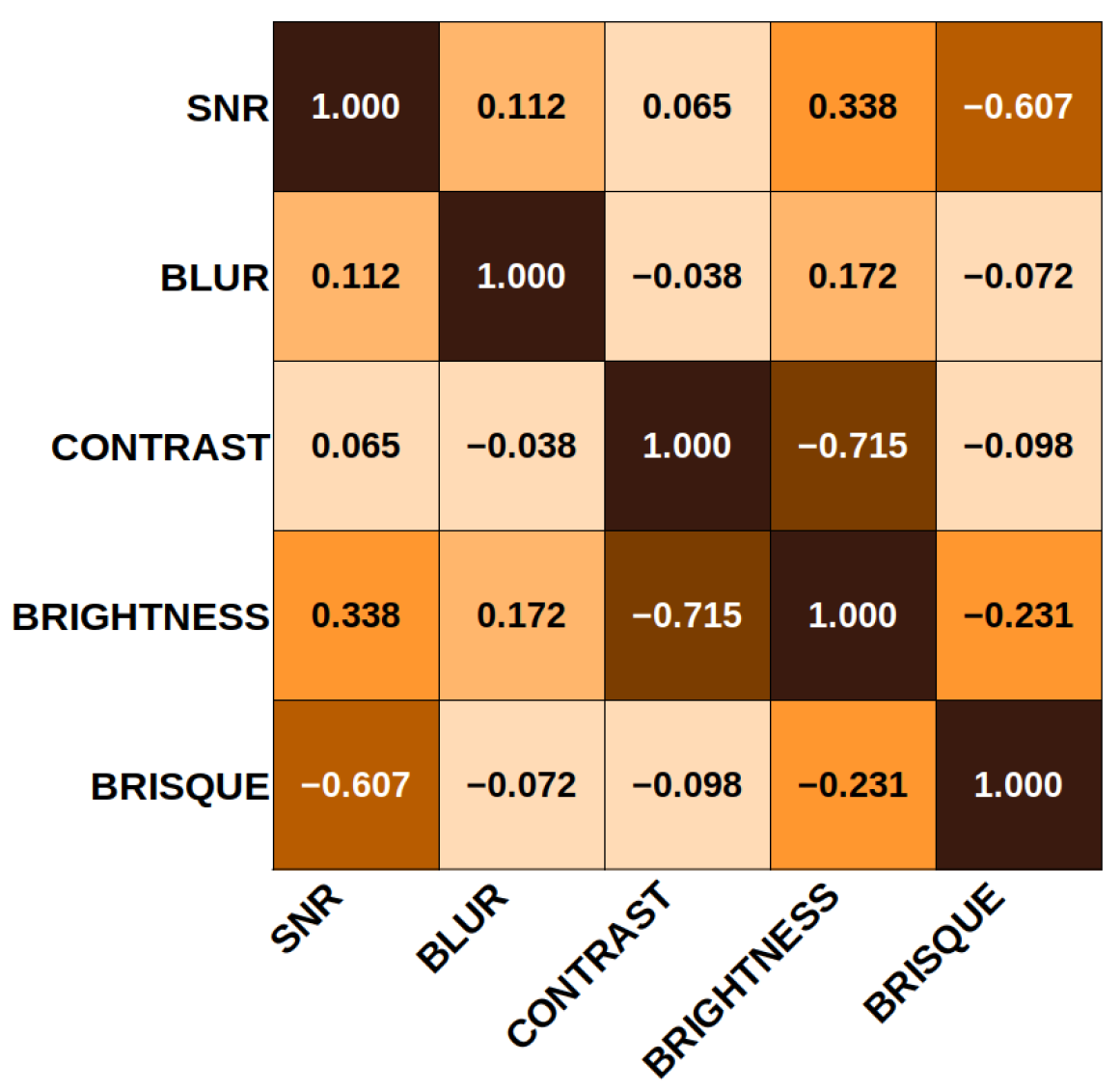
In that context, the correlation coefficients obtained from the correlation matrix are shown in Figure 9 to clarify the relationship between the SF and the quality evaluation score. It can be seen that the elements that correlate most closely are brightness and contrast. This result is expected, since the luminance and the contrast are related. In addition, the evaluation in BRISQUE is inversely related to the noise. By increasing the SNR value of an image, the BRISQUE score tends to decrease, translating into better quality. On the other hand, the feature least related to the others is the blur coefficient, which depends directly on the number of visible edges in the image and not on noise, contrast, or brightness levels.
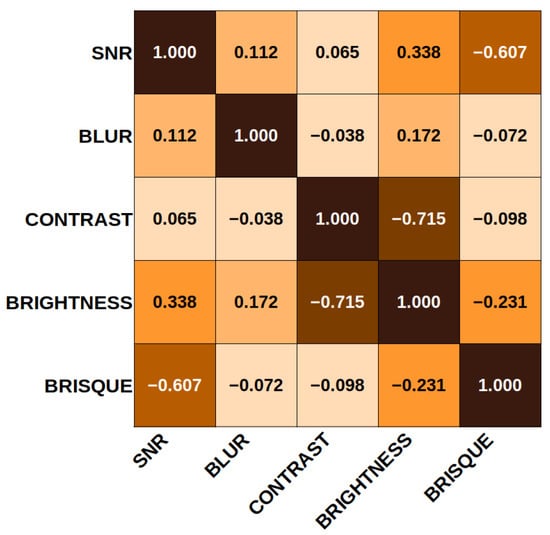
Figure 9.
Standard feature correlation matrix. The main diagonal corresponds to 1. Contrast and brightness are inversely related in proportion. The minor related features contrast vs. blur and contrast vs. SNR. There is an inverse relationship between SNR and BRISQUE.
3.4. KMeans
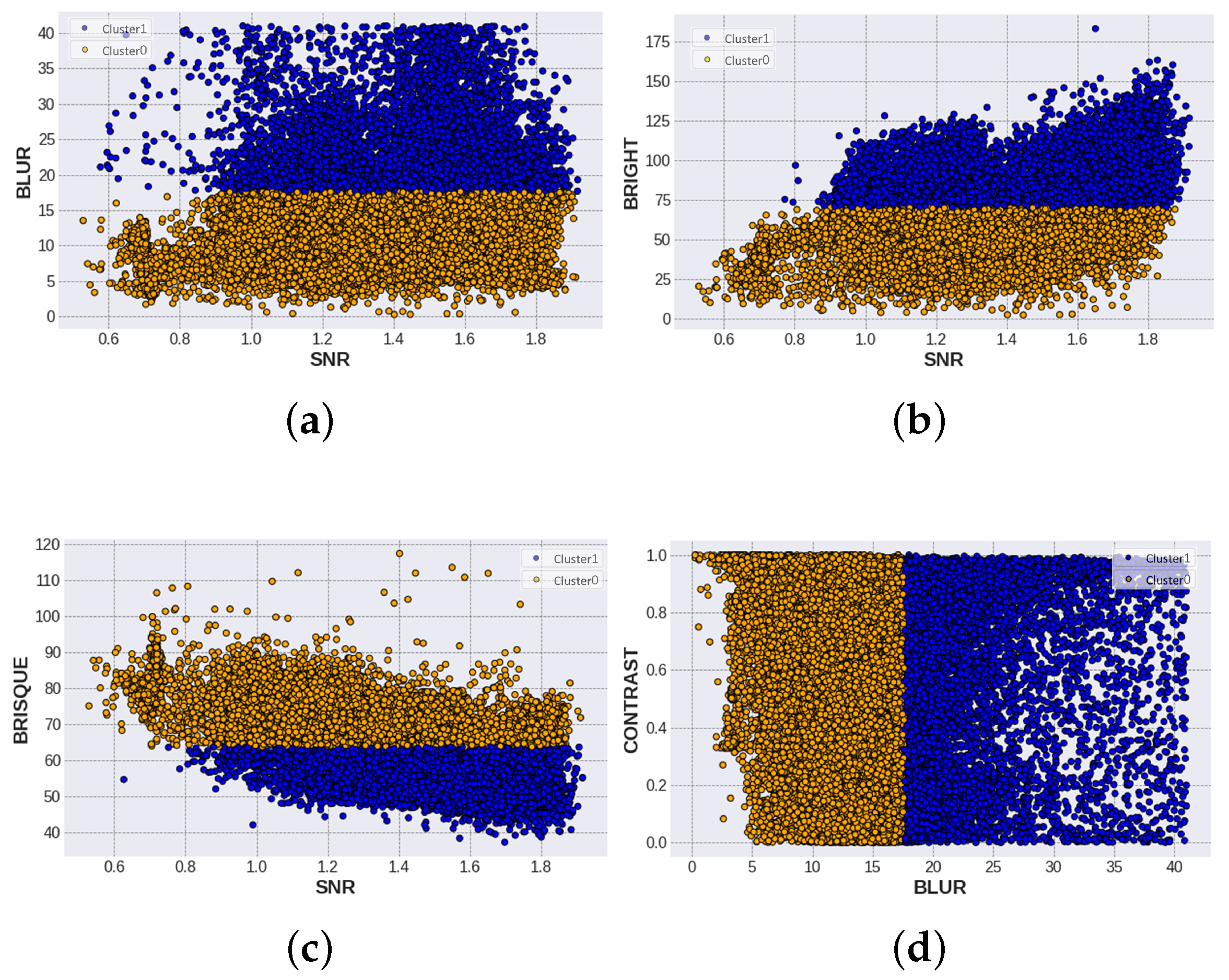
Once the SF and evaluation scores have been obtained, the KMeans cluster algorithm is used to find two groups with similar properties. The result of this algorithm is shown in Figure 10, where the following comparisons are possible: SNR vs. blur, SNR vs. bright, SNR vs. brisque, and blur vs. contrast. On the other hand, centroid values for each feature are shown in Table 4.
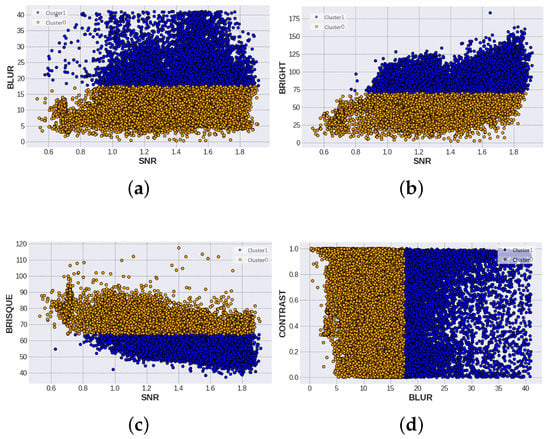
Figure 10.
Comparison of standard features according to their cluster label, cluster 0 and cluster 1. (a) SNR vs BLUR comparison with a separation value in BLUR. (b) SNR vs BRIGHTNESS comparison with value in BRIGHTESS. (c) SNR vs BRISQUE division in BRISQUE. (d) BLUR vs CONTRAST with threshold in BLUR.

Table 4.
KMeans centroids.
In the case of SNR vs. blur, the blue Cluster 1 corresponds to images with the blur value >17. It can also be seen that most of the data in Cluster 1 are found with SNR values greater than 1. So, the interpretation of this comparison is that less blurred and noisy images are grouped in Cluster 1 and tend to be of good quality. In addition, the SNR vs. BRISQUE plot clusters are separated by the BRISQUE value <75. By definition, the lower the threshold value, the better the quality of the images, so Cluster 1 inherits this characteristic. Nevertheless, the SNR vs. brightness graph separates bright and dark images, with the center at 75. Therefore, this separation is not as reliable as looking for images with a balanced brightness level. For this reason, it is necessary to perform the human equitation stage and discard those outlier images. In addition, the comparison between contrast and blur is displayed. A separation of clusters by the blur value can be seen. Like the first plot, values greater than 17 correspond to Cluster 1, and because the dataset has a uniform distribution of contrast levels, it is reflected in this result.
The clustering was performed on the entire available data set. The ratio between RFIs belonging to the low-quality and high-quality groups was obtained according to the graph above. Table 5 shows the result of the images labeled according to the group assigned by KMeans. K0 and K1 correspond to pre-selection low-quality and high-quality images, respectively. The MESSIDOR, IDRID, DRIMDB, and DRIVE datasets are primarily good quality images, since these images are obtained under controlled conditions and widely used in the state-of-the-art of diabetic retinopathy grading and segmentation tasks. On the other hand, the dataset of EyePACS and APTOS has a large amount of data with a more significant amount for both types of images.

Table 5.
Distribution of data sources. The K-labels are generated by KMeans clustering.
3.5. Classification
In this step, different models are trained to compare classification performance. There are models with initialization of ImageNet weights used for different applications in the state-of-the-art. These models are characterized by outstanding classification performance, some several parameters, which translates into high hardware requirements. On the other hand, we have the proposed CNN model in Table 2, a reduced version of a Swin [39], and another custom version of NAT [40] with the lowest number of parameters possible.
Once the SF measurements are obtained, the quality score and the labeling of the elements by clustering are obtained, and a sample of elements is taken to carry out labeling according to human opinion. The result of this step contains 2000 elements per class from the EyePACS [22] dataset.
The training is performed with the same data described in Table 6. The separation of the data is shown using the hold-out cross-validation technique, with a separation of 70% for training, 15% for validation, and 15% for testing. H0 corresponds to images of Cluster 0 and labeled as low quality by humans, and H1 is Cluster 1 and high quality by human opinion. In addition, some on-fly random data augmentation techniques have been applied. These include horizontal and vertical flips with probabilities of 0.5 for each one, rotations from −20° to 20°, and random contrast levels.

Table 6.
Human opinion labeling description and hold-out strategy for training dataset.
The hyper-parameters configuration consists of an Adam optimizer used with a learning rate of 0.001, 1 0.8, 2 0.999, weight decay of 1 × 10, 64 batch size, and 50 epochs. The loss function is Cross-Entropy, with a step learning scheduler to get out of possible local minima, with a gamma of 0.8 every ten epochs. The training under the PyTorch setup applies for this training stage.
The evaluation metrics of the models provide performance information for the classification. The average accuracy of all batches is computed during training and validation. In addition, the recall or sensitivity (Equation (8)) and specificity (Equation (9)) are calculated for the classification evaluation step. Finally, the characteristic curve area under the curve (AUC) is calculated as a classification performance metric.
where represents the true positives classification, represents the false positives, represents the false negatives, and represents the true negatives.
In Table 7, the results of the training and evaluation of the different classification models are shown. A similar performance between each model can be noted. For example, the model with the best result in the loss function is the NAT [40] Custom with a minimum value of 0.0029 in validation. It has a sensitivity of 0.9833 that ties with the transformer Swin [39]. For the case of specificity, the model proposed in this work DRNet-Q, VGG13 [36], and Resnet18 [35] has a result of 0.9833. Finally, the best result obtained from the area under the curve is the Resnet50 [35] model. Moreover, the difference between the results does not exceed 2% for the model proposed in this work. DRNet-Q has 740,000 parameters which, compared to the bigger models VGG13 [36] and InceptionV3 [34], is about 99.5% and 97% smaller, respectively. On the other hand, the custom transformer models NAT and Swin have similar performance with the same approximate number of parameters. With these results, the model proposed in this work offers the same performance as excellent state-of-the-art models [34,35,36,39,40], with a minimum number of parameters, which translates into a lower computational cost than these.

Table 7.
Models classification results.
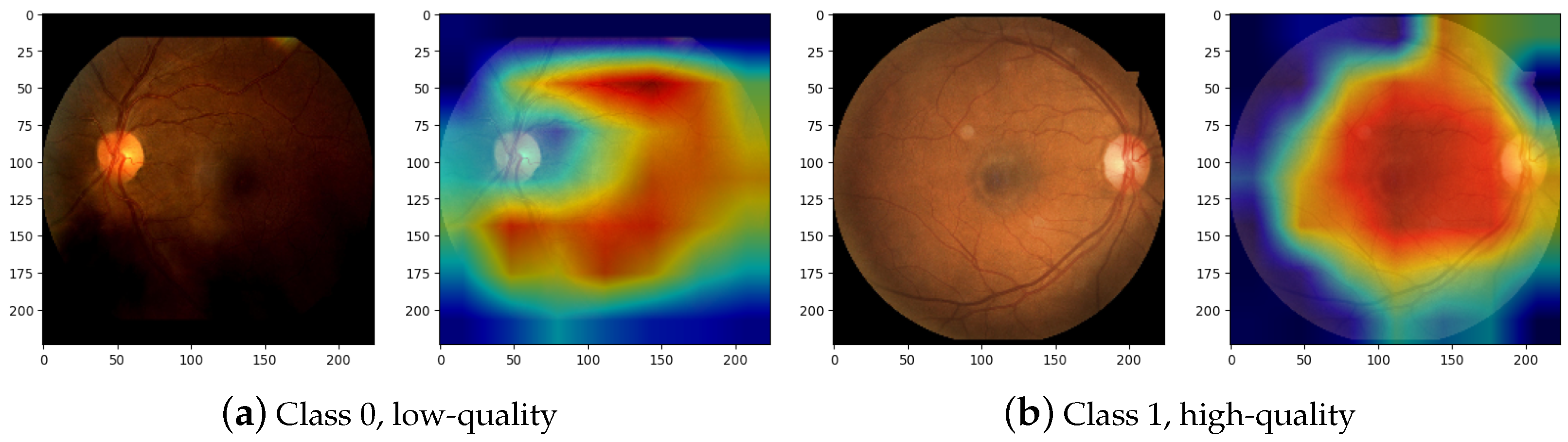
A point of interest in the training of CNNs is determining which visual elements are most relevant to classify an image by extracting its features. Therefore, the GradCAM [45] is applied to the last layer of DRNet-Q feature extraction, for example, class 0 for low quality and class 1 for high quality. The result of displaying the gradients in an image of the test dataset is shown in Figure 11. In Figure 11a, class 0 RFI classification is presented, where the gradient map shows an interest in the lower area of the image that corresponds to darkened and undesirable areas in RFIs. On the other hand, Figure 11b, where class 1 images are classified, shows an example with apparent good quality, and the CNN classification of a mapping of the main inner retinal elements are shown, which include the OD, macula, arcades, neovessels, and fovea. In that sense, it is safe to say that DRNet-Q classifies between low-quality and high-quality images according to their visible features.
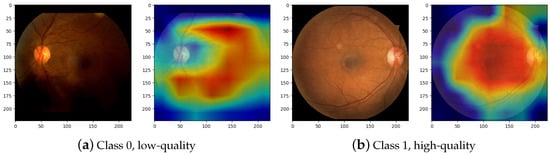
Figure 11.
Gradient map visualization for quality classification. Class 0 (a) corresponds to the low-quality classification according to DRNet-Q, and Class 1 (b) is the high-quality classification according to the CNN model.
3.6. Object Detection
EyePACS [22], the data used for this step, are taken from the label-1 classification dataset with 640 × 640 × 3 resolution and annotated manually by the macula and OD with a bound box localization. The data are separated using the cross-validation technique, with a ratio of 80% for training and 20% for validation, as shown in Table 8.

Table 8.
Human opinion labeling description and hold-out strategy for training dataset.
Several models are trained under the PyTorch setup. The hyper-parameter configuration includes an SGD optimizer, learning rate of , momentum of 0.937, weight decay of 5 × 10, 16 batch size, and 100 epochs. The loss function calculated is the multi-class mean average precision for the validation step shown in Equation (10):
where is the predicted probability value, y is the ground truth manual annotations, C is the number of classes, and AP is the average precision for a class defined in Equation (11)
where is the prediction sensitivity (8) and is the classification precision value expressed in Equation (12):
where and are the true positive and false positive rates.
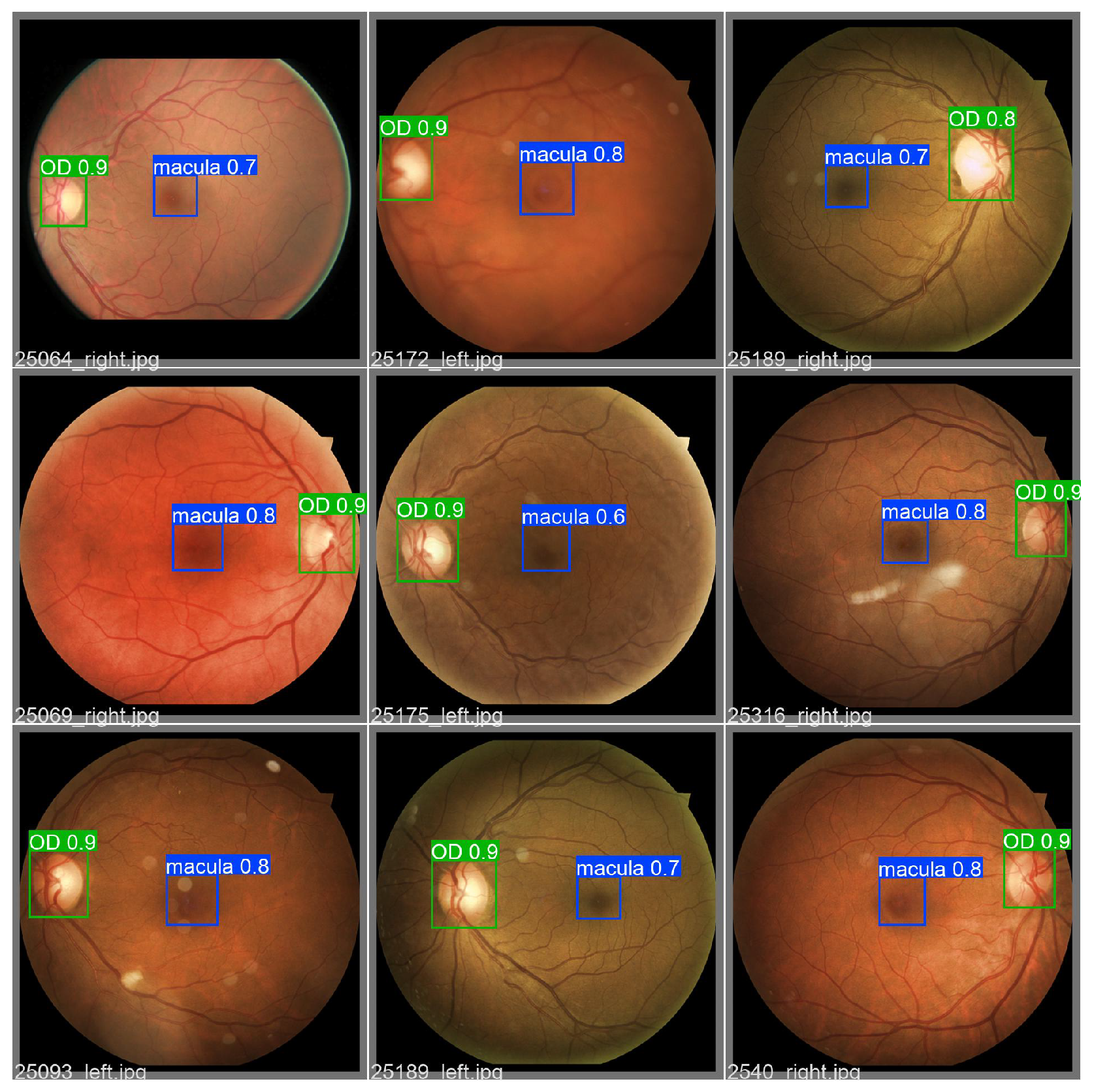
The results obtained are shown in Table 9. The training achieves 0.967 mAP as the best value for the model with 46.5 million parameters, 107.8 GFLOPs, and 10.1 ms of inference time. Moreover, the YOLOv5 nano version with 1.9 million parameters achieves 0.945 with 4.2 GFLOPs and 6.4 ms of inference time. In addition, an example of a validation bath is shown in Figure 12, showing the macula and the OD bounding boxes.

Table 9.
Object detector training results.
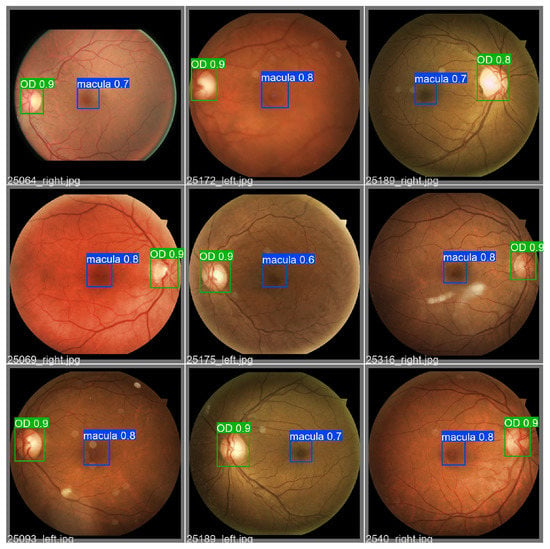
Figure 12.
Set of 9 validation RFIs object detection results obtained by the YOLOv5 model. The macula and OD are detected with their respective confidence value.
3.7. Suitable RFI for DR
For an RFI image to be suitable, it must be labeled as Q1 (high-quality image) from the CNN classification model and have positive detection of the anatomical elements (macula and OD). Table 10 presents the number of images labeled as Q0 or Q1 along with object detection labels. These results are obtained using both trained DL models (DRNet-Q and YOLOv5 nano). It is important to note that these models have the lowest number of trainable parameters. Thus, the training and inference time is faster than the other methods.

Table 10.
Number of suitability for each public dataset. Number of images where the Macula and Optic Disk (OD) are detected. The low-quality (Q0) and high-quality (Q1) RFI images.
Once the labels have been obtained on all available datasets using the CNN, the result can be compared with the labels obtained in the clustering stage. Table 11 compares the total available images labeled by KMeans (true) K0 and K1, as well as the result of DRNet-Q (prediction) Q0 and Q1. In addition, the accuracy, sensitivity, and specificity, which correspond to the CNN classification performance, are shown. It is important to note that the training was performed on data obtained from KMeans and validated by a human observer. Therefore, the result of 0.9523 in accuracy is similar to that obtained with the training, validation, and test datasets.

Table 11.
CNN performance compared to KMeans.
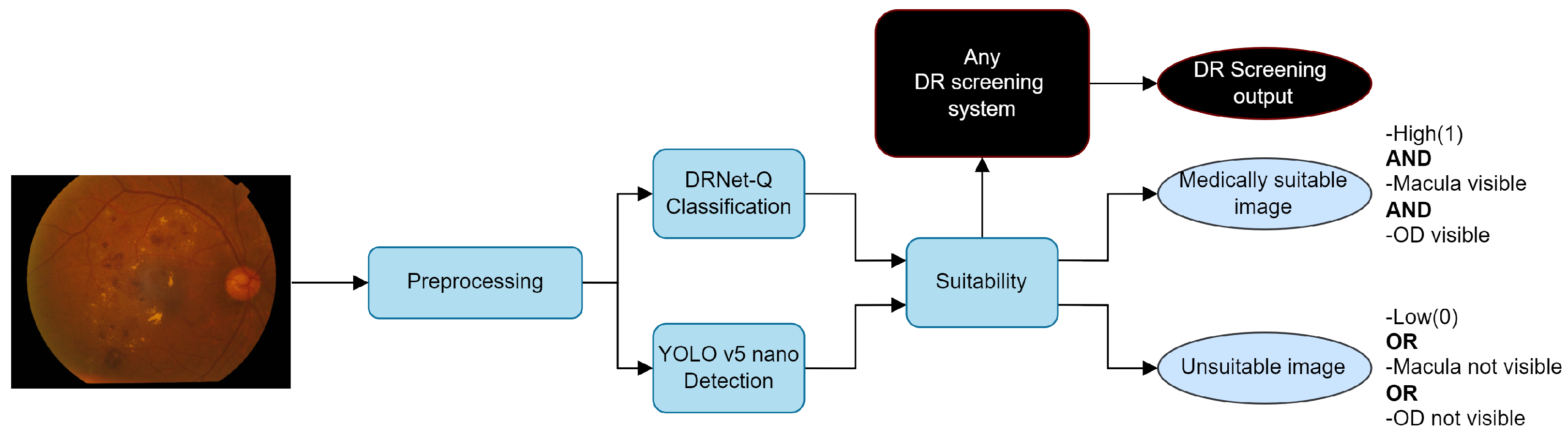
Figure 13 shows an integration of the proposed work into DR detection systems. The system obtains the medical suitability label with any given input that goes through pre-processing, quality, and anatomical detection. Just as in Dai et al. [46], the quality function provides feedback to the operator to identify quality problems in an image, thus reducing the rate of ungradable RFIs and increasing DR screening efficiency.
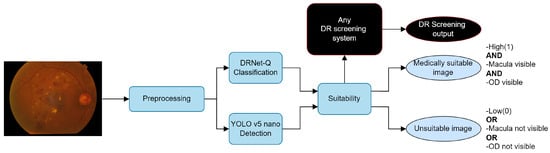
Figure 13.
Suitability integration example in a DR system using RFIs.
4. Discussion
According to this study, the RFI suitability could be assessed by quality metrics and a field definition of human perception, considering the essential anatomical features of DR. In this work, two models were trained: the first one classifies between low- and high-quality RFI, and the second one detects anatomical objects. Nevertheless, there may be degradations due to retinal anatomy due to conditions such as DR. Thus, the impact of the quality level for each DR level can be evaluated.
The ICO [2] as No Apparent Diabetic Retinopathy (NADR), Non-Proliferative Diabetic Retinopathy (NPDR), moderate NPDR, severe NPDR, and Proliferative DR4 [2,11], labeled from DR0 to DR4, respectively.
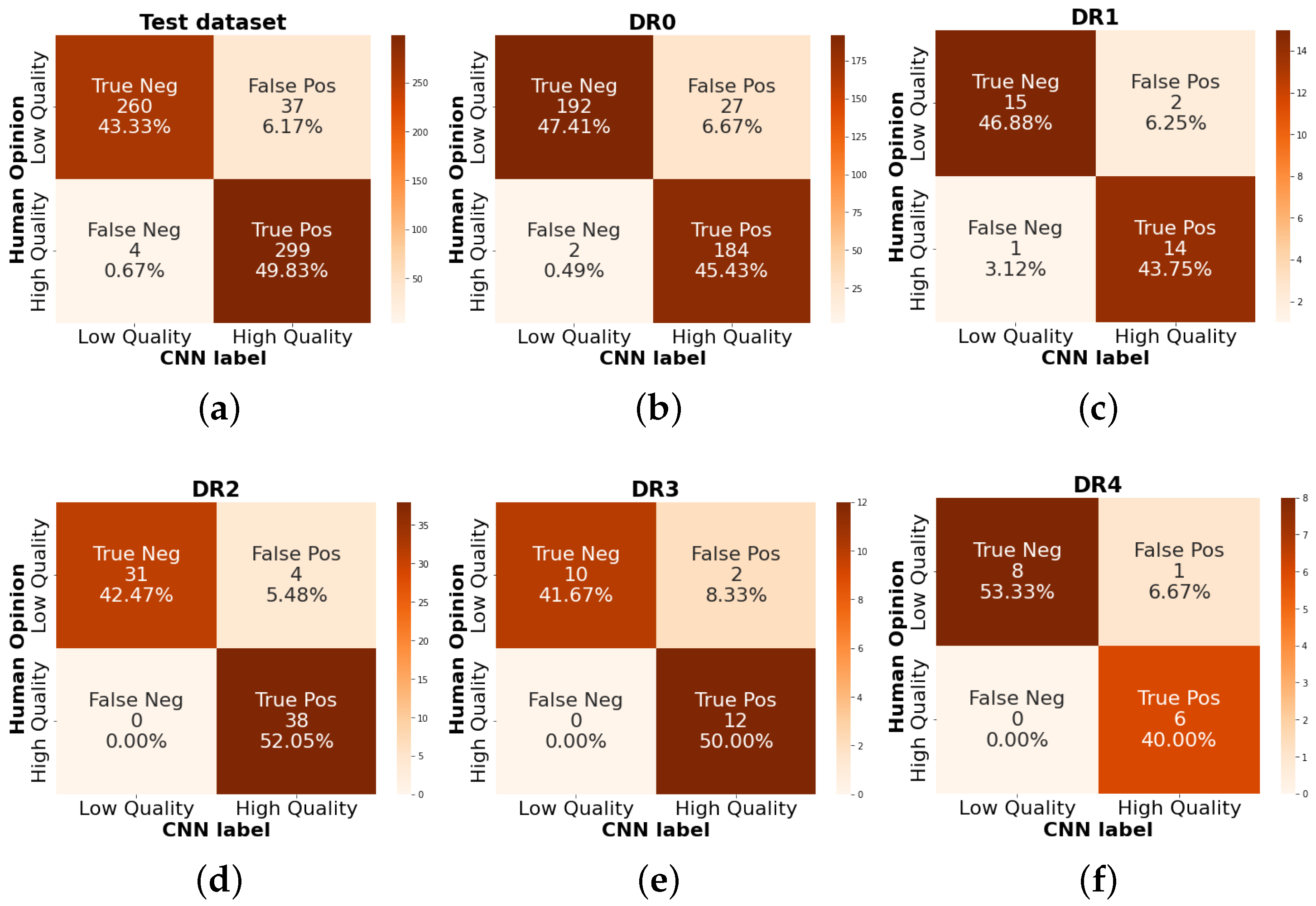
Figure 14 shows the number of images classified by CNN as low (0) and high (1) quality compared to human opinion. There are 300 images per class, with a total of 600. The DR0 grade has 405 examples, making up 67% of the dataset total since, in the available data, the vast majority correspond to patients with NADR. Of these examples, 45% correspond to images classified as high quality and 47.41% correspond to images classified as low quality. Otherwise, the number of images corresponding to DR4 is 15, of which eight correspond to low-quality images and six correspond to high-quality images. This may indicate that at a higher grade of DR, the quality level of the images may be lower due to different conditions in patients, such as hemorrhages or retinal detachment, which correspond to outliers of retinal anatomy.
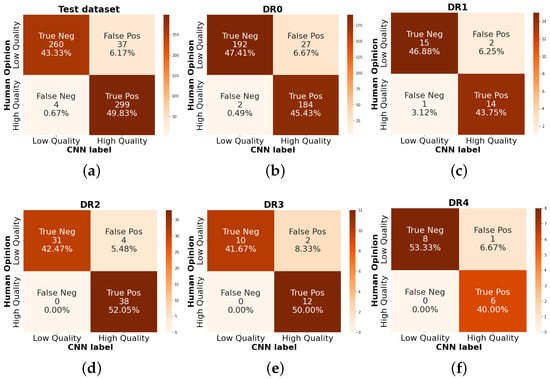
Figure 14.
Quality rating confusion matrix separated by DR grading. (a) It is the total of the test dataset 600 items, (b) 405 RFIs with DR grade 0, (c) 32 with DR1, (d) 73 items for DR2, (e) DR grade with 24 images and (f) 15 for proliferative DR.
5. Conclusions
This article presents a methodology for labeling RFI for quality classification. The clustering of Image Quality Standard Features and an Image Quality Evaluator has been combined in order to obtain pre-selected labeling. Additionally, these clusters have been inspected by a human observer taking into account specific anatomical features, allowing for the separation into label-0 (low) and label-1 (high) quality classifications. Furthermore, a novel lightweight CNN is proposed, which was designed specifically for low computational cost binary classification with 739,000 parameters. The training results are comparable to the state-of-the-art CNNs widely used in deep learning applications. The implemented architecture achieves a classification performance of 98.6% AUC, 97.66% sensitivity, and 98.33% specificity, respectively. The medical suitability of a retinal image is enhanced by implementing a Deep Learning object detection model. In order to find the optic disc and the macular region in RFI, an object detection model was trained, obtaining 94.5% of the mAP metric. In addition, the proposed approach (classification and detection) can be integrated into any automatic RFI workflow avoiding data drift or the out-of-distribution data problem. Remarkably, the models trained and implemented for suitability have low computational costs; this allows integration in any RFI analysis system for DR.
Author Contributions
Conceptualization, G.P.-D. and E.U.M.-S.; methodology, G.P.-D. and A.S.; software, G.P.-D. and A.S.; validation, E.U.M.-S., S.O.-C. and J.R.; formal analysis, G.P.-D. and E.U.M.-S.; investigation, S.O.-C. and F.J.R.-N.; data curation, G.P.-D. and A.S.; writing, review and editing, P.M.-A. and J.R.; visualization, F.J.R.-N.; supervision, S.O.-C.; project administration, S.O.-C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by CONACYT, grant number 923969.
Informed Consent Statement
The data used are for public use; thus, the study subjects are anonymous.
Data Availability Statement
The data that support the findings of this study are openly available in https://github.com/alonso59/fundus_suitable (accessed on 29 May 2022) https://drive.google.com/drive/folders/1_eRt44yiX_WTrSNv-K2bRFMnpI_xNBuY?usp=sharing (accessed on 29 May 2022). Tensorboard pre-processing: https://tensorboard.dev/experiment/XwlgpDp6QvqCxGNiBCDc1w/ (accessed on 29 May 2022); Tensorboard classification: https://tensorboard.dev/experiment/5J0QZJx4ToayKDd0REfgQA/ (accessed on 29 May 2022); Tensorboard detection: https://tensorboard.dev/experiment/aUpKg0aZSPCEBioXocYOww (accessed on 29 May 2022).
Acknowledgments
Special thanks to Cinvestav Unidad Guadalajara and the artificial intelligence department of the Government of Jalisco for their technological and administrative support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Teo, Z.L.; Tham, Y.C.; Yu, M.; Chee, M.L.; Rim, T.H.; Cheung, N.; Bikbov, M.M.; Wang, Y.X.; Tang, Y.; Lu, Y.; et al. Global Prevalence of Diabetic Retinopathy and Projection of Burden through 2045: Systematic Review and Meta-analysis. Ophthalmology 2021, 128, 1580–1591. [Google Scholar] [CrossRef] [PubMed]
- Guidelines for Diabetic Eye Care; International Council Ophthalmology: Brussels, Belgium, 2017.
- Cole, E.D.; Novais, E.A.; Louzada, R.N.; Waheed, N.K. Contemporary retinal imaging techniques in diabetic retinopathy: A review. Clin. Exp. Ophthalmol. 2016, 44, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Xue, D.; Feng, X. Automatic Diabetic Retinopathy Grading via Self-Knowledge Distillation. Electronics 2020, 9, 1337. [Google Scholar] [CrossRef]
- Paradisa, R.H.; Bustamam, A.; Mangunwardoyo, W.; Victor, A.A.; Yudantha, A.R.; Anki, P. Deep feature vectors concatenation for eye disease detection using fundus image. Electronics 2022, 11, 23. [Google Scholar] [CrossRef]
- Fan, R.; Liu, Y.; Zhang, R. Multi-Scale Feature Fusion with Adaptive Weighting for Diabetic Retinopathy Severity Classification. Electronics 2021, 10, 1369. [Google Scholar] [CrossRef]
- Pham, Q.T.; Ahn, S.; Song, S.J.; Shin, J. Automatic drusen segmentation for age-related macular degeneration in fundus images using deep learning. Electronics 2020, 9, 1617. [Google Scholar] [CrossRef]
- Ruamviboonsuk, P.; Tiwari, R.; Sayres, R.; Nganthavee, V.; Hemarat, K.; Kongprayoon, A.; Raman, R.; Levinstein, B.; Liu, Y.; Schaekermann, M.; et al. Real-time diabetic retinopathy screening by deep learning in a multisite national screening programme: A prospective interventional cohort study. Lancet Digit. Health 2022, 4, e235–e244. [Google Scholar] [CrossRef]
- Ting, D.S.W.; Cheung, C.Y.L.; Lim, G.; Tan, G.S.W.; Quang, N.D.; Gan, A.; Hamzah, H.; Garcia-Franco, R.; San Yeo, I.Y.; Lee, S.Y.; et al. Development and Validation of a Deep Learning System for Diabetic Retinopathy and Related Eye Diseases Using Retinal Images From Multiethnic Populations With Diabetes. J. Am. Med. Assoc. 2017, 318, 2211–2223. [Google Scholar] [CrossRef] [PubMed]
- Beede, E.; Baylor, E.; Hersch, F.; Iurchenko, A.; Wilcox, L.; Ruamviboonsuk, P.; Vardoulakis, L.M. A Human-Centered Evaluation of a Deep Learning System Deployed in Clinics for the Detection of Diabetic Retinopathy. In Proceedings of the 2020 CHI Conference on Human Factors in Computing Systems, Honolulu, HI, USA, 25–30 April 2020; pp. 1–12. [Google Scholar]
- Gonzalez-Briceno, G.; Sanchez, A.; Ortega-Cisneros, S.; Garcia Contreras, M.S.; Pinedo Diaz, G.A.; Moya-Sanchez, E.U. Artificial Intelligence-Based Referral System for Patients With Diabetic Retinopathy. Computer 2020, 53, 77–87. [Google Scholar] [CrossRef]
- Dodge, S.; Karam, L. Understanding how image quality affects deep neural networks. In Proceedings of the 2016 8th International Conference on Quality of Multimedia Experience (QoMEX), Lisbon, Portugal, 6–8 June 2016; pp. 1–6. [Google Scholar] [CrossRef]
- Zhai, G.; Min, X. Perceptual image quality assessment: A survey. Sci. China Inf. Sci. 2020, 63, 1–52. [Google Scholar] [CrossRef]
- Kamble, V.; Bhurchandi, K.M. Optik No-reference image quality assessment algorithms: A survey. Optik 2015, 126, 1090–1097. [Google Scholar] [CrossRef]
- Niu, Y.; Zhong, Y.; Guo, W.; Shi, Y.; Chen, P. 2D and 3D Image Quality Assessment: A Survey of Metrics and Challenges. IEEE Access 2019, 7, 782–801. [Google Scholar] [CrossRef]
- Stępień, I.; Oszust, M. A Brief Survey on No-Reference Image Quality Assessment Methods for Magnetic Resonance Images. J. Imaging 2022, 8, 160. [Google Scholar] [CrossRef]
- Xu, Z.; Zou, B.; Liu, Q. A Deep Retinal Image Quality Assessment Network with Salient Structure Priors. arXiv 2020, arXiv:2012.15575. [Google Scholar]
- Pires Dias, J.M.; Oliveira, C.M.; da Silva Cruz, L.A. Retinal image quality assessment using generic image quality indicators. Inf. Fusion 2014, 19, 73–90. [Google Scholar] [CrossRef]
- Yao, Z.; Zhang, Z.; Xu, L.Q.; Fan, Q.; Xu, L. Generic features for fundus image quality evaluation. In Proceedings of the 2016 IEEE 18th International Conference on e-Health Networking, Applications and Services (Healthcom), Munich, Germany, 14–17 September 2016; pp. 1–6. [Google Scholar] [CrossRef]
- Yu, F.; Sun, J.; Li, A.; Cheng, J.; Wan, C.; Liu, J. Image quality classification for DR screening using deep learning. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju Island, Korea, 11–15 July 2017; pp. 664–667. [Google Scholar] [CrossRef]
- Zago, G.T.; Andreão, R.V.; Dorizzi, B.; Teatini Salles, E.O. Retinal image quality assessment using deep learning. Comput. Biol. Med. 2018, 103, 64–70. [Google Scholar] [CrossRef]
- “EyePACS, Diabetic Retinopathy Detection competition” Kaggle. 2015. Available online: https://www.kaggle.com/c/diabetic-retinopathy-detection/ (accessed on 2 May 2022).
- Clinic, E. Diabetic Retinopathy Segmentation and Grading Challenge. 2012. Available online: https://idrid.grand-challenge.org/Data/ (accessed on 2 May 2022).
- Decencière, E.; Zhang, X.; Cazuguel, G.; Lay, B.; Cochener, B.; Trone, C.; Gain, P.; Ordonez, R.; Massin, P.; Erginay, A.; et al. Feedback on a publicly distributed database: The Messidor database. Image Anal. Stereol. 2014, 33, 231–234. [Google Scholar] [CrossRef]
- 4th Asia Pacific Tele-Ophthalmology Society (APTOS) Symposium. Available online: https://www.kaggle.com/competitions/aptos2019-blindness-detection/overview/description (accessed on 2 May 2022).
- Staal, J.; Abramoff, M.; Niemeijer, M.; Viergever, M.; van Ginneken, B. Ridge based vessel segmentation in color images of the retina. IEEE Trans. Med. Imaging 2004, 23, 501–509. [Google Scholar] [CrossRef]
- Sevik, U. DRIMDB (Diabetic Retinopathy Images Database) Database for Quality Testing of Retinal Images. 2014. Available online: https://academictorrents.com/details/99811ba62918f8e73791d21be29dcc372d660305 (accessed on 2 May 2022).
- Briceno, G.G.; Sánchez, A.; Sánchez, E.U.M.; Cisneros, S.O.; Pinedo, G.; Contreras, M.S.G.; Castillo, B.A. Automatic cropping of retinal fundus photographs using convolutional neural networks. Res. Comput. Sci. 2020, 149, 161–167. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Munich, Germany, 5–9 October 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer: Cham, Switzerland; pp. 234–241. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-Unet: Unet-like Pure Transformer for Medical Image Segmentation. arXiv 2021, arXiv:2105.05537. [Google Scholar]
- Mittal, A.; Moorthy, A.K.; Bovik, A.C. No-Reference Image Quality Assessment in the Spatial Domain. IEEE Trans. Image Process. 2012, 21, 4695–4708. [Google Scholar] [CrossRef] [PubMed]
- MacQueen, J. Some methods for classification and analysis of multivariate observations. In Proceedings of the 50th Berkeley Symposium on Mathematical Statistics and Probability, Volume 1: Statistics; University of California Press: Berkeley, CA, USA, 1967; pp. 281–297. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 2015 3rd IAPR Asian Conference on Pattern Recognition (ACPR), Kuala Lumpur, Malaysia, 3–6 November 2015; pp. 730–734. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.U.; Polosukhin, I. Attention is All you Need. In Proceedings of the Advances in Neural Information Processing Systems; Guyon, I., Luxburg, U.V., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R., Eds.; Curran Associates, Inc.: San Jose, CA, USA, 2017; Volume 30. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. In Proceedings of the 9th International Conference on Learning Representations, ICLR 2021, Virtual Event, 3–7 May 2021; Available online: https://openreview.net/forum?id=YicbFdNTTy (accessed on 12 May 2022).
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Hassani, A.; Walton, S.; Li, J.; Li, S.; Shi, H. Neighborhood Attention Transformer. arXiv 2022, arXiv:2204.07143. [Google Scholar]
- Jocher, G.; Stoken, A.; Borovec, J.; NanoCode012; ChristopherSTAN; Changyu, L.; Laughing; Tkianai; Hogan, A.; Lorenzomammana; et al. Ultralytics/yolov5: V3.1—Bug Fixes and Performance Improvements. Zenodo. 2020. Available online: https://zenodo.org/record/4154370 (accessed on 3 May 2022).
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Advances in Neural Information Processing Systems 32; Wallach, H., Larochelle, H., Beygelzimer, A., d’Alché-Buc, F., Fox, E., Garnett, R., Eds.; Curran Associates, Inc.: San Jose, CA, USA, 2019; pp. 8024–8035. [Google Scholar]
- Yakubovskiy, P. Segmentation Models Pytorch. 2020. Available online: https://github.com/qubvel/segmentation_models.pytorch (accessed on 5 May 2022).
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar] [CrossRef]
- Dai, L.; Wu, L.; Li, H.; Cai, C.; Wu, Q.; Kong, H.; Liu, R.; Wang, X.; Hou, X.; Liu, Y.; et al. A deep learning system for detecting diabetic retinopathy across the disease spectrum. Nat. Commun. 2021, 12, 3242. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).