Intelligent Hybrid Deep Learning Model for Breast Cancer Detection
Abstract
:1. Introduction
- In this research, a new hybrid DL(CNN-GRU) model is presented that automatically extracts BC-IDC (+,−) features and classifies them into IDC (+) and IDC (−) from histopathology images to reduce the pathologist’s error.
- The hybrid DL model (CNN-GRU) is proposed to efficiently classify IDC breast cancer detection in clinical research.
- In the evaluation process of the proposed CNN-GRU model, we have compared the key performance measure (Acc (%), Prec (%), Sens (%), and Spec (%), F1-score, and AUC with the current ML/DL model implemented the same dataset (Kaggle). In order to find the classification performance of the hybrid models. It is found that the proposed hybrid model has impressive classification outcomes compared to other hybrid DL models.
2. Related Works
3. Materials and Methods
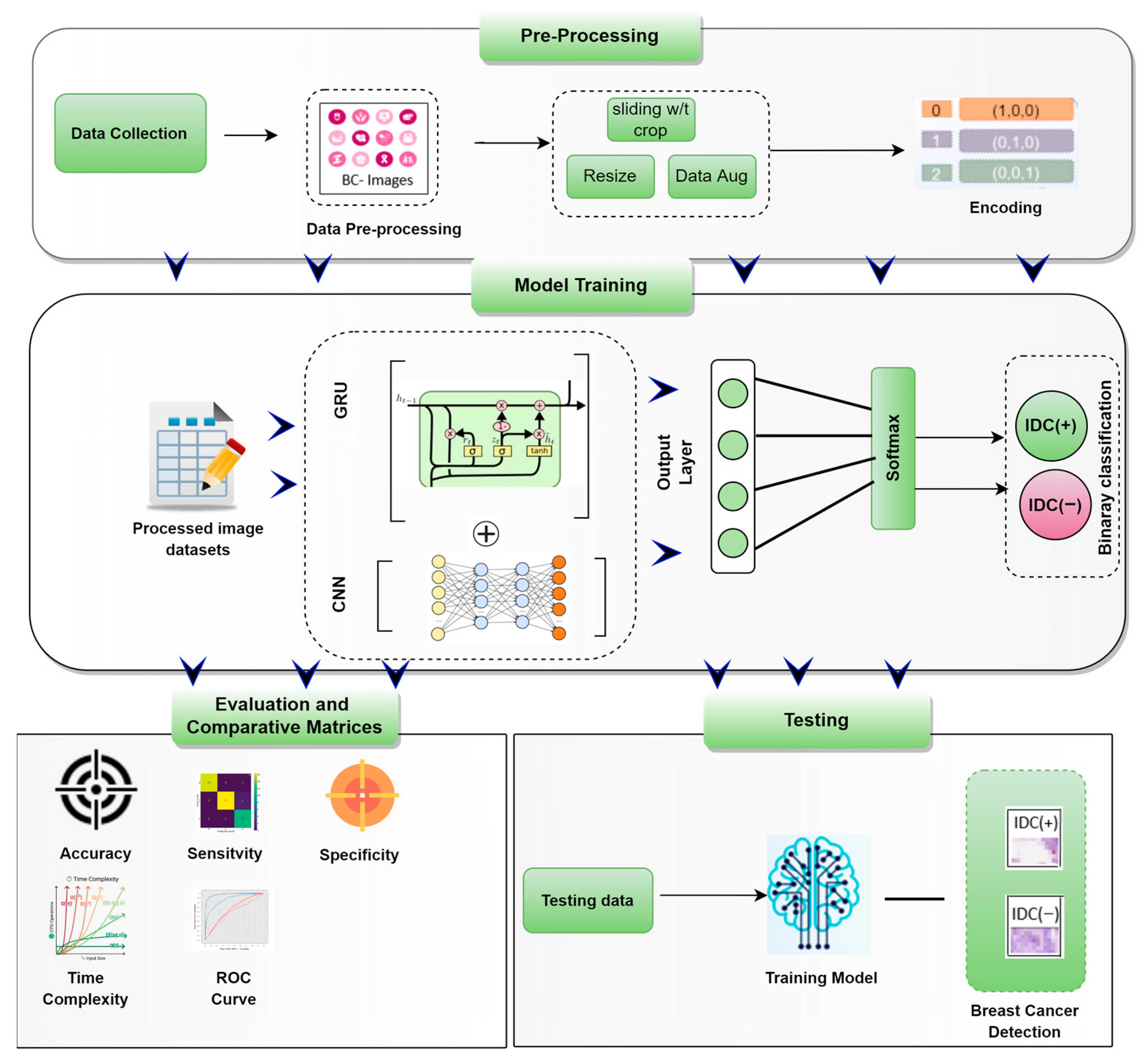
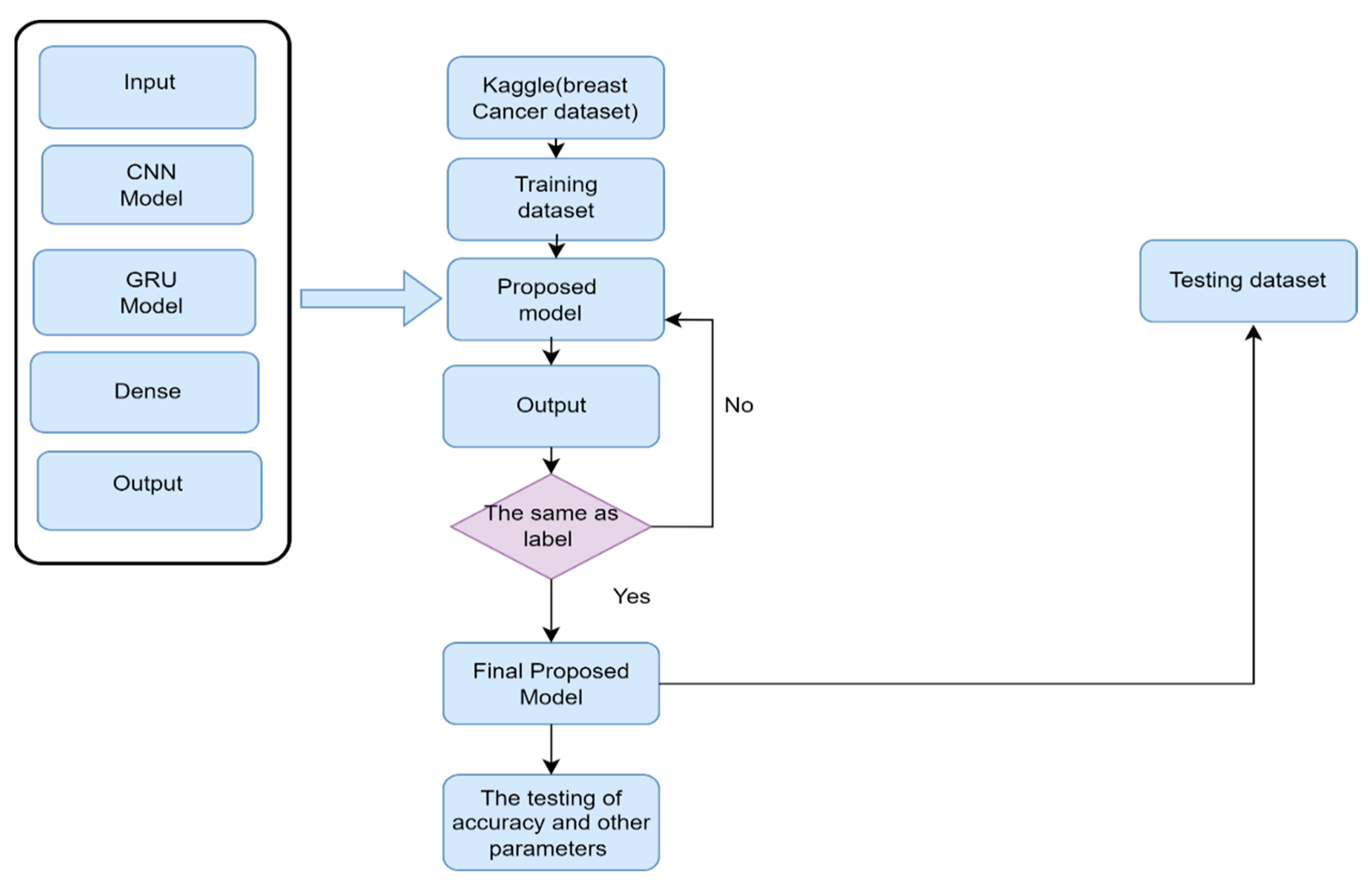
3.1. The Framework of Predicting BC-IDC Detection
3.2. Data Collection and Class Label
3.3. Data Pre-Processing
3.4. Random Cropping
3.5. Convolutional Neural Networks (CNN)
3.6. Gated Recurrent Unit Network (GRU)
3.7. CNN-GRU
4. Experimental Setup
5. Performance Metrics
- True positive (TP): positive IDC (+) samples were predicted.
- True negative (TN): refers to negative IDC (−) tissue samples found to be negative.
- False positive (FP): negative IDC (−) samples that are predicted to be positive IDC (+).
- False negative (FN): positive IDC (+) samples are predicted IDC (−).
6. Result and Discussion
6.1. Analysis of Performance Measure (Acc, Pres, Sens, Spec, F1 Score, and AUC)
6.2. Confusion Matrix
6.3. ROC Curve Analysis
6.4. FNR, FOR, FPR, and FDR Analysis
6.5. Evaluation of TNR, TPR and MCC
6.6. Model Efficiency
6.7. Comprative Anaylis Considering Proposed Hybird Alogerthm with ML/DL Exting Model
7. Conclusions and Future Work
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| IDC | Invasive ductal carcinoma |
| ML | Machine learning |
| DL | Deep learning |
| IDC | Invasive ductal carcinoma |
| DCIS | Ductal carcinoma in situ |
| BCW | Breast cancer Wisconsin |
| WSI | Whole slide images |
| CNN | Convolutional neural network |
| LSTM | Long short-term memory |
| GRU | Gated recurrent unit |
| BiLSTM | Bidirectional long short-term memory |
| DNN | Deep neural network |
| GPU | Graphics processing unit |
References
- Faruqui, N.; Yousuf, M.A.; Whaiduzzaman, M.; Azad, A.K.M.; Barros, A.; Moni, M.A. LungNet: A hybrid deep-CNN model for lung cancer diagnosis using CT and wearable sensor-based medical IoT data. Comput. Biol. Med. 2021, 139, 104961. [Google Scholar] [CrossRef] [PubMed]
- Adrienne, W.G.; Winer, E.P. Breast cancer treatment: A review. JAMA 2019, 321, 288–300. [Google Scholar]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.; Kimberly, D.M. Cancer statistics, 2018. CA A Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Aly, G.H.; Marey, M.; El-Sayed, S.A.; Tolba, M.F. YOLO based breast masses detection and classification in full-field digital mammograms. Comput. Methods Programs Biomed. 2021, 200, 105823. [Google Scholar] [CrossRef]
- Khamparia, A.; Bharati, S.; Podder, P.; Gupta, D.; Khanna, A.; Phung, T.K.; Thanh, D.N.H. Diagnosis of breast cancer based on modern mammography using hybrid transfer learning. Multidimens. Syst. Signal Processing 2021, 32, 747–765. [Google Scholar] [CrossRef]
- Naik, S.; Doyle, S.; Agner, S.; Madabhushi, A.; Feldman, M.; Tomaszewski, A.J. Automated Gland and Nuclei Segmentation for Grading of Prostate and Breast Cancer Histopathology. In Proceedings of the 2008 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Paris, France, 14–17 May 2008; pp. 284–287. [Google Scholar]
- Edge, S.; Byrd, D.R.; Compton, C.C.; Fritz, A.G.; Greene, F.; Trotti, A. AJCC Cancer Staging Handbook, 7th ed.; Springer: New York, NY, USA, 2010. [Google Scholar]
- Dundar, M.M.; Badve, S.; Bilgin, G.; Raykar, V.; Jain, R.; Sertel, O.; Gurcan, M.N. Computerized classification of intraductal breast lesions using histopathological images. IEEE Trans. Biomed. Eng. 2011, 58, 1977–1984. [Google Scholar] [CrossRef]
- Aloysius, N.; Geetha, M. A review on deep convolutional neural networks. In Proceedings of the International Conference on Communication and Signal Processing (ICCSP), Chennai, India, 6–8 April 2017; IEEE: Piscataway, NJ, USA; pp. 0588–0592. [Google Scholar]
- Ahmad, I.; Ullah, I.; Khan, W.U.; Rehman, A.U.; Adrees, M.S.; Saleem, M.Q.; Cheikhrouhou, O.; Hamam, H.; Shafiq, M. Efficient Algorithms for E-Healthcare to Solve Metaobject Fuse Detection Problem. J. Healthc. Eng. 2021, 2021, 9500304. [Google Scholar] [CrossRef]
- Chen, M.; Yang, J.; Hu, L.; Hossain, M.S.; Muhammad, G. Urban healthcare big data system based on crowdsourced and cloud-based air quality indicators. IEEE Commun. Mag. 2018, 56, 14–20. [Google Scholar] [CrossRef]
- Hossain, M.S. Cloud-supported cyber-physical localization framework for patients monitoring. IEEE Syst. J. 2017, 11, 118–127. [Google Scholar] [CrossRef]
- Alanazi, S.A.; Kamruzzaman, M.M.; Alruwaili, M.; Alshammari, N.; Alqahtani, S.A.; Karime, A. Measuring and preventing COVID-19 using the SIR model and machine learning in smart health care. J. Healthc. Eng. 2020, 2020, 8857346. [Google Scholar] [CrossRef] [PubMed]
- Benjelloun, M.; El Adoui, M.; Larhmam, M.A.; Mahmoudi, S.A. Automated breast tumor segmentation in DCE-MRI using deep learning. In Proceedings of the 2018 4th International Conference on Cloud Computing Technologies and Applications (Cloudtech), Brussels, Belgium, 26–28 November 2018; IEEE: Piscataway, NJ, USA, 2018. [Google Scholar]
- Tufail, A.B.; Ma, Y.K.; Kaabar, M.K.; Martínez, F.; Junejo, A.R.; Ullah, I.; Khan, R. Deep learning in cancer diagnosis and prognosis prediction: A minireview on challenges, recent trends, and future directions. Comput. Math. Methods Med. 2021, 2021, 9025470. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Yang, Q.; Ullah, I.; Rehman, A.U.; Tufail, A.B.; Noor, A.; Cengiz, K. 3D convolutional neural networks based automatic modulation classification in the presence of channel noise. IET Commun. 2021, 16, 497–509. [Google Scholar] [CrossRef]
- Tufail, A.B.; Ullah, I.; Khan, W.U.; Asif, M.; Ahmad, I.; Ma, Y.K.; Ali, M. Diagnosis of Diabetic Retinopathy through Retinal Fundus Images and 3D Convolutional Neural Networks with Limited Number of Samples. Wirel. Commun. Mob. Comput. 2021, 2021, 6013448. [Google Scholar] [CrossRef]
- Kamruzzaman, M.M. Architecture of smart health care system using artificial intelligence. In Proceedings of the 2020 IEEE International Conference on Multimedia & Expo Workshops (ICMEW), London, UK, 6–10 July 2020; pp. 1–6. [Google Scholar]
- Min, W.; Bao, B.-K.; Xu, C.; Hossain, M.S. Cross-platform multi-modal topic modeling for personalized inter-platform recommendation. IEEE Trans. Multimed. 2015, 17, 1787–1801. [Google Scholar] [CrossRef]
- Ahmad, I.; Liu, Y.; Javeed, D.; Shamshad, N.; Sarwr, D.; Ahmad, S. A review of artificial intelligence techniques for selection & evaluation. IOP Conf. Ser. Mater. Sci. Eng. 2020, 853, 012055. [Google Scholar]
- Hossain, M.S.; Amin, S.U.; Alsulaiman, M.; Muhammad, G. Applying deep learning for epilepsy seizure detection and brain mapping visualization. ACM Trans. Multimed. Comput. Appl. 2019, 15, 1–17. [Google Scholar] [CrossRef]
- Wang, J.L.; Ibrahim, A.K.; Zhuang, H.; Ali, A.M.; Li, A.Y.; Wu, A. A study on automatic detection of IDC breast cancer with convolutional neural networks. In Proceedings of the 2018 International Conference on Computational Science and Computational Intelligence (CSCI), Las Vegas, NV, USA, 12–14 December 2018; pp. 703–708. [Google Scholar]
- Aurna, N.F.; Abu Yousuf, M.; Abu Taher, K.; Azad, A.; Moni, M.A. A classification of MRI brain tumor based on two stage feature level ensemble of deep CNN models. Comput. Biol. Med. 2022, 146, 105539. [Google Scholar] [CrossRef]
- Shayma’a, A.H.; Sayed, M.S.; I Abdalla, M.I.; Rashwan, M.A. Breast cancer masses classification using deep convolutional neural networks and transfer learning. Multimed. Tools Appl. 2020, 79, 30735–30768. [Google Scholar]
- Hossain, M.S.; Muhammad, G. Emotion-aware connected healthcare big data towards 5G. IEEE Internet Things J. 2018, 5, 2399–2406. [Google Scholar] [CrossRef]
- Mahbub, T.N.; Yousuf, M.A.; Uddin, M.N. A Modified CNN and Fuzzy AHP Based Breast Cancer Stage Detection System. In Proceedings of the 2022 International Conference on Advancement in Electrical and Electronic Engineering (ICAEEE), Gazipur, Bangladesh, 24–26 February 2022; IEEE: Piscataway, NJ, USA, 2022. [Google Scholar]
- Pang, H.; Lin, W.; Wang, C.; Zhao, C. Using transfer learning to detect breast cancer without network training. In Proceedings of the 2018 5th IEEE International Conference on Cloud Computing and Intelligence Systems (CCIS), Nanjing, China, 23–25 November 2018; IEEE: Piscataway, NJ, USA, 2018. [Google Scholar]
- Cruz-Roa, A.; Basavanhally, A.; González, F.; Gilmore, H.; Feldman, M.; Ganesan, S.; Madabhushi, A. Automatic detection of invasive ductal carcinoma in whole slide images with convolutional neural networks. In Medical Imaging 2014: Digital Pathology; SPIE: Bellingham, WA, USA, 2014; Volume 9041, p. 904103. [Google Scholar]
- Amin, S.U.; Alsulaiman, M.; Muhammad, G.; Bencherif, M.A.; Hossain, M.S. Multilevel weighted feature fusion using convolutional neural networks for EEG motor imagery classification. IEEE Access 2019, 7, 18940–18950. [Google Scholar] [CrossRef]
- Sharma, S.; Aggarwal, A.; Choudary, T. Breast cancer detection using machine learning algorithms. In Proceedings of the International Conference on Computational Techniques, Electronics and Mechanical System (CTEMS), Belgaum, India, 21–22 December 2018; pp. 114–118. [Google Scholar]
- Jafarbigloo, S.K.; Danyali, H. Nuclear atypia grading in breast cancer histopathological images based on CNN feature extraction and LSTM classification. CAAI. Trans. Intell. Technol. 2021, 6, 426–439. [Google Scholar] [CrossRef]
- Nawaz, M.; Sewissy, A.A.; Soliman, T.H.A. Multi-class breast cancer classification using deep learning convolution neural network. Int. J. Adv. Comput. Sci. Appl. 2018, 9, 316–322. [Google Scholar]
- Wahab, N.; Khan, A. Multifaceted fused-CNN based scoring of breast cancer whole-slide histopathology images. Appl. Soft Comput. 2020, 97, 106808. [Google Scholar] [CrossRef]
- Gravina, M.; Marrone, S.; Sansone, M.; Sansone, C. DAE-CNN: Exploiting and disentangling contrast agent effects for breast lesions classification in DCE-MRI. Pattern Recognit. Lett. 2021, 145, 67–73. [Google Scholar] [CrossRef]
- Tsochatzidis, L.; Koutla, P.; Costaridou, L.; Pratikakis, I. Integrating segmentation information into CNN for breast cancer diagnosis of mammographic masses. Comput. Methods Programs Biomed. 2021, 200, 105913. [Google Scholar] [CrossRef]
- Malathi, M.; Sinthia, P.; Farzana, F.; Mary, G.A.A. Breast cancer detection using active contour and classification by deep belief network. Mater. Today Proc. 2021, 45, 2721–2724. [Google Scholar] [CrossRef]
- Desai, M.; Shah, M. An anatomization on breast cancer detection and diagnosis employing multi-layer perceptron neural network (MLP) and Convolutional neural network (CNN). Clin. Ehealth 2021, 4, 1–11. [Google Scholar] [CrossRef]
- Abdelhafiz, D.; Bi, J.; Ammar, R.; Yang, C.; Nabavi, S. Convolutional neural network for automated mass segmentation in mammography. BMC Bioinform. 2020, 21, 192. [Google Scholar] [CrossRef]
- Rezaeilouyeh, H.; Mollahosseini, A.; Mahoor, M.H. Microscopic medical image classification framework via deep learning and shearlet transform. J. Med. Imaging 2016, 3, 044501. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, G.; Shuib, L.; Wahab, A.W.A.; Mujtaba, G.; Nweke, H.F.; Al-Garadi, M.A.; Zulfiqar, F.; Raza, G.; Azmi, N.A. Deep learning-based breast cancer classification through medical imaging modalities: State of the art and research challenges. Artif. Intell. Rev. 2020, 53, 1655–1720. [Google Scholar] [CrossRef]
- Alhamid, M.F.; Rawashdeh, M.; Al Osman, H.; Hossain, M.S.; El Saddik, A. Towards context-sensitive collaborative media recommender system. Multimed. Tools Appl. 2015, 74, 11399–11428. [Google Scholar] [CrossRef]
- Qian, S.; Zhang, T.; Xu, C.; Hossain, M.S. Social event classification via boosted multimodal supervised latent dirichlet allocation. ACM Trans. Multimed. Comput. Commun. Appl. 2015, 11, 1–22. [Google Scholar] [CrossRef]
- Singh, D.; Singh, S.; Sonawane, M.; Batham, R.; Satpute, P.A. Breast cancer detection using convolution neural network. Int. Res. J. Eng. Technol. 2017, 5, 316–318. [Google Scholar]
- Javeed, D.; Gao, T.; Khan, M.T.; Shoukat, D. A hybrid intelligent framework to combat sophisticated threats in secure industries. Sensors 2022, 22, 1582. [Google Scholar] [CrossRef]
- Alhussein, M.; Muhammad, G.; Hossain, M.S.; Amin, S.U. Cognitive IoT-cloud integration for smart healthcare: Case study for epileptic seizure detection and monitoring. Mob. Netw. Appl. 2018, 23, 1624–1635. [Google Scholar] [CrossRef]
- Janowczyk, A. Use Case 6: Invasive Ductal Carcinoma (IDC) Segmentation. Available online: http://www.andrewjanowczyk.com/use-case-6-invasive-ductal-carcinoma-idc-segmentation/ (accessed on 10 March 2022).
- Khuriwal, N.; Mishra, N. Breast cancer detection from histopathological images using deep learning. In Proceedings of the 3rd International Conference and Workshops on Recent Advances and Innovations in Engineering, Jaipur, India, 22–25 November 2018. [Google Scholar]
- Kumar, A.; Sushil, R.; Tiwari, A.K. Comparative study of classification techniques for breast cancer diagnosis. Int. J. Comput. Sci. Eng. 2019, 7, 234–240. [Google Scholar] [CrossRef]
- Nallamala, S.H.; Mishra, P.; Koneru, S.V. Breast cancer detection using machine learning way. Int. J. Recent Technol. Eng. 2019, 8, 1402–1405. [Google Scholar]
- Abdolahi, M.; Salehi, M.; Shokatian, I.; Reiazi, R. Artificial intelligence in automatic classification of invasive ductal carcinoma breast cancer in digital pathology images. Med. J. Islamic Repub. Iran 2020, 34, 140. [Google Scholar] [CrossRef]
- Weal, E.F.; Amr, S.G. A deep learning approach for breast cancer mass detection. Int. J. Adv. Comput. Sci. Appl. 2019, 10, 175–182. [Google Scholar]
- Mekha, P.; Teeyasuksaet, N. Deep learning algorithms for predicting breast cancer based on tumor cells. In Proceedings of the 4th International Conference on Digital Arts, Media and Technology and 2nd ECTI Northern Section Conference on Electrical, Electronics, Computer and Telecommunication Engineering, Nan, Thailand, 30 January–2 February 2019; pp. 343–346. [Google Scholar]
- Kavitha, T.; Mathai, P.P.; Karthikeyan, C.; Ashok, M.; Kohar, R.; Avanija, J.; Neelakandan, S. Deep learning based capsule neural network model for breast cancer diagnosis using mammogram images. Interdiscip. Sci. Comput. Life Sci. 2022, 14, 113–129. [Google Scholar] [CrossRef] [PubMed]
- Jabeen, K.; Khan, M.A.; Alhaisoni, M.; Tariq, U.; Zhang, Y.-D.; Hamza, A.; Mickus, A.; Damaševičius, R. Breast cancer classification from ultrasound images using probability-based optimal deep learning feature fusion. Sensors 2022, 22, 807. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.kaggle.com/c/histopathologic-cancer-detection/data (accessed on 10 March 2022).
- Ramadan, S.Z. Using convolutional neural network with cheat sheet and data augmentation to detect breast cancer in mammograms. Comput. Math. Methods Med. 2020, 2020, 2020. [Google Scholar] [CrossRef]
- Mehmood, M.; Ayub, E.; Ahmad, F.; Alruwaili, M.; Alrowaili, Z.A.; Alanazi, S.; Rizwan, M.H.M.; Naseem, S.; Alyas, T. Machine learning enabled early detection of breast cancer by structural analysis of mammograms. Comput. Mater. Contin. 2021, 67, 641–657. [Google Scholar] [CrossRef]
- Isfahani, Z.N.; Jannat-Dastjerdi, I.; Eskandari, F.; Ghoushchi, S.J.; Pourasad, Y. Presentation of Novel Hybrid Algorithm for Detection and Classification of Breast Cancer Using Growth Region Method and Probabilistic Neural Network. Comput. Intell. Neurosci. 2021, 2021, 1–14. [Google Scholar] [CrossRef]
- Addeh, J.; Ata, E. Breast cancer recognition using a novel hybrid intelligent method. J. Med. Signals Sens. 2012, 2, 95. [Google Scholar]
- Waddell, M.; Page, D.; Shaughnessy, J., Jr. Predicting cancer susceptibility from single-nucleotide polymorphism data: A case study in multiple myeloma. In Proceedings of the 5th International Workshop on Bioinformatics, Chicago, IL, USA, 21 August 2005; pp. 21–28. [Google Scholar] [CrossRef]
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2015, 13, 8–17. [Google Scholar] [CrossRef] [Green Version]
- Kanavati, F.; Ichihara, S.; Tsuneki, M. A deep learning model for breast ductal carcinoma in situ classification in whole slide images. Virchows Arch. 2022, 480, 1009–1022. [Google Scholar] [CrossRef]
- Gupta, I.; Nayak, S.R.; Gupta, S.; Singh, S.; Verma, K.; Gupta, A.; Prakash, D. A deep learning based approach to detect IDC in histopathology images. Multimed. Tools Appl. 2022, 1–22. [Google Scholar] [CrossRef]
- Snigdha, V.; Nair, L.S. Hybrid Feature-Based Invasive Ductal Carcinoma Classification in Breast Histopathology Images. Machine Learning and Autonomous Systems; Springer: Singapore, 2022; pp. 515–525. [Google Scholar]













| References | Dataset | Model | Achievement |
|---|---|---|---|
| [49] | Kaggle | CNN, LSTM | CNN achieved higher accuracy (81%) and sensitivity (78.5%) than LSTM for the binary classification tasks, |
| [50] | BreakHis | CNN, DCNN | CNN has the best accuracy than DCNN, achieving 80% accuracy. |
| [51] | MIAS | CNN | The proposed model has a high accuracy of 70.9% for binary classification. |
| [52] | BCW (Breast Cancer Wisconsin) | DNN | Obtained an accuracy of 79.01%. |
| [53] | Kaggle | VGG-16, CNN | Achieved 80% Accuracy, Sens 79.9%, and Spec 78%. |
| [54] | UCI-cancer | R.N.N., GRU. | Proposed approaches performed better in the three toys problem and have 78.90% accuracy. |
| [55] | BCW (Breast Cancer Wisconsin) | CNN | Obtained 73% accuracy compared to four cancer classifications and 70.50% for distinguishing two mixed groupings of classes. |
| Proposed Layers | Stride | Padding | Kernel_Size | Input Data | Act_Funcion | Output |
|---|---|---|---|---|---|---|
| Con2D_Layer_1 | S = 1 | P = Same | 3 × 3 | (50,50,3) | Relu_Func | (50,50,128) |
| Max_pooling_1 | S = 1 | P = Same | 2 × 2 | (48,48,128) | ----- | (48,48,128) |
| Drop_out = 0.3 | ------- | ------ | ---- | (48,48,128) | ----- | (48,48,128) |
| Con2D_Layer_2 | S = 1 | P = Same | 3 × 3 | (48,48,128) | Relu_Func | (46,46,256) |
| Max_pooling_2 | S = 1 | P = Same | 2 × 2 | (46,46,256) | ---- | (44,44,256) |
| Drop_out = 0.9 | ------ | ----- | ---- | (44,44,256) | ----- | (44,44,256) |
| Con2D_Layer_3 | S = 1 | P = Same | 3 × 3 | (44,44,256) | Relu_Func | (42,42,256) |
| Max_pooling_3 | S = 1 | P = Same | 2 × 2 | (42,42,256) | ----- | (41,41,256) |
| Dropout = 0.5 | ------ | ----- | ---- | (41,41,256) | ---- | (41,41,256) |
| Con2D_Layer_4 | S = 1 | P = Same | 3 × 3 | (41,41,256) | Relu_Func | (39,39,256) |
| Dropout = 0.9 | ------- | ------ | ---- | (39,39,256) | ----- | (39,39,256) |
| Flatten | ------ | ----- | ---- | (32,32,512) | ---- | (524,288) |
| Dense1 | ----- | ----- | ---- | (524,288) | ----- | (1024) |
| Drop_out = 0.3 | ------ | ------ | ---- | (1024) | --- | (1024) |
| Dense2 | ------ | ----- | ---- | (1024) | --- | (2000) |
| GRU | ----- | ------ | None,512 | -------- | ------- | |
| Dense3 | ------- | ------ | (2000) | ----- | (2000) |
| RAM | 8 GB. |
| CPU | 2.80 GHz processor, Core-i7, 7th Gen |
| GPU | Nvidia, 1060, 8 GB |
| Languages | Version 3.8 Python |
| OS | 64-bit Window |
| Libraries | Scikitlearn, NumPy, Pandas, Koras, Tensor Flow |
| Publication | Cancer Type | Models | Dataset | Acc (%) | Sens (%) | Spec (%) | F1-Score (%) |
|---|---|---|---|---|---|---|---|
| Proposed Model | BC | CNN-GRU | Kaggle | 86.21% | 85% | 84.60% | 86% |
| [58] | Breast cancer | DCNNs | BreakHis | 80% | 79.90% | 79% | 79% |
| [59] | IDC (+,−) | CNN | Kaggle | 75.70% | 74.50% | 74% | 76% |
| [60] | Breast cancer | FCM-GA | Breast cancer Wisconsin (BCW) | 76% | 75.50% | 75.10% | 78% |
| [61] | Breast cancer | SVM | Kaggle | 65% | 64.90% | 63.50% | 66% |
| [62] | Colon carcinomatosis | BN | Kaggle | 78% | 76.40% | 75% | 80% |
| [63] | BC-IDC (+,−) | DCNNs | BreakHis | 80% | 78.90% | 78% | 82% |
| [64] | BC-IDC (+,−) | CNN, SVM | Breast cancer Wisconsin (BCW) | 76% | 75.20% | 73.80% | 78.80% |
| [65] | BC-IDC (+,−) | ML | Kaggle | 70% | 68% | 67.50% | 72.80% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Ahmad, I.; Javeed, D.; Zaidi, S.A.; Alotaibi, F.M.; Ghoneim, M.E.; Daradkeh, Y.I.; Asghar, J.; Eldin, E.T. Intelligent Hybrid Deep Learning Model for Breast Cancer Detection. Electronics 2022, 11, 2767. https://doi.org/10.3390/electronics11172767
Wang X, Ahmad I, Javeed D, Zaidi SA, Alotaibi FM, Ghoneim ME, Daradkeh YI, Asghar J, Eldin ET. Intelligent Hybrid Deep Learning Model for Breast Cancer Detection. Electronics. 2022; 11(17):2767. https://doi.org/10.3390/electronics11172767
Chicago/Turabian StyleWang, Xiaomei, Ijaz Ahmad, Danish Javeed, Syeda Armana Zaidi, Fahad M. Alotaibi, Mohamed E. Ghoneim, Yousef Ibrahim Daradkeh, Junaid Asghar, and Elsayed Tag Eldin. 2022. "Intelligent Hybrid Deep Learning Model for Breast Cancer Detection" Electronics 11, no. 17: 2767. https://doi.org/10.3390/electronics11172767
APA StyleWang, X., Ahmad, I., Javeed, D., Zaidi, S. A., Alotaibi, F. M., Ghoneim, M. E., Daradkeh, Y. I., Asghar, J., & Eldin, E. T. (2022). Intelligent Hybrid Deep Learning Model for Breast Cancer Detection. Electronics, 11(17), 2767. https://doi.org/10.3390/electronics11172767






