Fast 3D Liver Segmentation Using a Trained Deep Chan-Vese Model
Abstract
:1. Introduction
- -
- -
- It shows how to improve the segmentation accuracy by employing liver probability maps as 3D CVNN-UNet data terms instead of the CT intensity. The probability maps and the initializations are obtained from the output of a pixel-wise organ detection algorithm.
- -
- It introduces novel types of perturbations based on connected components that induce variability in the initialization and help avoid overfitting, a problem that severely impacts the 3D CVNN-UNet accuracy even when using perturbations that are 3D extensions of [2].
- -
- It presents a full multi-resolution 3D liver segmentation application, where a computationally intensive 3D CVNN algorithm based on U-Net is used at low resolution, and a computationally efficient 3D CVNN algorithm is used to refine the low resolution result at the higher resolutions. The proposed method obtains results competitive with the state of the art liver segmentation methods.
Related Work
Chan-Vese Overview
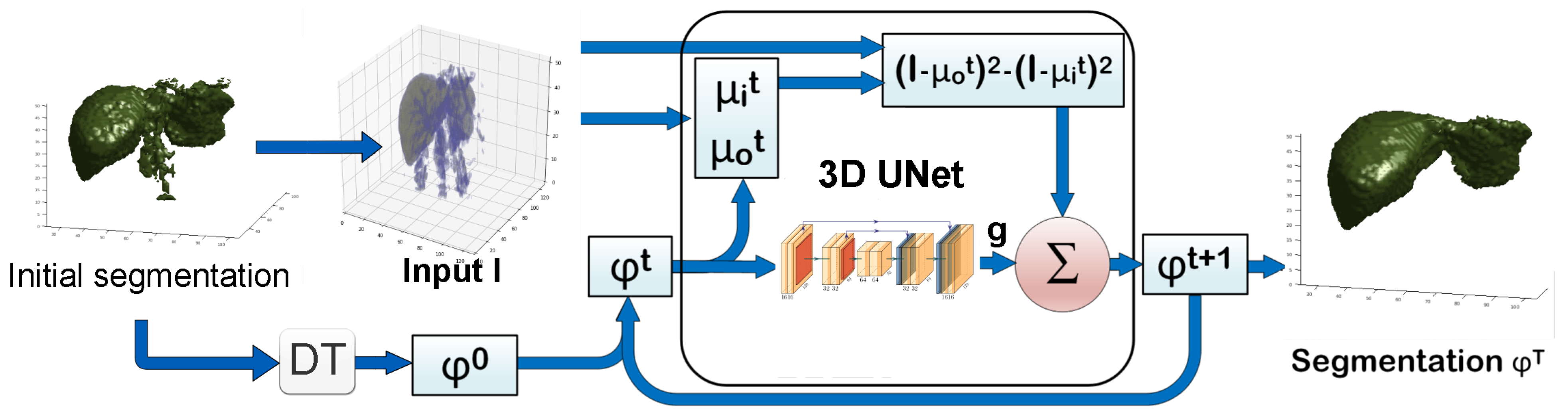
2. Proposed Method
| Algorithm 1 Deep Chan-Vese 3D Organ Segmentation |
|
2.1. Pre-Processing
2.1.1. Pixelwise Organ Detection
2.1.2. Constructing Probability Maps
| Algorithm 2 Probability Map Computation |
|
2.2. 2D Approach
2.2.1. 2D CVNN Architecture
2.2.2. 2D CVNN-UNet
2.3. 3D Approach
2.3.1. 3D CVNN-UNet with Low-Resolution Input
2.3.2. 3D CVNN with Medium-Resolution Input
- 1.
- To further improve the accuracy given the new detection and probability maps.
- 2.
- To obtain finer medium resolution segmentations, since the low-resolution segmentation would look coarse when upsampled.
- 3.
- To show that by combining the 3D CVNN-UNet and the 3D CVNN, one can achieve high accuracy for medium resolution input with a reduced computation complexity.
2.4. Implementation Details
- 1.
- Thresholding the probability map with a random threshold t. For the 3D CVNN-UNet, t is randomly chosen from . These values could range a larger span but for this work these values were enough to sustain generality. Then of the smallest connected components of the thresholded probability map are deleted at random. Those values are picked so that the Dice coefficient of for each t and the Dice of the detection map have roughly the same value, yet each initialization would have different false positives and false negatives. This would help train the CVNN to recover the correct shape from many scenarios and thus improve generalization. It is similar to having a golf player practice hitting the hole in 4 shots from a large number of locations roughly at the same distance from the hole.
- 2.
- When we do not use the above connected component-based initialization, for of the time, the initialization was obtained the same way as at test time, namely through the detection map . Another and the remaining of the time, the initializations were obtained from the detection map and ground truth Y, respectively, by the following distortions: first, semi-spheres with a random radius were added, or holes were punched at random locations on the boundary of the detection map or Y, then Gaussian noise was added to the distorted map around the boundary.
3. Experiments
3.1. Data
| Algorithm 3 Input preprocessing |
|
3.2. Metrics
3.3. Results
3.4. Ablation Study
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chan, T.F.; Vese, L.A. Active Contours without Edges. IEEE Trans. Image Process. 2001, 10, 266–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akal, O.; Barbu, A. Learning Chan-Vese. In Proceedings of the ICIP, Taipei, Taiwan, 22–25 September 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1590–1594. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the MICCAI, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the MICCAI, Athens, Greece, 17–21 October 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 424–432. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Berlin/Heidelberg, Germany, 2018; pp. 3–11. [Google Scholar]
- Akal, O.; Peng, Z.; Hermosillo Valadez, G. ComboNet: Combined 2D and 3D architecture for aorta segmentation. arXiv 2020, arXiv:2006.05325. [Google Scholar]
- Guerrero, R.; Qin, C.; Oktay, O.; Bowles, C.; Chen, L.; Joules, R.; Wolz, R.; Valdés-Hernández, M.d.C.; Dickie, D.; Wardlaw, J.; et al. White matter hyperintensity and stroke lesion segmentation and differentiation using convolutional neural networks. Neuroimage Clin. 2018, 17, 918–934. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the CVPR, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Heinrich, M.P.; Oktay, O.; Bouteldja, N. OBELISK-Net: Fewer layers to solve 3D multi-organ segmentation with sparse deformable convolutions. Med. Image Anal. 2019, 54, 1–9. [Google Scholar] [CrossRef]
- Isensee, F.; Jäger, P.F.; Full, P.M.; Vollmuth, P.; Maier-Hein, K.H. nnU-Net for brain tumor segmentation. In Proceedings of the International MICCAI Brainlesion Workshop, Lima, Peru, 4 October 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 118–132. [Google Scholar]
- Kenton, J.D.M.W.C.; Toutanova, L.K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. In Proceedings of the NAACL-HLT, Minneapolis, Minnesota, 2–7 June 2019; pp. 4171–4186. [Google Scholar]
- Carion, N.; Massa, F.; Synnaeve, G.; Usunier, N.; Kirillov, A.; Zagoruyko, S. End-to-end object detection with transformers. In Proceedings of the European Conference on Computer Vision, Glasgow, UK, 23–28 August 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 213–229. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Zheng, S.; Lu, J.; Zhao, H.; Zhu, X.; Luo, Z.; Wang, Y.; Fu, Y.; Feng, J.; Xiang, T.; Torr, P.H.; et al. Rethinking semantic segmentation from a sequence-to-sequence perspective with transformers. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 6881–6890. [Google Scholar]
- Hatamizadeh, A.; Tang, Y.; Nath, V.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.R.; Xu, D. Unetr: Transformers for 3d medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2022; pp. 574–584. [Google Scholar]
- Xie, Y.; Zhang, J.; Shen, C.; Xia, Y. Cotr: Efficiently bridging cnn and transformer for 3d medical image segmentation. In Proceedings of the MICCAI, Strasbourg, France, 27 September–1 October 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 171–180. [Google Scholar]
- Ngo, T.A.; Lu, Z.; Carneiro, G. Combining deep learning and level set for the automated segmentation of the left ventricle of the heart from cardiac cine magnetic resonance. Med. Image Anal. 2017, 35, 159–171. [Google Scholar] [CrossRef]
- Mohamed, A.r.; Dahl, G.E.; Hinton, G. Acoustic modeling using deep belief networks. IEEE Trans. Audio Speech Lang. Process. 2011, 20, 14–22. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Xu, C.; Gui, C.; Fox, M.D. Distance regularized level set evolution and its application to image segmentation. IEEE Trans. Image Process. 2010, 19, 3243–3254. [Google Scholar] [CrossRef]
- Hu, P.; Shuai, B.; Liu, J.; Wang, G. Deep level sets for salient object detection. In Proceedings of the CVPR, Honolulu, HI, USA, 21–26 July 2017; pp. 2300–2309. [Google Scholar]
- Hu, P.; Wang, G.; Kong, X.; Kuen, J.; Tan, Y.P. Motion-guided cascaded refinement network for video object segmentation. In Proceedings of the CVPR, Salt Lake City, UT, USA, 18–23 June 2018; pp. 1400–1409. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Hancock, M.C.; Magnan, J.F. Lung nodule segmentation via level set machine learning. arXiv 2019, arXiv:1910.03191. [Google Scholar]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Homayounfar, N.; Xiong, Y.; Liang, J.; Ma, W.C.; Urtasun, R. Levelset r-cnn: A deep variational method for instance segmentation. In Proceedings of the European Conference on Computer Vision, Glasgow, UK, 23–28 August 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 555–571. [Google Scholar]
- Raju, A.; Miao, S.; Jin, D.; Lu, L.; Huang, J.; Harrison, A.P. Deep implicit statistical shape models for 3d medical image delineation. In Proceedings of the AAAI Conference on Artificial Intelligence, Virtual, 22 February–1 March 2022; Volume 36, pp. 2135–2143. [Google Scholar]
- Tripathi, S.; Singh, S.K. An Object Aware Hybrid U-Net for Breast Tumour Annotation. arXiv 2022, arXiv:2202.10691. [Google Scholar]
- Mumford, D.; Shah, J. Optimal approximations by piecewise smooth functions and associated variational problems. Commun. Pure Appl. Math. 1989, 42, 577–685. [Google Scholar] [CrossRef] [Green Version]
- Paszke, A.; Gross, S.; Chintala, S.; Chanan, G.; Yang, E.; DeVito, Z.; Lin, Z.; Desmaison, A.; Antiga, L.; Lerer, A. Automatic differentiation in PyTorch. In NeurIPS Autodiff Workshop; 2017; Available online: https://openreview.net/pdf?id=BJJsrmfCZ (accessed on 13 September 2022).
- Taghanaki, S.A.; Zheng, Y.; Zhou, S.K.; Georgescu, B.; Sharma, P.; Xu, D.; Comaniciu, D.; Hamarneh, G. Combo loss: Handling input and output imbalance in multi-organ segmentation. Comput. Med. Imaging Graph. 2019, 75, 24–33. [Google Scholar] [CrossRef] [Green Version]
- Sudre, C.H.; Li, W.; Vercauteren, T.; Ourselin, S.; Cardoso, M.J. Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Berlin/Heidelberg, Germany, 2017; pp. 240–248. [Google Scholar]
- Barbu, A. Training an active random field for real-time image denoising. IEEE Trans. Image Process. 2009, 18, 2451–2462. [Google Scholar] [CrossRef]
- Huang, G.; Li, Y.; Pleiss, G.; Liu, Z.; Hopcroft, J.E.; Weinberger, K.Q. Snapshot ensembles: Train 1, get M for free. arXiv 2017, arXiv:1704.00109. [Google Scholar]
- Gibson, E.; Giganti, F.; Hu, Y.; Bonmati, E.; Bandula, S.; Gurusamy, K.; Davidson, B.; Pereira, S.P.; Clarkson, M.J.; Barratt, D.C. Automatic multi-organ segmentation on abdominal CT with dense v-networks. IEEE Trans. Med. Imaging 2018, 37, 1822–1834. [Google Scholar] [CrossRef] [Green Version]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [Green Version]
- Roth, H.R.; Farag, A.; Turkbey, E.B.; Lu, L.; Liu, J.; Summers, R.M. Data from pancreas-CT. Cancer Imaing Arch. 2016. [Google Scholar] [CrossRef]
- Roth, H.R.; Lu, L.; Farag, A.; Shin, H.C.; Liu, J.; Turkbey, E.B.; Summers, R.M. Deeporgan: Multi-level deep convolutional networks for automated pancreas segmentation. In Proceedings of the MICCAI, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 556–564. [Google Scholar]
- Landman, B.; Xu, Z.; Igelsias, J.; Styner, M.; Langerak, T.; Klein, A. MICCAI Multi-Atlas Labeling Beyond the Cranial Vault–Workshop and Challenge. 2015. Available online: https://www.synapse.org/#!Synapse:syn3193805/files/ (accessed on 13 September 2022).
- Xu, Z.; Lee, C.P.; Heinrich, M.P.; Modat, M.; Rueckert, D.; Ourselin, S.; Abramson, R.G.; Landman, B.A. Evaluation of six registration methods for the human abdomen on clinically acquired CT. IEEE Trans. Biomed. Eng. 2016, 63, 1563–1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heinrich, M.P.; Jenkinson, M.; Brady, M.; Schnabel, J.A. MRF-based deformable registration and ventilation estimation of lung CT. IEEE Trans. Med. Imaging 2013, 32, 1239–1248. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Suh, J.W.; Das, S.R.; Pluta, J.B.; Craige, C.; Yushkevich, P.A. Multi-atlas segmentation with joint label fusion. IEEE Trans. PAMI 2012, 35, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Dou, Q.; Yu, L.; Heng, P.A. Voxresnet: Deep voxelwise residual networks for volumetric brain segmentation. arXiv 2016, arXiv:1608.05895. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 565–571. [Google Scholar]
- Simpson, A.L.; Antonelli, M.; Bakas, S.; Bilello, M.; Farahani, K.; Van Ginneken, B.; Kopp-Schneider, A.; Landman, B.A.; Litjens, G.; Menze, B.; et al. A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv 2019, arXiv:1902.09063. [Google Scholar]
- Akal, O. Deep Learning Based Generalization of Chan-Vese Level Sets Segmentation. Ph.D. Thesis, Florida State University, Tallahassee, FL, USA, 2020. Order No. 28022313. [Google Scholar]







| Architecture | Inference | Upsampled | Metric | Dice | Boundary | 95% Hausdorff | Segment. |
|---|---|---|---|---|---|---|---|
| Size | to | Region | Err (mm) | Distance (mm) | Time (s) | ||
| 3D CVNN-UNet | - | whole | 95.58 | 1.77 | 4.45 | 1.25 | |
| - | ROI | 95.56 | 1.67 | 4.42 | 0.53 | ||
| Deep Chan-Vese 3D | - | whole | 95.59 | 1.71 | 4.45 | 0.26 | |
| whole | 95.07 | 1.59 | 4.53 | 0.26 | |||
| - | ROI | 95.39 | 1.58 | 4.42 | 0.11 | ||
| ROI | 95.24 | 1.49 | 4.40 | 0.11 |
| x-val | Volumes | Boundary | 95% Haussdorf | Segmentation | |||
|---|---|---|---|---|---|---|---|
| Arhitecture | res | Folds | Tested | Dice | Err (mm) | Distance (mm) | Time (s) |
| DEEDS [42]+JLF [43] | 144 | 9 | 90 | 94 | 2.1 | 6.2 | 4740 |
| VoxResNet [44] | 144 | 9 | 90 | 95 | 2.0 | 5.2 | |
| VNet [45] | 144 | 9 | 90 | 94 | 2.2 | 6.4 | |
| DenseVNet [36] | 144 | 9 | 90 | 96 | 1.6 | 4.9 | 12 |
| ObeliskNet [10] | 144 | 4 | 43 | 95.4 | - | - | |
| SETR [15] | 96 | 5 | 30 | 95.4 | - | - | 25 |
| CoTr [17] | 96 | 5 | 30 | 96.3 | - | - | 19 |
| UNETR [16] | 96 | 5 | 30 | 97.1 | - | - | 12 |
| nnU-Net [11] | 128 | 1 | 13 | 96.4 | 1.7 | - | 10 |
| DISSM [28] | - | 1 | 13 | 96.5 | 1.1 | - | 12 |
| 3D CVNN-UNet (ours) | 128 | 4 | 90 | 95.6 | 1.67 | 4.42 | 0.53 |
| Deep Chan-Vese 3D (ours) | 256 | 4 | 90 | 95.2 | 1.49 | 4.40 | 0.64 |
| 3D | U-Net | 1-it | 2-it | 3-it | 4-it | ||
|---|---|---|---|---|---|---|---|
| 2D CVNN | - | - | 87.58 | 92.75 | 93.66 | 93.63 | 93.68 |
| 2D CVNN-UNet | - | + | 87.58 | 92.61 | 93.72 | 93.63 | 93.75 |
| 3D CVNN | + | - | 87.58 | 88.29 | 90.23 | 91.43 | 91.74 |
| 3D CVNN-UNet | + | + | 87.58 | 92.83 | 94.41 | 95.09 | 95.52 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akal, O.; Barbu, A. Fast 3D Liver Segmentation Using a Trained Deep Chan-Vese Model. Electronics 2022, 11, 3323. https://doi.org/10.3390/electronics11203323
Akal O, Barbu A. Fast 3D Liver Segmentation Using a Trained Deep Chan-Vese Model. Electronics. 2022; 11(20):3323. https://doi.org/10.3390/electronics11203323
Chicago/Turabian StyleAkal, Orhan, and Adrian Barbu. 2022. "Fast 3D Liver Segmentation Using a Trained Deep Chan-Vese Model" Electronics 11, no. 20: 3323. https://doi.org/10.3390/electronics11203323
APA StyleAkal, O., & Barbu, A. (2022). Fast 3D Liver Segmentation Using a Trained Deep Chan-Vese Model. Electronics, 11(20), 3323. https://doi.org/10.3390/electronics11203323






