Multiscale Brain Network Models and Their Applications in Neuropsychiatric Diseases
Abstract
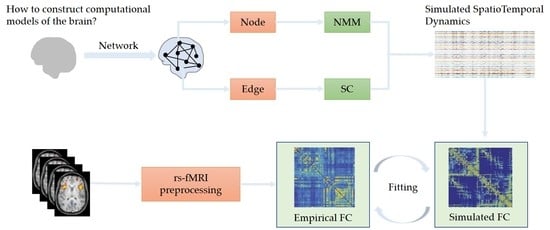
:1. Introduction
2. Methods for Implementing Multiscale BNMs
2.1. Estimation of Structural Network Connections
2.1.1. Brain Parcellation
2.1.2. Inferring of Connections
2.2. Selection of Node Dynamics
2.2.1. Computational Models of Node Dynamics
- Biophysical neural mass models
- Simplified abstract models
2.2.2. Heterogeneity between Node Dynamics
2.2.3. Dynamical System Theory of Node Dynamics
- Multistability: In some systems, two or more stable states coexist for the same parameter. Such a system is called a multistable system. The multistability of node dynamics can be used to explain the slow fluctuations underlying dynamical FC. Specifically, if there are only two stable states (a fixed-point and a limit cycle), the system can be assumed to be bistable.
- Metastability: In a metastability regime, no stable states exist. Additionally, the system moves along only intermittently stable state sequences.
- Bifurcation: The system changes from a stable state to another and this qualitative change is caused by a parameter of the system. In terms of the stability property after bifurcation, bifurcation can be classified as supercritical when stable objects appear and as subcritical when unstable objects emerge after bifurcation.
- Fixed-point: It refers to a steady state in a system or a stable equilibrium.
- Limit-cycle: It is a simple closed orbit, which means the system is periodically oscillating.
2.3. Model Fitting
2.3.1. Time-Averaged Functional Connectivity
2.3.2. Dynamic Functional Connectivity
2.3.3. Direct fMRI BOLD Time-Series
3. Comparison between Multiscale BNMs and Other BNMs
3.1. Comparison between Multiscale BNMs and Descriptive BNMs
3.1.1. Descriptive BNMs
3.1.2. Comparison
3.2. Comparison between Multiscale BNMs and Dynamic Causal Modeling
3.2.1. Dynamical Causal Modeling
3.2.2. Comparison
4. Applications of Multiscale BNMs in Understanding Neuropsychiatric Disorders
4.1. Parkinson’s Disease
4.2. Alzheimer’s Disease
4.3. Schizophrenia
| References | Node Dynamics | Parcellation | Model Fitting | SC Data | Diseases |
|---|---|---|---|---|---|
| Saenger et al. [102] | Normal form of Hopf | AAL atlas 90 regions | Static and dynamical FC, fMRI data | DTI | Parkinson |
| Van Nifterick et al. [105] | NMMs | AAL atlas 78 regions | Power spectrum, MEG data | DTI | Alzheimer |
| Sanchez-Rodriguez et al. [106] | Duffing oscillator | DK atlas 78 regions | Spectral measures, EEG data | DWI, T1-weighted MRIs | Alzheimer |
| Cabral et al. [111] | Mean-field approximation of IF models * | AAL atlas 90 regions | Static FC fMRI data | DTI | Schizophrenia |
| Yang et al. [112,113] Anticevic et al. [114], Cole et al. [115] | Mean-field models | 66 regions | Static FC fMRI data | DWI | Schizophrenia |
5. Discussion
5.1. Limitations
5.2. Future Directions
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Glomb, K.; Ponce-Alvarez, A.; Gilson, M.; Ritter, P.; Deco, G. Resting state networks in empirical and simulated dynamic functional connectivity. Neuroimage 2017, 159, 388–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deco, G.; Jirsa, V.K.; Robinson, P.A.; Breakspear, M.; Friston, K. The dynamic brain: From spiking neurons to neural masses and cortical fields. PLoS Comput. Biol. 2008, 4, e1000092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hodgkin, A.L.; Huxley, F. A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. 1952, 117, 500–544. [Google Scholar] [CrossRef] [PubMed]
- Izhikevich, E.M. Dynamical Systems in Neuroscience: The Geometry of Excitability and Bursting; The MIT Press: Cambridge, MA, USA, 2007. [Google Scholar]
- Morris, C.; Lecar, H. Voltage oscillations in the barnacle giant muscle fiber. Biophys. J. 1981, 35, 193–213. [Google Scholar] [CrossRef] [Green Version]
- Terman, D.; Rubin, J.E.; Yew, A.C.; Wilson, C.J. Activity patterns in a model for the subthalamopallidal network of the basal ganglia. J. Neurosci. 2002, 22, 2963–2976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- So, R.Q.; Kent, A.R.; Grill, W.M. Relative contributions of local cell and passing fiber activation and lesioning: A computational modeling study. J. Comput. Neurosci. 2012, 32, 499–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiff, S.J. Towards model-based control of Parkinson’s disease. Philos. Trans. A Math Phys. Eng. Sci. 2010, 368, 2269–2308. [Google Scholar] [CrossRef] [Green Version]
- Lu, M.; Wei, X.; Che, Y.; Wang, J.; Loparo, K.A. Application of reinforcement learning to deep brain stimulation in a computational model of Parkinson’s disease. IEEE Trans. Neural Syst. Rehabil. Eng. 2020, 28, 339–349. [Google Scholar] [CrossRef]
- Liu, C.; Wang, J.; Li, H.; Lu, M.; Deng, B.; Yu, H.; Wei, X.; Fietkiewicz, C.; Loparo, K.A. Closed-loop modulation of the pathological disorders of the basal ganglia network. IEEE Trans. Neural Netw. Learn Syst. 2017, 28, 371–382. [Google Scholar] [CrossRef]
- Depannemaecker, D.; Destexhe, A.; Jirsa, V.; Bernard, C. Modeling seizures: From single neurons to networks. Seizure 2021, 90, 4–8. [Google Scholar] [CrossRef]
- Boustani, E.I.; Destexhe, A. A master equation formalism for macroscopic modeling of asynchronous irregular activity states. Neural Comput. 2009, 21, 46–100. [Google Scholar] [CrossRef] [PubMed]
- Wilson, H.R.; Cowan, J.D. Excitatory and inhibitory interactions in localized populations of model neuron. Biophys. J. 1972, 12, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Jansen, B.H.; Rit, V.G. Electroencephalogram and visual evoked potential generation in a mathematical model of coupled cortical columns. Biol. Cybern. 1995, 73, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Kuramoto, Y.; Nishikawa, I. Statistical macrodynamics of large dynamical systems. Case of a phase transition in oscillator communities. J. Stat. Phys. 1987, 49, 569–605. [Google Scholar] [CrossRef]
- Breakspear, M. Dynamic models of large-scale brain activity. Nat. Neurosci. 2017, 20, 340–352. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Todo, R.; Salvador, R.; Santarnecchi, E.; Wendling, F.; Deco, G.; Ruffini, G. Personalization of hybrid brain models from neuroimaging and electrophysiology data. bioRxiv 2018, 461350. [Google Scholar] [CrossRef] [Green Version]
- Bullmore, E.; Sporns, O. Complex brain networks: Graph theoretical analysis of structure and functional systems. Nat. Rev. Neurosci. 2009, 10, 186–198. [Google Scholar] [CrossRef]
- Deco, G.; Kringelbach, M.L. Great expectations: Using whole-brain computational connectomics for understanding neuropsychiatric disorders. Neuron 2014, 84, 892–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutchison, R.M.; Womelsdorf, T.; Allen, E.A.; Bandettini, P.A.; Calhoun, V.D.; Corbetta, M.; Gonzalez-Castillo, J. Dynamic functional connectivity: Promise, issues, and interpretations. NeuroImage 2013, 80, 360–378. [Google Scholar] [CrossRef] [Green Version]
- Heitmann, S.; Breakspear, M. Putting the “dynamic” back into dynamic functional connectivity. Net. Neurosci. 2018, 2, 150–174. [Google Scholar] [CrossRef]
- Giacopelli, G.; Tegolo, D.; Spera, E.; Migliore, M. On the structural connectivity of large-scale models of brain networks at cellular level. Sci. Rep. 2021, 11, 4345. [Google Scholar] [CrossRef] [PubMed]
- Markov, N.T.; Ercsey-Ravasz, M.M.; Ribeiro Gomes, A.R.; Lamy, C.; Magrou, L.; Vezoli, J.; Misery, P.; Falchier, A.; Quilodran, R.; Gariel, M.A. A weighted and directed interareal connectivity matrix for macaque cerebral cortex. Cereb. Cortex 2014, 24, 17–36. [Google Scholar] [CrossRef] [PubMed]
- Deco, G.; Ponce-Alvarez, A.; Mantini, D.; Romani, G.L.; Hagmann, P.; Corbetta, M. Resting-state functional connectivity emerges from structurally and dynamically shaped slow linear fluctuations. J. Neurosci. 2013, 33, 11239–11252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deco, G.; Jirsa, V.K. Ongoing cortical activity at rest: Criticality, multistability, and ghost attractors. J. Neurosci. 2012, 32, 3366–3375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hagmann, P.; Cammoun, L.; Gigandet, X.; Meuli, R.; Honey, C.J.; Wedeen, V.J.; Sporns, O. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008, 6, e159. [Google Scholar] [CrossRef] [PubMed]
- Zalesky, A.; Fornito, A.; Harding, I.H.; Cocchi, L.; Yucel, M.; Pantelis, C.; Bullmore, E.T. Whole-brain anatomical networks: Does the choice of nodes matter? NeuroImage 2020, 50, 970–983. [Google Scholar] [CrossRef]
- Eickhoff, S.B.; Constable, R.T.; Yeo, B.T.T. Topographic organization of the cerebral cortex and brain cartography. Neuroimage 2018, 170, 332–347. [Google Scholar] [CrossRef] [Green Version]
- Popovych, O.V.; Manos, T.; Hoffstaedter, F.; Eickhoff, S.B. What can computational models contribute to the neuroimaging data analytics. Front. Syst. Neurosci. 2019, 12, 68. [Google Scholar] [CrossRef] [Green Version]
- Yeh, C.H.; Jones, D.K.; Liang, X.; Descoteaux, M.; Connelly, A. Mapping structural connectivity using diffusion MRI: Challenges and opportunities. J. Magn. Reson. Imaging 2021, 53, 1666–1682. [Google Scholar] [CrossRef]
- Eickhoff, S.B.; Yeo, B.T.T.; Genon, S. Imaging-based parcellations of the human brain. Nat. Rev. Neurosci. 2018, 19, 672–686. [Google Scholar] [CrossRef]
- Pathak, A.; Roy, D.; Banerjee, A. Whole-brain network models: From physics to bedside. Front. Comput. Neurosci. 2022, 16, 866517. [Google Scholar] [CrossRef] [PubMed]
- Nowinski, W.L. Evolution of human brain atlases in terms of content, applications, functionality, and availability. Neuroinformatics 2021, 19, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Kale, P.; Zalesky, A.; Gollo, L.L. Estimating the impact of structural directionality: How reliable are undirected connectomes? Netw. Neurosci. 2018, 2, 259–284. [Google Scholar] [CrossRef] [PubMed]
- Friston, K.J. Functional and effective connectivity: A review. Brain Connect 2011, 1, 13–36. [Google Scholar] [CrossRef]
- Gilson, M.; Moreno-Bote, R.; Ponce-Alvarez, A.; Ritter, P.; Deco, G. Estimation of directed effective connectivity from fMRI functional connectivity hints at asymmetries of cortical connectome. PLoS Comput. Biol. 2016, 12, e1004762. [Google Scholar] [CrossRef] [Green Version]
- Murray, J.D.; Demirtas, M.; Anticevic, A. Biophysical modeling of large-scale brain dynamics and applications for computational psychiatry. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 2018, 3, 777–787. [Google Scholar] [CrossRef]
- Modha, D.S.; Singh, R. Network architecture of the long-distance pathways in the macaque brain. Proc. Natl. Acad. Sci. USA 2010, 107, 13485–13490. [Google Scholar] [CrossRef] [Green Version]
- Forrester, M.; Crofts, J.J.; Sotiropoulos, S.N.; Coombes, S.; O’Dea, R.D. The role of node dynamics is shaping emergent functional connectivity patterns in the brain. Netw. Neurosci. 2020, 4, 467–483. [Google Scholar] [CrossRef] [Green Version]
- Stam, C.J.; van Straaten, E.C.; Van Dellen, E.; Tewarie, P.; Gong, G.; Hillebrand, A.; Meier, J.; Van Mieghem, P. The relation between structural and functional connectivity patterns in complex brain networks. Int. J. Psychophysiol. 2016, 103, 149–160. [Google Scholar] [CrossRef]
- Hlinka, J.; Coombes, S. Using computational models to relate structural and functional Brain Connectivity. Eur. J. Neurosci. 2012, 36, 2137–2145. [Google Scholar] [CrossRef]
- Honey, C.J.; Kötter, R.; Breakspear, M.; Sporns, O. Network structure of cerebral cortex shapes functional connectivity on multiple time scale. Proc. Natl. Acad. Sci. USA 2007, 104, 10240–10245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breakspear, M.; Terry, J.R.; Friston, K.J. Modulation of excitatory synaptic coupling facilitates synchronization and complex dynamics in a biophysical model of neuronal dynamics. Network 2003, 14, 703–732. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Rho, Y.; McIntosh, A.R.; Kötter, R.; Jirsa, V.K. Noise during rest enables the exploration of the brain’s dynamic repertoire. PLoS Comput. Biol. 2008, 4, e1000196. [Google Scholar] [CrossRef] [PubMed]
- Deco, G.; Jirsa, V.K.; McIntosh, A.R. Emerging concepts for the dynamical organization of resting-state activity in the brain. Nat. Rev. Neurosci. 2011, 12, 43–56. [Google Scholar] [CrossRef]
- Anticevic, A.; Murray, J.D.; Narch, D.M. Bridging levels of understanding in schizophrenia through computational modeling. ClinPsychol. Sci. 2015, 3, 433–459. [Google Scholar] [CrossRef] [Green Version]
- Murray, J.D.; Wang, X.J. Cortical circuit models in psychiatry: Linking disrupted excitation-inhibition balance to cognitive deficits associated with schizophrenia. In Computational Psychiatry: Mathematical Modeling of Mental Illness; Academin Press: London, UK, 2017; pp. 3–25. [Google Scholar]
- Deco, G.; Jirsa, V.; McIntosh, A.R.; Sporn, O.; Kötter, R. Key role of coupling, delay, and noise in resting brain fluctuations. Proc. Natl. Acad. Sci. USA 2009, 106, 10302–10307. [Google Scholar] [CrossRef] [Green Version]
- Coronel-Oliveros, C.; Castro, S.; Cofre, R.; Orio, P. Structural features of the human connectome that facilitate the switching of brain dynamics via noradrenergic neuromodulation. Front. Comput. Neurosci. 2021, 15, 687075. [Google Scholar] [CrossRef]
- Hellyer, P.J.; Scott, G.; Shanahan, M.; Sharp, D.J.; Leech, R. Cognitive flexibility through metastable neural dynamics is disrupted by damage to the structural connectome. J. Neurosci. 2015, 35, 9050–9063. [Google Scholar] [CrossRef] [Green Version]
- Ponce-Alvarez, A.; Deco, G.; Hagmann, P.; Romani, G.L.; Mantini, D.; Corbetta, M. Resting-state temporal synchronization networks emerge from connectivity topology and heterogeneity. PLoS Comput. Biol. 2015, 11, e1004100. [Google Scholar] [CrossRef] [Green Version]
- Bettinardi, R.G.; Deco, G.; Karlaftis, V.M.; Van Hartevelt, T.J.; Fernandes, H.M.; Kourtzi, Z.; Kringelbach, M.L.; Zamora-López, G. How structure sculpts function: Unveiling the contribution of anatomical connectivity to the brain’s spontaneous correlation structure. Chaos 2017, 27, 047409. [Google Scholar] [CrossRef]
- Demirtas, M.; Falcon, C.; Tucholka, A.; Gispert, J.D.; Molinuevo, J.L.; Deco, G. A whole-brain computational modeling approach to explain the alterations in resting-state functional connectivity during progression of Alzheimer’s disease. Neuroimage Clin. 2017, 16, 343–354. [Google Scholar] [CrossRef] [PubMed]
- López-González, A.; Panda, R.; Ponce-Alvarez, A.; Zamora-López, G.; Escrichs, A.; Martial, C.; Thibaut, A.; Gosseries, O.; Kringelbach, M.L.; Annen, J.; et al. Loss of consciousness reduces the stability of brain hubs and the heterogeneity of brain dynamics. Commun. Biol. 2021, 4, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Lord, L.D.; Stevner, A.B.; Deco, G.; Kringelbach, M.L. Understanding principles of integration and segregation using whole-brain computational connectomics: Implications for neuropsychiatric disorders. Philos. Trans. R Soc. A Math Phys. Eng. Sci. 2017, 375, 20160283. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, R.; LaFleur, K.J.R.; De Reus, M.A.; van den Berg, L.H.; van den Heuvel, M.P. Kuramoto model simulation of neural hubs and dynamic synchrony in the human cerebral connectome. BMC Neurosci. 2015, 16, 54. [Google Scholar] [CrossRef] [Green Version]
- Breakspear, M.; Heitmann, S.; Daffertshofer, A. Generative models of cortical oscillations: Neurobiological implications of the kuramoto model. Front. Hum. Neurosci. 2010, 4, 190. [Google Scholar] [CrossRef] [Green Version]
- Sanz Perl, Y.; Pallavicini, C.; Pérez lpiña, I.; Demertzi, A.; Bonhomme, V.; Martial, C.; Panda, R.; Annen, J.; lbañez, A.; Kringelbach, M.; et al. Perturbations in dynamical models of whole-brain activity dissociate between the level and stability of consciousness. PLoS Comput. Biol. 2021, 17, e1009139. [Google Scholar] [CrossRef] [PubMed]
- Glasser, M.F.; Goyal, M.S.; Preuss, T.M.; Raichle, M.E.; Van Essen, D.C. Trends and properties of human cerebral cortex: Correlations with cortical myelin content. Neuroimage 2014, 93, 165–175. [Google Scholar] [CrossRef] [Green Version]
- Deco, G.; Cabral, J.; Woolrich, M.W.; Stevner, A.B.; Van Hartevelt, T.J.; Kringelbach, M.L. Single or multiple frequency generators in ongoing brain activity: A mechanistic whole-brain model of empirical MEG data. Neuroimage 2017, 152, 538–550. [Google Scholar] [CrossRef]
- Demirtaş, M.; Burt, J.B.; Helmer, M.; Ji, J.L.; Adkinson, B.D.; Glasser, M.F.; Van Essen, D.C.; Sotiropoulos, S.N.; Anticevic, A.; Murray, J.D. Hierarchical heterogeneity across human cortex shapes large-scale neural dynamics. Neuron 2019, 101, 1181–1194.e13. [Google Scholar] [CrossRef] [Green Version]
- Schirner, M.; McIntosh, A.R.; Jirsa, V.; Deco, G.; Ritter, P. Inferring multi-scale neural mechanisms with brain network modelling. Elife 2018, 8, e28927. [Google Scholar] [CrossRef]
- Lee, W.H.; Frangou, S. Emergence of metastable dynamics in functional brain organization via spontaneous fMRI signal and whole-brain computational modeling. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2017, 2017, 4471–4474. [Google Scholar]
- Cabral, J.; Kringelbach, M.L.; Deco, G. Functional graph alterations in schizophrenia: A result from a global anatomic decoupling? Pharmacopsychiatry 2012, 45, S57–S64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaboodvand, N.; van den Heuvel, M.P.; Fransson, P. Adaptive frequency-based modeling of whole-brain oscillations: Predicting regional vulnerability and hazardousness rates. Netw. Neurosci. 2019, 3, 1094–1120. [Google Scholar] [CrossRef] [PubMed]
- Jobst, B.M.; Hindriks, R.; Laufs, H.; Tagliazucchi, E.; Hahn, G.; Ponce-Alvarez, A.; Stevner, A.B.A.; Kringelbach, M.L.; Deco, G. Increased stability and breakdown of brain effective connectivity during slow-brain computational modeling. Sci. Rep. 2017, 7, 4634. [Google Scholar] [CrossRef] [Green Version]
- Wischnewski, K.J.; Eickhoff, S.B.; Jirsa, V.K.; Popovych, O.V. Towards an efficient validation of dynamical whole-brain models. Sci. Rep. 2022, 12, 4331. [Google Scholar] [CrossRef]
- Cabral, J.; Kringelbach, M.L.; Deco, G. Exploring the network dynamics underlying brain activity during rest. Prog. Neurobiol. 2014, 114, 102–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabral, J.; Hugues, E.; Sporns, O.; Deco, G. Role of local network oscillations in resting-state functional connectivity. NeuroImage 2011, 57, 130–139. [Google Scholar] [CrossRef]
- Deco, G.; Jirsa, V.K.; McIntosh, A.R. Resting brains never rest: Computational insights into potential cognitive architectures. Trends Neurosci. 2013, 36, 268–274. [Google Scholar] [CrossRef] [Green Version]
- Shakil, S.; Lee, C.H.; Keilholz, S.D. Evaluation of sliding window correlation performance for characterizing dynamic functional connectivity and brain states. NeuroImage 2016, 133, 111–128. [Google Scholar] [CrossRef] [Green Version]
- Kashyap, A.; Keilholz, S. Dynamic properties of simulated brain network models and empirical resting-state data. Netw. Neurosci. 2019, 3, 405–426. [Google Scholar] [CrossRef]
- Allen, E.A.; Damaraju, E.; Plis, S.M.; Erhardt, E.B.; Eichele, T.; Calhoun, V.D. Tracking whole-Brain Connectivity dynamics in the resting state. Cereb. Cortex 2014, 24, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Cabral, J.; Vidaurre, D.; Marques, P.; Magalhães, R.; Silva Moreira, P.; Miguel Soares, J.; Deco, G.; Sousa, N.; Kringelbach, M. Cognitive performance in health older adults relates to spontaneous switching between states of functional connectivity during rest. Sci. Rep. 2017, 7, 5135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deco, G.; Kringelbach, M.L.; Jirsa, V.K.; Ritter, P. The dynamics of resting fluctuations in the brain: Metastability and its dynamical cortical core. Sci. Rep. 2017, 7, 3095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakoğlu, U.; Pearlson, G.D.; Kiehl, K.A.; Wang, Y.M.; Michael, A.M.; Calhoun, V.D. A method for evaluating dynamic functional network connectivity and task-modulation: Application to schizophrenia. MAGMA 2010, 23, 351–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Wen, H.; Hu, X.; Xie, P.; Qiu, S.; Qian, Y.; Qiu, J.; He, H. Transition and dynamics reconfiguration of whole-brain network in major depressive disorder. Mol. Neurobiol. 2020, 57, 4031–4044. [Google Scholar] [CrossRef]
- Friston, K.J.; Harrison, L.; Penny, W. Dynamic causal modelling. Neuroimage 2003, 19, 1273–1302. [Google Scholar] [CrossRef]
- Razi, A.; Seghier, M.L.; Zhou, Y.; McColgan, P.; Zeidman, P.; Park, H.J.; Sporns, O.; Rees, G.; Friston, K.J. Large-scale DCMs for resting-state fMRI. Netw. Neurosci. 2017, 1, 222–241. [Google Scholar] [CrossRef]
- Kashyap, A.; Keilholz, S. Brain network constraints and recurrent neural networks reproduce unique trajectories and state transitions seen over the span of minutes in resting-state fMRI. Netw. Neurosci. 2020, 4, 448–466. [Google Scholar] [CrossRef] [Green Version]
- Kashyap, A.; Plis, S.; Schirner, M.; Ritter, P.; Keilholz, S. A deep learning approach to estimating initial conditions of brain network models in reference to measured fMRI data. bioRxiv 2021. [Google Scholar] [CrossRef]
- Chen, R.; Rubanova, Y.; Bettencourt, J.; Duvenaud, D. Neural ordinary differential equations. arXiv 2018, arXiv:1806. 07366v5. [Google Scholar]
- Bassett, D.S.; Xia, C.H.; Satterthwaite, T.D. Understanding the emergence of neuropsychiatric disorders with network neuroscience. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 2018, 3, 742–753. [Google Scholar] [CrossRef] [PubMed]
- Bassett, D.S.; Zurn, P.; Gold, J.I. On the nature and use of models in network neuroscience. Nat. Rev. Neurosci. 2018. Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Bassett, D.S.; Sporns, O. Network neuroscience. Nat. Neurosci. 2017, 20, 353–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valdes-Sasa, P.A.; Roebroeck, A.; Daunizeau, J.; Friston, K. Effective connectivity: Influence, causality and biophysical modeling. Neuroimage 2011, 58, 339–361. [Google Scholar] [CrossRef] [PubMed]
- Friston, K.J.; Litvka, V.; Oswal, A.; Razi, A.; Stephan, K.E.; van Wijk, B.C.M.; Ziegler, G.; Zeidman, P. Bayesian model reduction and empirical bayes for group (DCM) studies. Neuroimage 2016, 128, 413–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friston, K.J.; Mattout, J.; Trujillo-Barreto, N.; Ashburner, J.; Penny, W. Variational free energy and the Laplace approximation. Neuroimage 2007, 34, 220–234. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Yap, P.T. From descriptive connectome to mechanistic connectome: Generative modeling in functional magnetic resonance imaging analysis. Front. Hum. Neurosci. 2022, 16, 940842. [Google Scholar] [CrossRef]
- Friston, K.; Moran, R.; Seth, A.K. Analysing connectivity with Granger causality and dynamic causal modelling. Curr. Opin. Neurobiol. 2013, 23, 172–178. [Google Scholar] [CrossRef] [Green Version]
- Buxton, R.; Wong, E.; Frank, L. Dynamics of blood flow and oxygenation changes during brain activation: The balloon model. Magn. Reson. Med. 1998, 39, 855–864. [Google Scholar] [CrossRef]
- Stephan, K.; Kasper, L.; Harrison, L.; Daunizeau, J.; den Ouden, H.; Breakspear, M.; Friston, K. Nonlinear dynamic causal models for fMRI. Neuroimage 2008, 42, 649–662. [Google Scholar] [CrossRef] [Green Version]
- Bishop, C.M. Pattern Recognition and Machine Learning; Springer: New York, NY, USA, 2006; Volume 12, p. 105. [Google Scholar]
- Frässel, S.; Lomakina, E.I.; Kasper, L.; Manjaly, Z.M.; Leff, A.; Pruessmann, K.P. A generative model of whole-brain effective connectivity. Neuroimage 2018, 179, 505–529. [Google Scholar] [CrossRef] [PubMed]
- Frässel, S.; Manjaly, Z.M.; Do, C.T.; Kasper, L.; Pruessmann, K.P.; Stephan, K.E. Whole-brain estimates of directed connectivity for human connectomics. NeuroImage 2021, 225, 117491. [Google Scholar] [CrossRef] [PubMed]
- Frässel, S.; Lomakina, E.I.; Razi, A.; Friston, K.J.; Buhmann, J.M.; Stephan, K.E. Regression DCM for fMRI. NeuroImage 2017, 155, 406–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilson, M.; Deco, G.; Friston, K.J.; Hagmann, P.; Mantini, D.; Betti, V.; Romani, G.L.; Corbetta, M. Effective connectivity inferred from fMRI transition dynamics during movie viewing points to a balanced reconfiguration of cortical interactions. Neuroimage 2017, 180, 534–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilson, M.; Zamora-López, G.; Pallarés, V.; Adhikari, M.H.; Senden, M.; Campo, A.T.; Mantini, D.; Corbetta, M.; Deco, G.; Insabato, A. Model-based whole-brain effective connectivity to study distributed cognition in health and disease. Netw. Neurosci. 2020, 4, 338–373. [Google Scholar] [CrossRef] [Green Version]
- Lebouvier, T.; Chaumette, T.; Paillusson, S.; Duyckaerts, C.; Bruley des Varannes, S.; Neunlist, M.; Derkinderen, P. The second brain and Parkinson’s disease. Eur. J. Neurosci. 2009, 30, 735–741. [Google Scholar] [CrossRef]
- Van Eimeren, T.; Monchi, O.; Ballanger, B.; Strafella, A.P. Dysfunction of the default mode network in Parkinson’s disease. Arch. Neurol. 2009, 66, 877–883. [Google Scholar] [CrossRef] [Green Version]
- Van Hartevelt, T.J.; Cabral, J.; Deco, G.; Moller, A.; Green, A.L.; Aziz, T.Z.; Kringelbach, M.L. Neural plasticity in human Brain Connectivity: The effects of long term deep brain stimulation of the subthalamic nucleus in Parkinson’s disease. PLoS ONE 2014, 9, e86496. [Google Scholar] [CrossRef]
- Saenger, V.M.; Kahan, J.; Foltynie, T.; Friston, K.; Aziz, T.Z.; Green, A.L.; Hartevelt, T.J.; Cabral, J.; Stevner, A.B.A.; Fernandes, H.M.; et al. Uncovering the underlying mechanisms and whole-brain dynamics of deep brain stimulation for Parkinson’s disease. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Brier, M.R.; Thomas, J.B.; Ances, B.M. Network dysfunction in Alzheimer’s disease: Refining the disconnection hypothesis. Brain Connect 2014, 4, 299–311. [Google Scholar] [CrossRef] [Green Version]
- Dennis, E.L.; Thompson, P.M. Functional Brain Connectivity using fMRI in aging and Alzheimer’s disease. Neuropsychol. Rev. 2014, 24, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Van Nifterick, A.M.; Gouw, A.A.; van Kesteren, R.E.; Scheltens, P.; Stam, C.J.; de Haan, W. A multiscale brain network model links Alzheimer’s disease-mediated neuronal hyperactivity to large-scale oscillatory slowing. Alzheimers Res. Ther. 2022, 14, 101. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Rodriguez, L.M.; Iturria-Medina, Y.; Baines, E.A.; Mallo, S.C.; Dousty, M.; Sotero, R.C. Design of optimal nonlinear network controllers for Alzheimer’s disease. PLoS Comput. Biol. 2018, 14, e1006136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephan, K.E.; Baldeweg, T.; Friston, K.J. Synaptic plasticity and disconnection in schizophrenia. Biol. Psychiatry 2006, 59, 929–939. [Google Scholar] [CrossRef]
- Uhlhaas, P.J. Dysconnectivity, large-scale networks and neuronal dynamics in schizophrenia. Curr. Opin. Neurobiol. 2013, 23, 283–290. [Google Scholar] [CrossRef]
- Lewis, D.A.; Hashimoto, T.; Volk, D.W. Cortical inhibitory neurons and schizophrenia. Nat. Rev. Neurosci. 2005, 6, 312–324. [Google Scholar] [CrossRef]
- Nakazawa, K.; Zsiros, V.; Jiang, Z.; Nakao, K.; Kolata, S.; Zhang, S.; Belforte, J.E. GABAergic interneuron origin of schizophrenia pathophysiology. Neuropharmacology 2012, 62, 1574–1583. [Google Scholar] [CrossRef] [Green Version]
- Cabral, J.; Fernandes, H.M.; Van Hartevelt, T.J.; James, A.C.; Kringelbach, M.L.; Deco, G. Structural connectivity in schizophrenia and its impact on the dynamics of spontaneous functional networks. Chaos 2013, 23, 046111. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.J.; Murray, J.D.; Repovs, G.; Cole, M.W.; Savic, A.; Glasser, M.F.; Pittenger, C.; Krystal, J.H.; Wang, X.; Pearlson, G.D.; et al. Altered global brain signal in schizophrenia. Proc. Natl. Acad. Sci. USA 2014, 111, 7438–7443. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.J.; Murray, J.D.; Wang, X.J.; Glahn, D.C.; Pearlson, G.D.; Repovs, G.; Krystal, J.H.; Anticevic, A. Functional hierarchy underlies preferential connectivity disturbances in schizophrenia. Proc. Natl. Acad. Sci. USA 2016, 113, E219–E228. [Google Scholar] [CrossRef] [Green Version]
- Anticevic, A.; Hu, X.; Xiao, Y.; Hu, J.; Li, F.; Bi, F.; Cole, M.W.; Savic, A.; Yang, G.J.; Repovs, G.; et al. Early-course unmedicated schizophrenia patients exhibit elevated prefrontal connectivity associated with longitudinal change. J. Neurosci. 2015, 35, 267–886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cole, M.W.; Yang, G.J.; Murry, J.D.; RepovŠ, G.; Anticevic, A. Functional connectivity change as shared signal dynamics. J. Neurosci. Methods 2016, 259, 22–39. [Google Scholar] [CrossRef] [PubMed]
- Anticevic, A.; Gancsos, M.; Murray, J.D.; Repovs, G.; Driesen, N.R.; Ennis, D.J.; Niciu, M.J.; Morgan, P.T.; Surti, T.S.; Bloch, M.H.; et al. NMDA receptor function in large-scale anticorrelated neural systems with implications for cognition and schizophrenia. Proc. Natl. Acad. Sci. USA 2012, 109, 16720–16725. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Braun, U.; Harneit, A.; Zang, Z.; Geiger, L.S.; Betzel, R.F.; Chen, J.; Schweiger, J.L.; Schwarz, K.; Reinwald, J.R.; et al. Generative network models of altered structural Brain Connectivity in schizophrenia. Neuroimage 2021, 225, 117510. [Google Scholar] [CrossRef] [PubMed]
- Bansal, K.; Nakuci, J.; Muldoon, S.F. Personalized brain network models for assessing structure-functional relationships. Curr. Opin. Neurobiol. 2018, 52, 42–47. [Google Scholar] [CrossRef] [Green Version]
- Muldoon, S.F.; Pasqualetti, F.; Gu, S.; Cieslak, M.; Grafton, S.T.; Vettel, J.M.; Bassett, D.S. Stimulation-based control of dynamical brain networks. PLoS Comput. Biol. 2016, 12, e1005076. [Google Scholar] [CrossRef] [Green Version]
- Desikan, R.S.; Ségonne, F.; Fischl, B.; Quinn, B.T.; Dickerson, B.C.; Blacker, D.; Buckner, R.L.; Dale, A.M.; Maguire, R.P.; Hyman, B.T.; et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 2006, 31, 968–980. [Google Scholar] [CrossRef]
- Tzourio-Mazoyer, N.; Landeau, B.; Papathanassiou, D.; Crivello, F.; Etard, E.; Delcroix, N.; Mazoyer, B.; Joliot, M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002, 15, 273–289. [Google Scholar] [CrossRef]
- Proix, T.; Spiegler, A.; Schirner, M.; Rothmeier, S.; Ritter, P.; Jirsa, V.K. How do parcellation size and short-range connectivity affect dynamics in large-scale brain network models? Neuroimage 2016, 142, 135–149. [Google Scholar] [CrossRef]
- Deco, G.; Tononi, G.; Boly, M.; Kringelbach, M.L. Rethinking segregation and integration: Contributions of whole-brain modelling. Nat. Rev. Neurosci. 2015, 16, 430–439. [Google Scholar] [CrossRef]
- Markov, N.T.; Misery, P.; Falchier, A.; Lamy, C.; Vezoli, J.; Quilodran, R.; Gariel, M.A.; Giroud, P.; Ercsey-Ravasz, M.; Pilaz, L.J.; et al. Weight consistency specifies regularities of macaque cortical networks. Cereb. Cortex 2011, 21, 1254–1272. [Google Scholar] [CrossRef] [PubMed]
- Dimitriadis, S.I.; Drakesmith, M.; Bells, S.; Parker, G.D.; Linden, D.E.; Jones, D.K. Improving the reliability of network metrics in structural brain networks by integrating different network weighting strategies into a single graph. Front. Neurosci. 2017, 11, 736. [Google Scholar] [CrossRef] [PubMed]
- Messaritaki, E.; Dimitriadis, S.I.; Jones, D.K. Optimization of graph construction can significantly increase the power of structural brain network studies. Neuroimage 2019, 199, 495–511. [Google Scholar] [CrossRef]
- Conti, E.; Mitra, J.; Calderoni, S.; Pannek, K.; Shen, K.K.; Pagnozzi, A.; Rose, S.; Mazzotti, S.; Scelfo, D.; Tosetti, M.; et al. Network over-connectivity differentiates autism spectrum disorder from other developmental disorders in toddlers: A diffusion MRI study. Hum. Brain Mapp. 2017, 38, 2333–2344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oxtoby, N.P.; Garbarino, S.; Firth, N.C.; Warren, J.D.; Schott, J.M.; Alexander, D.C. Data-driven sequence of changes to anatomical Brain Connectivity in sporadic Alzheimer’s disease. Front. Neurol. 2017, 8, 580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betzel, R.F.; Bassett, D.S. Specificity and robustness of long-distance connections in weighted, interareal connectomes. Proc. Natl. Acad. Sci. USA 2018, 115, E4880–E4889. [Google Scholar] [CrossRef] [Green Version]
- Singh, M.F.; Braver, T.S.; Cole, M.W.; Ching, S. Estimation and validation of individualized dynamic brain models with resting state fMRI. Neuroimage 2020, 221, 117046. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Liu, Y.; Zheng, Y.; Wu, Y.; Li, D.; Liang, X.; Chen, Y.; Cui, Y.; Yap, P.T.; Qiu, S.; et al. Multiscale neural modeling of resting-state fMRI reveals executive-limbic malfunction as a core mechanism in major depressive disorder. Neuroimage 2021, 31, 102758. [Google Scholar] [CrossRef]
- Sip, V.; Petkoski, S.; Hashemi, M.; Dickscheid, T.; Amunts, K.; Jirsa, V. Parameter inference on brain network models with unknown node dynamics and spatial heterogeneity. bioRxiv 2022. [Google Scholar] [CrossRef]
- Hipp, J.F.; Hawellek, D.J.; Corbetta, M.; Siegel, M.; Engel, A.K. Large-scale cortical correlation structural of spontaneous oscillatory activity. Nat. Neurosci. 2012, 15, 884–890. [Google Scholar] [CrossRef] [Green Version]
- Brookes, M.J.; Hale, J.R.; Zumer, J.M.; Stevenson, C.M.; Francis, S.T.; Barnes, G.R.; Owen, J.P.; Morris, P.G.; Nagarajan, S.S. Measuring functional connectivity using meg: Methodology and comparison with fcMRI. NeuroImage 2011, 56, 1082–1104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabuffo, G.; Fousek, J.; Bernard, C.; Jirsa, V. Neuronal cascades shape whole-brain functional dynamics at rest. eNeuro 2021, 8, 0283-21. [Google Scholar] [CrossRef] [PubMed]
- Warbrick, T. Simultaneous EEG-fMRI: What have we learned and what does the future hold? Sensors 2022, 22, 2262. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Wu, L.; Bridwell, D.A.; Erhardt, E.B.; Du, Y.; He, H.; Chen, J.; Liu, P.; Sui, J.; Pearlson, G.; et al. Building an EEG-fMRI multi-modal brain graph: A concurrent EEG-fMRI study. Front. Hum. Neurosci. 2016, 10, 476. [Google Scholar] [CrossRef] [Green Version]
- Prokopiou, P.C.; Xifra-Porxas, A.; Kassinopoulos, M.; Boudriad, M.H.; Mitsis, G.D. Modeling the hemodynamic response function using EEG-fMRI data during eyes-open resting-state conditions and motor task execution. Brain Topogr. 2022, 35, 302–321. [Google Scholar] [CrossRef] [PubMed]
- Santanielloa, S.; Gale, J.T.; Sarma, S. Systems approaches to optimizing deep brain stimulation therapies in Parkinson’s disease. WIREs Syst. Biol. Med. 2018. [Google Scholar] [CrossRef] [Green Version]
- Fisher, R.S.; Velasco, A.L. Electrical brain stimulation for epilepsy. Nat. Rev. Neurol. 2014, 10, 261–270. [Google Scholar] [CrossRef]
- Johnson, M.D.; Lim, H.H.; Netoff, T.I.; Connolly, A.T.; Johnson, N.; Roy, A.; Holt, A.; Lim, K.O.; Carey, J.R.; Vitek, J.L.; et al. Neuromodulation for brain disorders: Challenges and opportunities. IEEE Trans. Biomed. Eng. 2013, 60, 610–624. [Google Scholar] [CrossRef]
- Srivastava, P.; Fotiadis, P.; Parkes, L.; Bassett, D.S. The expanding horizons of network neuroscience: From description to prediction and control. Neuroimage 2022, 258, 119250. [Google Scholar] [CrossRef]
- Pasqualetti, F. Controllability metrics, limitations and algorithms for complex networks. Trans. Control Netw. Syst. 2012, 1, 40–52. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, P.; Nozari, E.; Kim, J.Z.; Ju, H.; Zhou, D.; Becker, C.; Pasqualetti, F.; Pappas, G.J.; Bassett, D.S. Models of communication and control for brain networks: Distinctions, convergence, and future outlook. Netw. Neurosci. 2020, 4, 1122–1159. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Pasqualetti, F.; Cieslak, M.; Telesford, Q.K.; Yu, A.B.; Kahn, A.E.; Medaglia, J.D.; Vettel, J.M.; Miller, M.B.; Grafton, S.T.; et al. Controllability of structural brain networks. Nat. Commun. 2015, 6, 8414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karrer, T.M.; Kim, J.Z.; Stiso, J.; Kahn, A.E.; Pasqualetti, F.; Habel, U.; Bassett, D.S. A practical guide to methodological considerations in the controllability of structural brain networks. J. Neural Eng. 2020, 17, 026031. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.Y.; Slotine, J.J.; Barabasi, A.L. Controllability of complex networks. Nature 2011, 473, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Cole, M.; Braver, T.; Ching, S. Developing control-theoretic objectives for large-scale brain dynamics and cognitive enhancement. Annu. Rev. Control. 2022. [Google Scholar] [CrossRef]
- Singh, M.; Wang, M.; Cole, M.; Ching, S. Efficient Identification for Modeling High-Dimensional Brain Dynamics; IEEE: Piscataway, NJ, USA, 2022; pp. 1353–1358. [Google Scholar]
- Ljung, L. System Identification: Theory for the User; Prentice Hall: Hoboken, NJ, USA, 1987. [Google Scholar]
- Yang, Y.; Connolly, A.T.; Shanechi, M.M. A control-theoretic system identification framework and a real-time closed-loop clinical simulation testbed for electrical brain stimulation. J. Neural Eng. 2018, 15, 066007. [Google Scholar] [CrossRef]




| Network Properties | Multiscale BNMs | Descriptive BNMs | DCM |
|---|---|---|---|
| Node Dynamics | Differential equations | NA | Differential equations |
| Network Connections | SC | SC FC EC | EC |
| Model Fitting | Static FC Dynamic FC BOLD signals | Static SC Static FC Static EC | Static FC Dynamic FC BOLD signals |
| Network Size | Large-scale | Large-scale | Relatively small networks |
| Biophysical Foundation | Firing rate Membrane potential Excitability Inhibitory | NA | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, M.; Guo, Z.; Gao, Z.; Cao, Y.; Fu, J. Multiscale Brain Network Models and Their Applications in Neuropsychiatric Diseases. Electronics 2022, 11, 3468. https://doi.org/10.3390/electronics11213468
Lu M, Guo Z, Gao Z, Cao Y, Fu J. Multiscale Brain Network Models and Their Applications in Neuropsychiatric Diseases. Electronics. 2022; 11(21):3468. https://doi.org/10.3390/electronics11213468
Chicago/Turabian StyleLu, Meili, Zhaohua Guo, Zicheng Gao, Yifan Cao, and Jiajun Fu. 2022. "Multiscale Brain Network Models and Their Applications in Neuropsychiatric Diseases" Electronics 11, no. 21: 3468. https://doi.org/10.3390/electronics11213468