Incorporation of Synthetic Data Generation Techniques within a Controlled Data Processing Workflow in the Health and Wellbeing Domain
Abstract
:1. Introduction
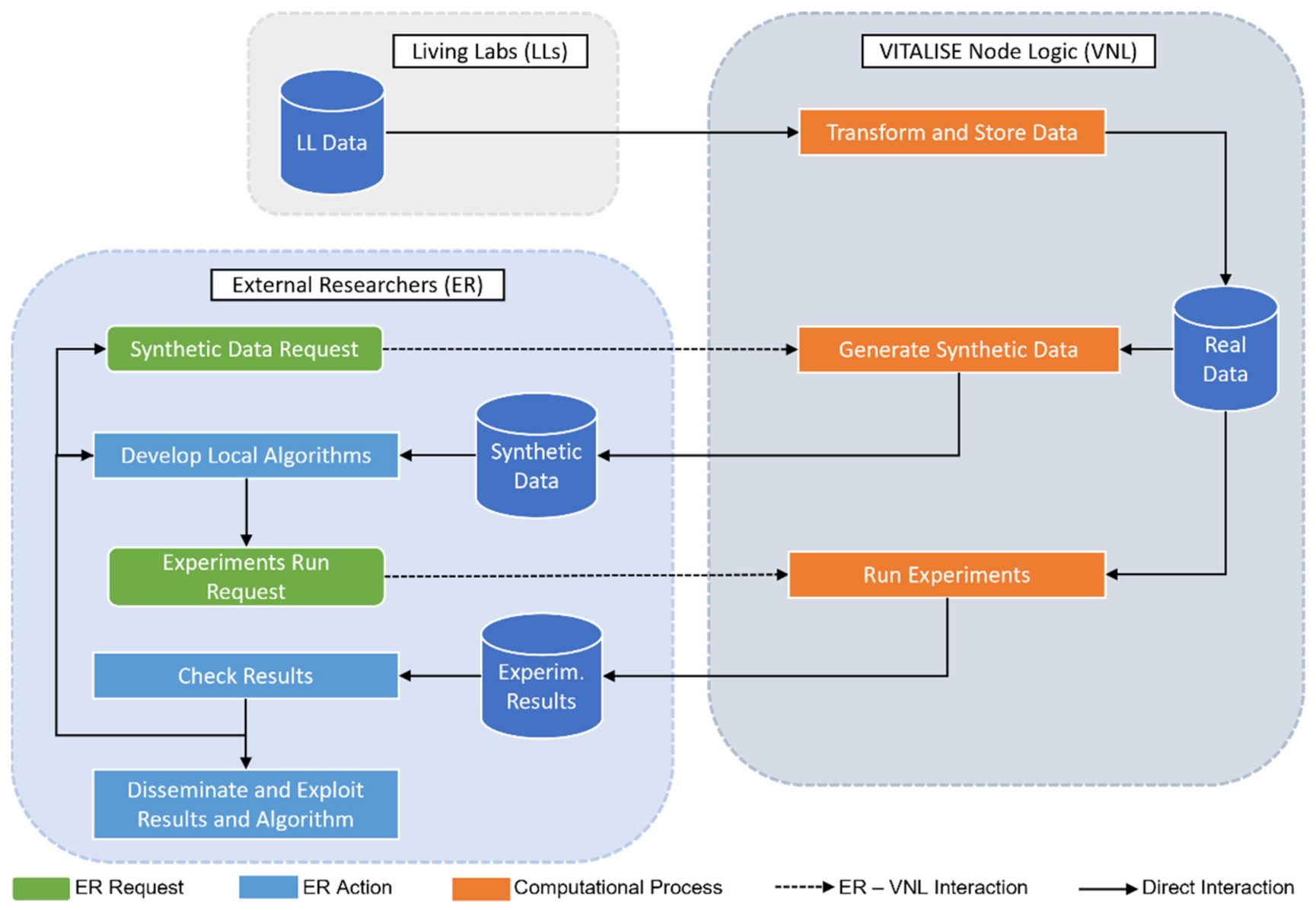
- We present a controlled data processing workflow for a secure data exchange and analysis without compromising data privacy. The workflow involves the generation of SD based on previously uploaded RD and the remote execution of experiments with RD on LLs premises.
- To the best of our knowledge, this work is the first attempt to propose the incorporation and automation of SDG models within a controlled data processing workflow whose objective is to ensure compliance with personal data protection laws.
- We have conducted a real-world usage example to demonstrate the usefulness and efficiency of the proposed workflow. To conduct the experiments, we have used heart rate data measured from Fitbit smart wristbands.
- Additionally, we have performed an experiment with the SD obtained from the heart rate values to analyze the performance on the resemblance and utility dimensions. For this analysis, we have used some metrics to evaluate the resemblance of SD to RD, and we have performed some forecasting analyses locally with different SD assets and executed them remotely with RD.
2. Materials and Methods
2.1. VITALISE LL Controlled Data Processing Workflow
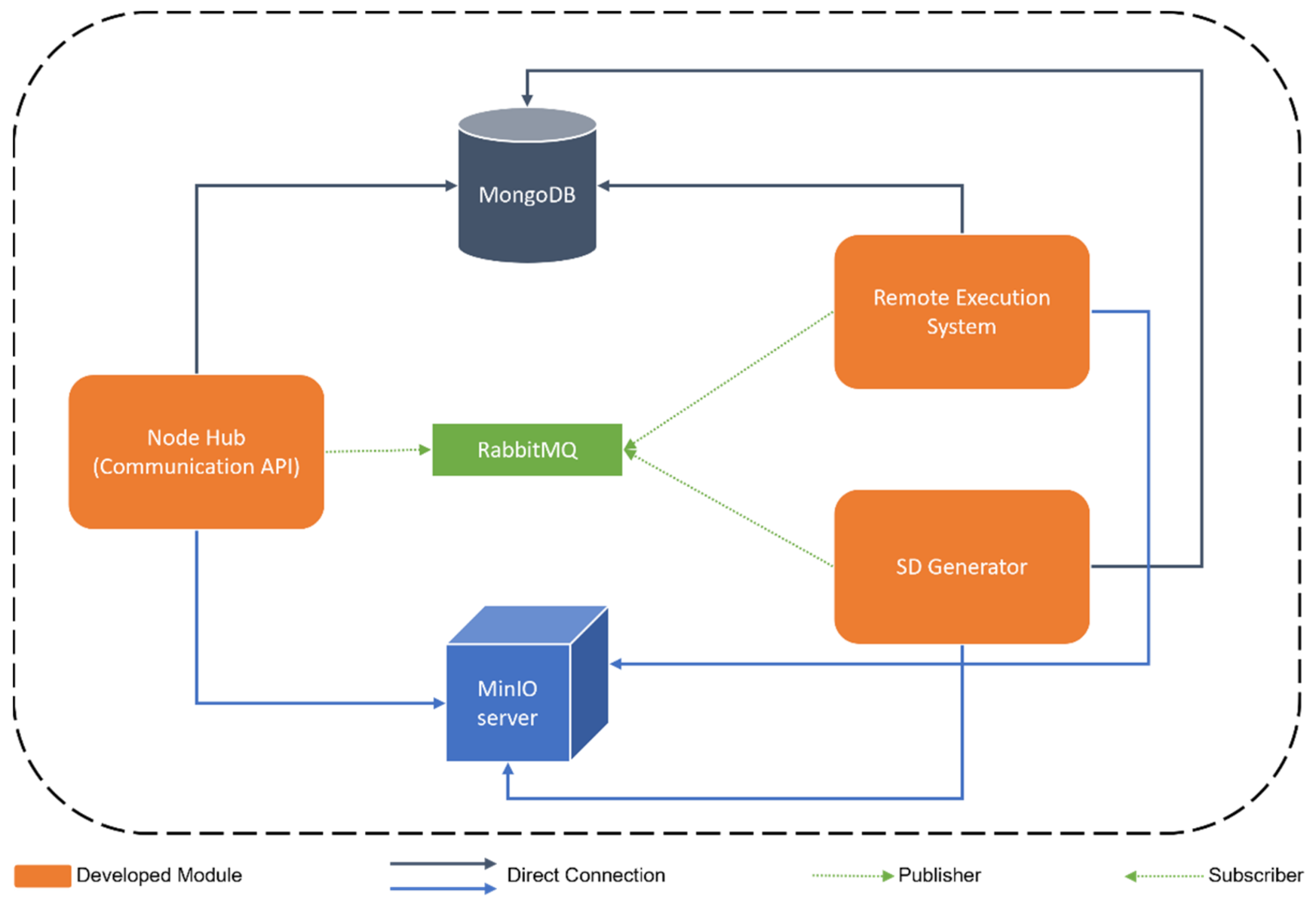
VITALISE Node Logic
- MongoDB is a distributed NoSQL database system [33] to store real data from LLs and generated SD in the defined Vitalise Data Format.
- RabbitMQ is an open-source message broker [34] used to communicate the modules of the environment and queue tasks.
- MinIO server is an object storage server [35] that stores trained SDG models, generated SD in CSV format, the necessary files for the remote execution of experiments with RD, and the results produced as part of remote execution of experiments with RD.
- The Node Hub is a Communication Application Programming Interface (API) developed in Python using the FastAPI framework [36] that handles the requests coming from ER through the web portal and works as an intermediary between the other two modules (SD Generator and Remote Execution Engine). Thereby, it has access to RabbitMQ (to queue tasks for the other modules), MongoDB (to store and query available RD and SD), and MinIO server (to write input files for the remote execution of experiments with RD).
- The SD Generator is an MQTT client subscribed to the SD topic of RabbitMQ and developed in Python. This module is responsible for training SDG models and generating SD. Thus, it has access to RabbitMQ (to subscribe to the SD topic), MongoDB (to query available RD and store the generated SD), and MinIO server (to store the trained SDG models and the generated SD in CSV format).
- The Remote Execution Engine is a distributed system, which is developed using Celery [37] and Python, to process and queue the tasks regarding the remote execution of analysis with RD that are sent through RabbitMQ by the Node Hub. It has access to RabbitMQ (to see the queued tasks regarding the remote execution), MongoDB (to query available RD and SD), and MinIO server (to access the necessary files for the remote execution of each experiment and to store the results of them).
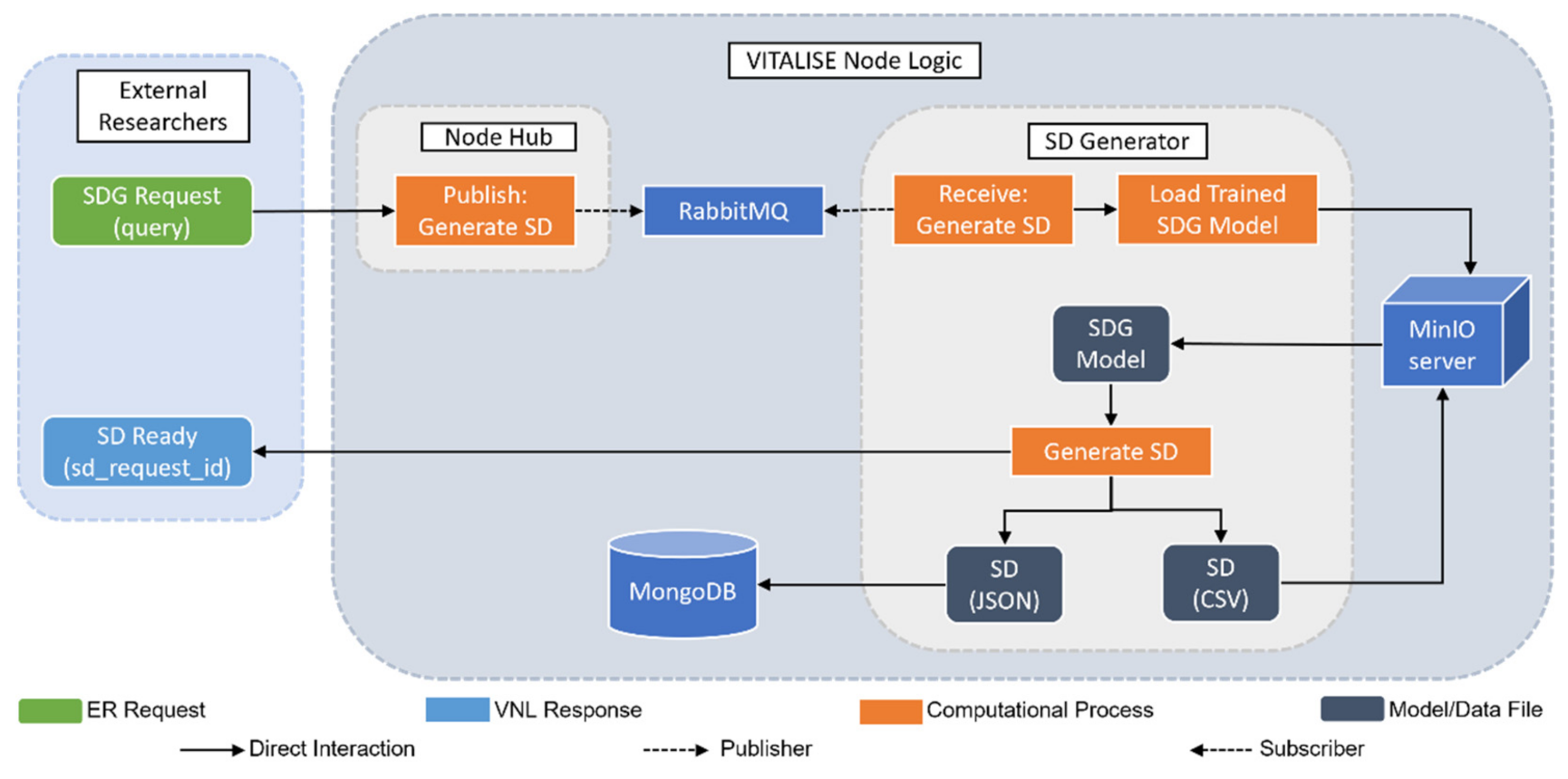
2.2. Synthetic Data Generation Module Integration
2.2.1. Synthetic Data Generation Approaches
2.2.2. Synthetic Data Generation Model Training
2.2.3. Synthetic Data Generation with Trained Models
2.2.4. Generated Synthetic Data Retrieval
3. Results
3.1. Used Data
3.2. Workflow Execution
- By simulating the role of an LLs manager, the VNL has been used to upload the heart rate measurements to the VNL. At this moment, the SDG approach has been trained with the uploaded data.
- By simulating the role of ER, a petition has been made to the VNL to request SD of the first five hours of the previously uploaded heart rate data.
- Taking advantage of the obtained SD request ID, the ER has been able to obtain SD in the desired format, either JSON or CSV. In both cases, a zip file has been downloaded with two files, one with the information of SD (fields shown in Table 1) and the other with the SD itself.
- Using the obtained SD in CSV format, a forecasting model has been trained with measurements from four hours and tested, making predictions for the next hour.
- The remote execution of the locally developed algorithm has been requested by the VNL to obtain the evaluation results of applying the same forecasting model to RD.
| Field | Description |
| sd_request_id | Unique identifier of the SDG request. |
| hash_query_real description timestamp | Hash to identify query of real data results. Description of the query used for SDG. Date of when the SD has been generated. |
3.2.1. Resemblance Evaluation of Generated SD
3.2.2. Data Forecasting Analyses
4. Discussion
4.1. SDG Integrated Workflow Execution Results
4.2. Main Findings
4.3. Limitations and Future Work
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- El Emam, K.; Hoptroff, R. The Synthetic Data Paradigm for Using and Sharing Data. Data Anal. Digit. Technol. 2019, 19, 12. [Google Scholar]
- Hernandez-Matamoros, A.; Fujita, H.; Perez-Meana, H. A novel approach to create synthetic biomedical signals using BiRNN. Inf. Sci. 2020, 541, 218–241. [Google Scholar] [CrossRef]
- Piacentino, E.; Guarner, A.; Angulo, C. Generating Synthetic ECGs Using GANs for Anonymizing Healthcare Data. Electronics 2021, 10, 389. [Google Scholar] [CrossRef]
- Hazra, D.; Byun, Y.-C. SynSigGAN: Generative Adversarial Networks for Synthetic Biomedical Signal Generation. Biology 2020, 9, 441. [Google Scholar] [CrossRef] [PubMed]
- Andreini, P.; Ciano, G.; Bonechi, S.; Graziani, C.; Lachi, V.; Mecocci, A.; Sodi, A.; Scarselli, F.; Bianchini, M. A Two-Stage GAN for High-Resolution Retinal Image Generation and Segmentation. Electronics 2022, 11, 60. [Google Scholar] [CrossRef]
- Porcu, S.; Floris, A.; Atzori, L. Evaluation of Data Augmentation Techniques for Facial Expression Recognition Systems. Electronics 2020, 9, 1892. [Google Scholar] [CrossRef]
- Han, C.; Hayashi, H.; Rundo, L.; Araki, R.; Shimoda, W.; Muramatsu, S.; Furukawa, Y.; Mauri, G.; Nakayama, H. GAN-based synthetic brain MR image generation. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 734–738. [Google Scholar]
- Stephens, M.; Estepar, R.S.J.; Ruiz-Cabello, J.; Arganda-Carreras, I.; Macía, I.; López-Linares, K. MRI to CTA Translation for Pulmonary Artery Evaluation Using CycleGANs Trained with Unpaired Data. In Thoracic Image Analysis, Proceedings of the Thoracic Image Analysis, Lima, Peru, 8 October 2020; Petersen, J., San José Estépar, R., Schmidt-Richberg, A., Gerard, S., Lassen-Schmidt, B., Jacobs, C., Beichel, R., Mori, K., Eds.; Springer International Publishing: Cham, Germany, 2020; pp. 118–129. [Google Scholar]
- Dahmen, J.; Cook, D. SynSys: A Synthetic Data Generation System for Healthcare Applications. Sensors 2019, 19, 1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norgaard, S.; Saeedi, R.; Sasani, K.; Gebremedhin, A.H. Synthetic Sensor Data Generation for Health Applications: A Supervised Deep Learning Approach. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 1164–1167. [Google Scholar]
- Li, Z.; Ma, C.; Shi, X.; Zhang, D.; Li, W.; Wu, L. TSA-GAN: A Robust Generative Adversarial Networks for Time Series Augmentation. In Proceedings of the 2021 International Joint Conference on Neural Networks (IJCNN), Shenzhen, China, 18–22 July 2021; IEEE: Shenzhen, China, 2021; pp. 1–8. [Google Scholar]
- Wang, J.; Chen, Y.; Gu, Y.; Xiao, Y.; Pan, H. SensoryGANs: An Effective Generative Adversarial Framework for Sensor-based Human Activity Recognition. In Proceedings of the 2018 International Joint Conference on Neural Networks (IJCNN), Rio de Janeiro, Brazil, 8–13 July 2018; pp. 1–8. [Google Scholar]
- Rankin, D.; Black, M.; Bond, R.; Wallace, J.; Mulvenna, M.; Epelde, G. Reliability of Supervised Machine Learning Using Synthetic Data in Health Care: Model to Preserve Privacy for Data Sharing. JMIR Med. Inform. 2020, 8, e18910. [Google Scholar] [CrossRef] [PubMed]
- Yale, A.; Dash, S.; Dutta, R.; Guyon, I.; Pavao, A.; Bennett, K.P. Generation and evaluation of privacy preserving synthetic health data. Neurocomputing 2020, 416, 244–255. [Google Scholar] [CrossRef]
- Beaulieu-Jones Brett, K.; Wu, Z.S.; Williams, C.; Lee, R.; Bhavnani, S.P.; Byrd, J.B.; Greene, C.S. Privacy-Preserving Generative Deep Neural Networks Support Clinical Data Sharing. Circ. Cardiovasc. Qual. Outcomes 2019, 12, e005122. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Myles, P.; Tucker, A. Generating and Evaluating Synthetic UK Primary Care Data: Preserving Data Utility Patient Privacy. In Proceedings of the 2019 IEEE 32nd International Symposium on Computer-Based Medical Systems (CBMS), Cordoba, Spain, 5–7 June 2019; pp. 126–131. [Google Scholar]
- Rashidian, S.; Wang, F.; Moffitt, R.; Garcia, V.; Dutt, A.; Chang, W.; Pandya, V.; Hajagos, J.; Saltz, M.; Saltz, J. SMOOTH-GAN: Towards Sharp and Smooth Synthetic EHR Data Generation. In Artificial Intelligence in Medicine, Proceedings of the Artificial Intelligence in Medicine, Minneapolis, MN, USA, 25–28 August 2020; Michalowski, M., Moskovitch, R., Eds.; Springer International Publishing: Cham, Germany, 2020; pp. 37–48. [Google Scholar]
- Yoon, J.; Drumright, L.N.; van der Schaar, M. Anonymization Through Data Synthesis Using Generative Adversarial Networks (ADS-GAN). IEEE J. Biomed. Health Inform. 2020, 24, 2378–2388. [Google Scholar] [CrossRef] [PubMed]
- Baowaly, M.K.; Lin, C.-C.; Liu, C.-L.; Chen, K.-T. Synthesizing electronic health records using improved generative adversarial networks. J. Am. Med. Inform. Assoc. 2019, 26, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Che, Z.; Cheng, Y.; Zhai, S.; Sun, Z.; Liu, Y. Boosting Deep Learning Risk Prediction with Generative Adversarial Networks for Electronic Health Records. In Proceedings of the 2017 IEEE International Conference on Data Mining (ICDM), New Orleans, LA, USA, 18–21 November 2017; pp. 787–792. [Google Scholar]
- Goncalves, A.; Ray, P.; Soper, B.; Stevens, J.; Coyle, L.; Sales, A.P. Generation and evaluation of synthetic patient data. BMC Med. Res. Methodol. 2020, 20, 108. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, M.; Epelde, G.; Alberdi, A.; Cilla, R.; Rankin, D. Standardised Metrics and Methods for Synthetic Tabular Data Evaluation. 2021. [CrossRef]
- Dankar, F.K.; Ibrahim, M.K.; Ismail, L. A Multi-Dimensional Evaluation of Synthetic Data Generators. IEEE Access 2022, 10, 11147–11158. [Google Scholar] [CrossRef]
- SYNTHO. Available online: https://www.syntho.ai/ (accessed on 13 January 2022).
- The Medkit-Learn(ing) Environment. Available online: https://github.com/vanderschaarlab/medkit-learn (accessed on 24 January 2022).
- Soni, R.; Zhang, M.; Avinash, G.B.; Saripalli, V.R.; Guan, J.; Pati, D.; Ma, Z. Medical Machine Synthetic Data and Corresponding Event Generation 2020. U.S. Patent Application No. 16/689,798, 29 October 2020. [Google Scholar]
- Build Better Datasets for AI with Synthetic Data. Available online: https://ydata.ai (accessed on 24 January 2022).
- The Synthetic Data Vault. Put Synthetic Data to Work! Available online: https://sdv.dev/ (accessed on 24 January 2022).
- VITALISE Project. Available online: https://vitalise-project.eu/ (accessed on 24 January 2022).
- Why VITALISE. Available online: https://vitalise-project.eu/why-vitalise/ (accessed on 13 January 2022).
- Rieke, N.; Hancox, J.; Li, W.; Milletarì, F.; Roth, H.R.; Albarqouni, S.; Bakas, S.; Galtier, M.N.; Landman, B.A.; Maier-Hein, K.; et al. The future of digital health with federated learning. NPJ Digit. Med. 2020, 3, 119. [Google Scholar] [CrossRef] [PubMed]
- Hallock, H.; Marshall, S.E.; Peter, A.C.’t.H.; Nygård, J.F.; Hoorne, B.; Fox, C.; Alagaratnam, S. Federated Networks for Distributed Analysis of Health Data. Front. Public Health 2021, 9, 712569. [Google Scholar] [CrossRef] [PubMed]
- MongoDB Documentation. Available online: https://docs.mongodb.com/ (accessed on 24 January 2022).
- Messaging That Just Works—RabbitMQ. Available online: https://www.rabbitmq.com/ (accessed on 24 January 2022).
- MinIO, Inc. MinIO|High Performance, Kubernetes Native Object Storage. Available online: https://min.io (accessed on 24 January 2022).
- FastAPI. Available online: https://fastapi.tiangolo.com/ (accessed on 24 January 2022).
- Celery-Distributed Task Queue—Celery 5.2.3 Documentation. Available online: https://docs.celeryproject.org/en/stable/ (accessed on 24 January 2022).
- Dalsania, N.; Patel, Z.; Purohit, S.; Chaudhury, B. An Application of Machine Learning for Plasma Current Quench Studies via Synthetic Data Generation. Fusion Eng. Des. 2021, 171, 112578. [Google Scholar] [CrossRef]
- Zhang, C.; Kuppannagari, S.R.; Kannan, R.; Prasanna, V.K. Generative Adversarial Network for Synthetic Time Series Data Generation in Smart Grids. In Proceedings of the 2018 IEEE International Conference on Communications, Control and Computing Technologies for Smart Grids (SmartGridComm), Aalborg, Denmark, 29–31 October 2018; pp. 1–6. [Google Scholar]
- Alqahtani, H.; Kavakli-Thorne, M.; Kumar, G. Applications of Generative Adversarial Networks (GANs): An Updated Review. Arch. Comput. Methods Eng. 2019, 28, 525–552. [Google Scholar] [CrossRef]
- Patki, N.; Wedge, R.; Veeramachaneni, K. The Synthetic Data Vault. In Proceedings of the 2016 IEEE International Conference on Data Science and Advanced Analytics (DSAA), Montreal, QC, Canada, 17–19 October 2016; pp. 399–410. [Google Scholar]
- Hittmeir, M.; Mayer, R.; Ekelhart, A. A Baseline for Attribute Disclosure Risk in Synthetic Data. In Proceedings of the Tenth ACM Conference on Data and Application Security and Privacy, New Orleans, LA, USA, 16–18 March 2020; Association for Computing Machinery: New York, NY, USA, 2020; pp. 133–143. [Google Scholar]
- Mayer, R.; Hittmeir, M.; Ekelhart, A. Privacy-Preserving Anomaly Detection Using Synthetic Data. In Data and Applications Security and Privacy XXXIV; Singhal, A., Vaidya, J., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Germany, 2020; Volume 12122, pp. 195–207. ISBN 978-3-030-49668-5. [Google Scholar]
- Hittmeir, M.; Ekelhart, A.; Mayer, R. Utility and Privacy Assessments of Synthetic Data for Regression Tasks. In Proceedings of the 2019 IEEE International Conference on Big Data (Big Data), Los Angeles, CA, USA, 9–12 December 2019; pp. 5763–5772. [Google Scholar]
- Hittmeir, M.; Ekelhart, A.; Mayer, R. On the Utility of Synthetic Data: An Empirical Evaluation on Machine Learning Tasks. In Proceedings of the 14th International Conference on Availability, Reliability and Security, Canterbury, UK, 26–29 August 2019; ACM: Canterbury, UK, 2019; pp. 1–6. [Google Scholar]
- Hyndman, R.J.; Athanasopoulos, G. Forecasting: Principles and Practice; OTexts: Melbourne, VIC, Australia, 2018; ISBN 978-0-9875071-1-2. [Google Scholar]
- SARIMAX: Introduction-Statsmodels. Available online: https://www.statsmodels.org/dev/examples/notebooks/generated/statespace_sarimax_stata.html (accessed on 24 January 2022).





| It. | Data Size | Mean | std | |
|---|---|---|---|---|
| 1 | RD | 2440 | 74.37 | ±11.76 |
| SD | 79.64 | ±8.8 | ||
| 2 | RD | 4392 | 71.07 | ±11.64 |
| SD | 76.11 | ±8.86 | ||
| 3 | RD | 6344 | 74.33 | ±12.37 |
| SD | 74.91 | ±9.9 | ||
| 4 | RD | 8296 | 75.66 | ±11.71 |
| SD | 75.91 | ±10.02 | ||
| 5 | RD | 10,248 | 77.54 | ±12.04 |
| SD | 76.71 | ±9.54 | ||
| 6 | RD | 11,724 | 77.36 | ±11.66 |
| SD | 75.16 | ±9.09 |
| It. | Train Size | Test Size | MFE | MAE | MSE | RMSE | |
|---|---|---|---|---|---|---|---|
| 1 | RD (RE) * | 1952 | 488 | 3.7848 * | 4.6536 * | 38.4651 * | 6.2020 * |
| SD (LE) | 13.3709 | 13.3873 | 233.1556 | 15.2691 | |||
| 2 | RD (RE) | 3904 | 488 | 5.1335 | 6.9897 | 72.8914 | 8.5376 |
| SD (LE) | 4.8278 | 8.0245 | 112.4181 | 10.6027 | |||
| 3 | RD (RE) | 5856 | 488 | 8.4036 | 10.2930 | 191.5266 | 13.8393 |
| SD (LE) | 1.4877 | 8.4385 | 100.6106 | 10.0304 | |||
| 4 | RD (RE) | 7808 | 488 | 9.8114 | 10.6352 | 189.7254 | 13.7740 |
| SD (LE)* | 2.1701 * | 6.6331 * | 62.1659 * | 7.8845 * | |||
| 5 | RD (RE) | 9760 | 488 | 6.2950 | 9.2991 | 149.4631 | 12.225 |
| SD (LE) | 2.7377 | 7.3770 | 76.2827 | 8.7341 | |||
| 6 | RD (RE) | 11,224 | 500 | 8.2860 | 8.6740 | 105.9700 | 10.2941 |
| SD (LE) | 5.3780 | 7.3140 | 89.0420 | 9.4362 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernandez, M.; Epelde, G.; Beristain, A.; Álvarez, R.; Molina, C.; Larrea, X.; Alberdi, A.; Timoleon, M.; Bamidis, P.; Konstantinidis, E. Incorporation of Synthetic Data Generation Techniques within a Controlled Data Processing Workflow in the Health and Wellbeing Domain. Electronics 2022, 11, 812. https://doi.org/10.3390/electronics11050812
Hernandez M, Epelde G, Beristain A, Álvarez R, Molina C, Larrea X, Alberdi A, Timoleon M, Bamidis P, Konstantinidis E. Incorporation of Synthetic Data Generation Techniques within a Controlled Data Processing Workflow in the Health and Wellbeing Domain. Electronics. 2022; 11(5):812. https://doi.org/10.3390/electronics11050812
Chicago/Turabian StyleHernandez, Mikel, Gorka Epelde, Andoni Beristain, Roberto Álvarez, Cristina Molina, Xabat Larrea, Ane Alberdi, Michalis Timoleon, Panagiotis Bamidis, and Evdokimos Konstantinidis. 2022. "Incorporation of Synthetic Data Generation Techniques within a Controlled Data Processing Workflow in the Health and Wellbeing Domain" Electronics 11, no. 5: 812. https://doi.org/10.3390/electronics11050812
APA StyleHernandez, M., Epelde, G., Beristain, A., Álvarez, R., Molina, C., Larrea, X., Alberdi, A., Timoleon, M., Bamidis, P., & Konstantinidis, E. (2022). Incorporation of Synthetic Data Generation Techniques within a Controlled Data Processing Workflow in the Health and Wellbeing Domain. Electronics, 11(5), 812. https://doi.org/10.3390/electronics11050812









