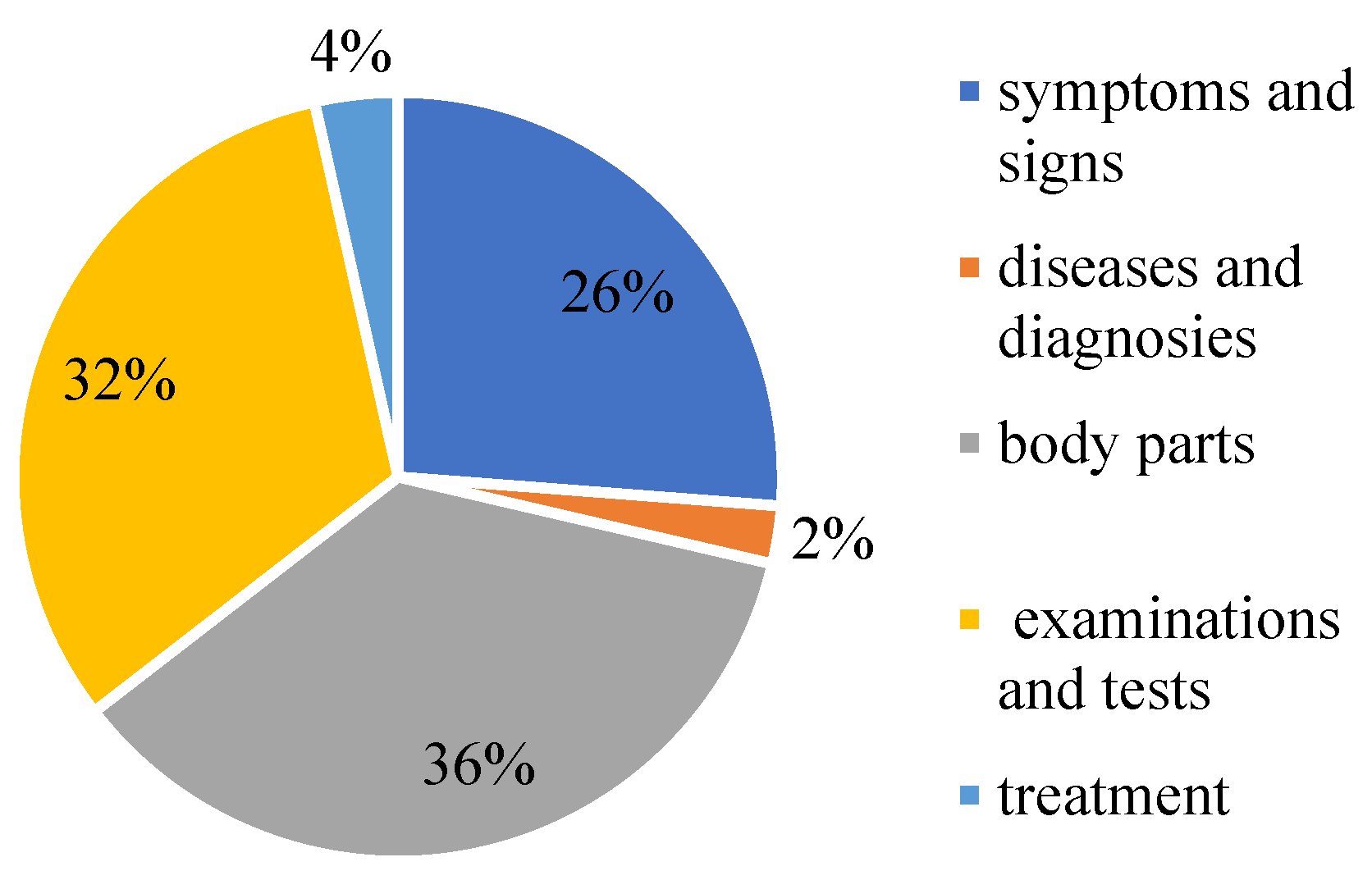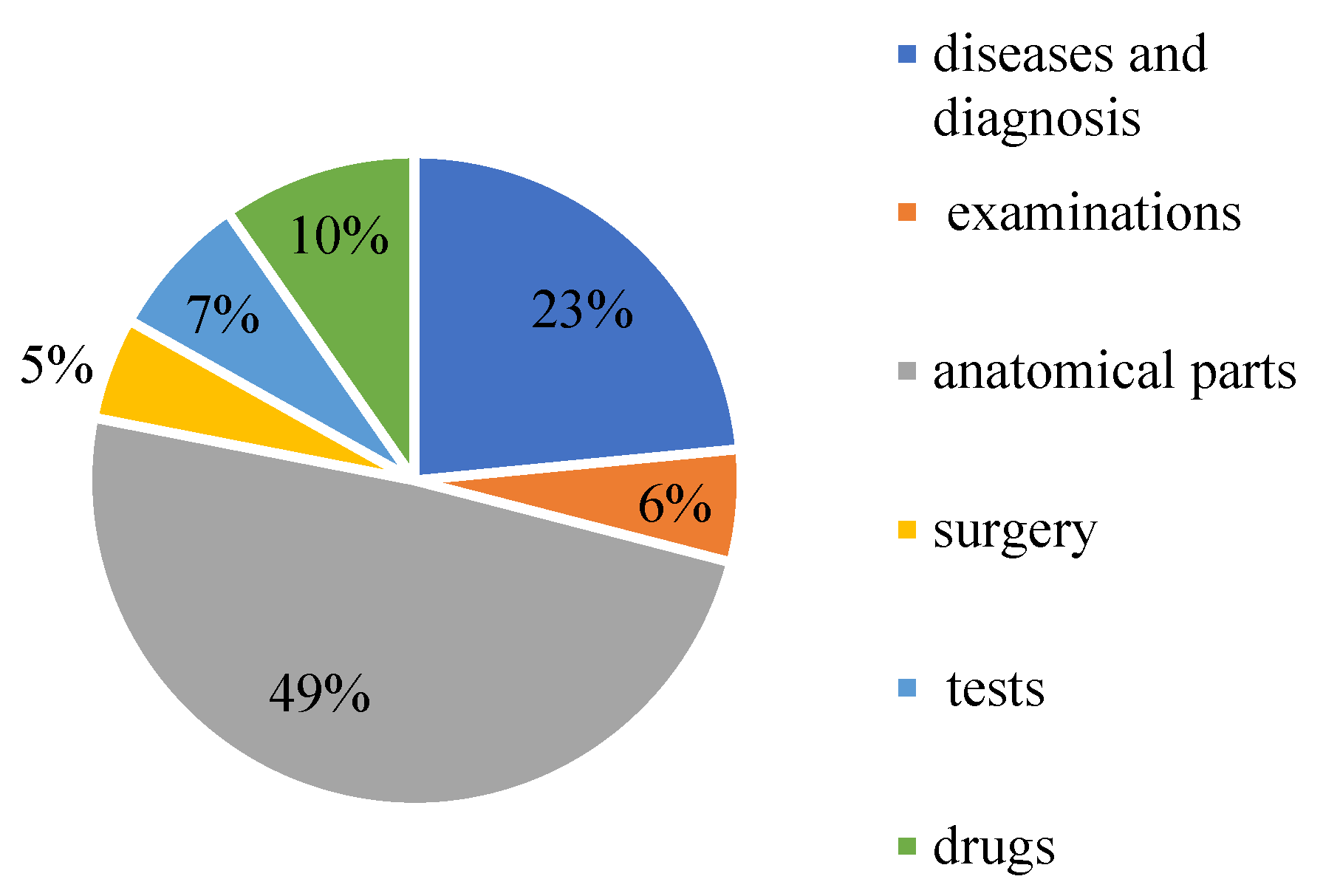Abstract
The goal of Clinical Named Entity Recognition (CNER) is to identify clinical terms from medical records, which is of great importance for subsequent clinical research. Most of the current Chinese CNER models use a single set of features that do not consider the linguistic characteristics of the Chinese language, e.g., they do not use both word and character features, and they lack morphological information and specialized lexical information on Chinese characters in the medical field. We propose a RoBerta Glyce-Flat Lattice Transformer-CRF (RG-FLAT-CRF) model to address this problem. The model uses a convolutional neural network to discern the morphological information hidden in Chinese characters, and a pre-trained model to obtain vectors with medical features. The different vectors are stitched together to form a multi-feature vector. To use lexical information and avoid the problem of word separation errors, the model uses a lattice structure to add lexical information associated with each word, which can be used to avoid the problem of word separation errors. The RG-FLAT-CRF model scored 95.61%, 85.17%, and 91.2% for F1 on the CCKS 2017, 2019, and 2020 datasets, respectively. We used statistical tests to compare with other models. The results show that most p-values less than 0.05 are statistically significant.
1. Introduction
Informatization has penetrated all aspects of social life. In the medical field, more and more hospitals are building information systems to improve their service level and core competitiveness, effectively use limited medical resources, and provide patients with high-quality treatment. These information systems can not only improve doctors’ efficiency but also enhance internal management, making information communication among departments more efficient and simplifying and standardizing the medical treatment process. Medical staff can be released from tedious and repetitive work, with extra time and energy being used to provide better patient services.
Existing medical systems have generated countless medical data, and if the data cannot be used effectively, it will be a waste of professional knowledge. As a medical record, Electronic Medical Record (EMR) has received great attention in scientific research [1] because it contains complete and detailed clinical information generated by patients during each visit. EMR refers to the digital information such as words, symbols, charts, graphics, data, images, and so on, generated by medical personnel using the information system of medical institutions in medical activities. EMR contains various information such as text and medical images. Medical images are mainly the results of laboratory tests of patients, such as CT and B-ultrasound. These medical images can currently be analyzed using pattern recognition and machine learning methods, but EMR also contains much textual data. To make use of the text data, Natural Language Processing (NLP) technology is essential. Electronic medical records cover all patient information from admission to discharge, including admission time, symptoms, body parts, examination methods, medication, and other physical information [2]. Medical services may consider providing patients with the facility to submit inquiries in the form of comments [3].
EMR information extraction is to identify various medical entities from texts and establish relationships among them. The information extraction of EMR was first carried out on English medical records, and many achievements have been achieved, while domestic research on Chinese EMR is still in its infancy. Therefore, it is our top priority.
Named Entity Recognition (NER) is the foundation of text data mining and information processing. For entity recognition in the medical field, it refers to identifying entities such as symptoms, body parts, examinations, etc. Identifying this information and analyzing the relationship among different entity information plays an indispensable role in establishing a knowledge map in the medical field, building an auxiliary diagnosis model, and providing data support for clinical decision-making.
Early NER systems are mainly rule-based approaches. This method extracts the target entity through the preset rule template and has achieved certain results. Although for some uncommon fields, experts need to write rules, which is demanding, time-consuming, and limited, rule-based approaches are not outdated but are still an important complement to other approaches.
Feature-based Supervised Learning Approaches transform NER tasks into classification tasks or sequence labeling tasks. Conditional Random Fields (CRF) and Hidden Markov Models (HMM) [4] are two common algorithms.
With the rapid expansion of deep learning, Gated Recurrent Unit (GRU) and Long Short-Term Memory (LSTM) are applied to the CNER tasks [4]. Alam et al. [5] proposed a new framework based on association rule mining for prognostic factor identification in malignant mesothelioma. At present, the integration of LSTM and CRF is a common method. However, there are limitations. Transformer [6] proposes self-attention, enabling the LSTM networks to solve long-distance dependencies. Transformers gradually replaced LSTM as the mainstream feature extractor in NLP.
Unsupervised pre-trained models are suitable for general domains but not appropriate for the medical domain.
In addition, Chinese NER is related to word segmentation. Since Chinese entities are generally composed of words, word segmentation errors will lead to errors in Chinese NER. The character-based Chinese NER model cannot fully utilize the information of words. The Lattice-LSTM proposed by Zhang et al. [7] improves the accuracy of this task by adding dictionary information to the model. However, due to the complexity of the lattice structure, it does not support parallel computing. Li et al. [8] proposed a Flat Lattice Transformer (FLAT), which uses a flatten lattice structure and transformer to realize parallel processing. At the same time, FLAT uses the calculation method of relative position in the Transformer-XL model [9], and by adding additional position information in the Transformer structure, it solves the modeling of long text and captures ultra-long distance dependencies.
Also, unlike other languages, Chinese is a pictograph. Chinese characters contain rich semantic information. Many words with similar meanings are similar in composition and structure of Chinese characters, which is especially obvious in the medical field. The glyph information of Chinese characters is also of significant reference value. Glyce, proposed by Meng [10], can extract the glyph vectors of Chinese characters. It attempts to extract the semantics of Chinese characters from various ancient and modern Chinese characters and various writing styles, and the performance is improved.
To solve problems, we propose a RoBerta Glyce-Flat Lattice Transformer-CRF (RG-FLAT-CRF) model suitable for Chinese CNER tasks. First, the glyph vector is obtained by Glyce, the character vector and word vector are obtained by Word2vec [11], and the character vector obtained by RoBerta is spliced with the glyph vector and the word vector obtained by Word2vec. At the same time, the Flat-lattice structure is used, word information is added, the head position code and tail position code are constructed for each character and vocabulary, and the relative position code is calculated. The concatenation of vectors and the corresponding position encoding are sent to a transformer to extract the context information of every Chinese character. Finally, we jointly decode the labels of the entire sentence using CRF. Our main contributions are as follows:
Contribution
In our contributions, we have:
- A RoBerta Glyce-Flat Lattice Transformer-CRF model is proposed, which can make full use of the glyph information and language features of Chinese medical texts, has strong coding and text representation capabilities, and can accurately identify various types of Chinese electronic clinical Entity records.
- According to the particularity of medical entities and the language characteristics of Chinese, a multi-feature fusion vector is constructed. The pre-trained model is used to obtain vectors that conform to medical characteristics. At the same time, to strengthen the semantic representation of medical entities, convolutional neural networks are used to extract the glyph features of medical entities, and different character vectors are spliced together to form a composite character vector.
- Use of a lattice structure to add potential lexical information to each word to avoid word segmentation errors. The relative position vector in the improved transformer directly captures the dependencies between words and vocabulary, makes full use of case information, and can be implemented in parallel.
2. Related Work
We include the following studies: (1) How to enhance the semantic representation of Chinese word vectors. (2) Feature extraction networks more applicable to the Chinese language. (3) The characteristics and difficulties of named entity recognition in Chinese electronic medical records. (4) Related Evaluation Metrics [12]. We used multiple strings such as “Chinese electronic medical record named entity recognition”, “Chinese named entity recognition”, and “medical named entity recognition” to retrieve peer-reviewed articles using Multiple databases, including Scopus, ACM Digital Library, IEEE Xplore, ScienceDirect, SpringerLink, and Google Scholar [13].
This section primarily provides a brief introduction to rule-based and dictionary-based methods, machine learning-based methods, and deep learning-based methods. Then, the representation method of the word vector is introduced.
2.1. Rule-and-Dictionary-Based Clinical Named Entity Recognition
Nowadays, Rule-and-Dictionary-Based CNER is commonly used, and these methods benefit from the development of professional medical dictionaries. Researchers complete the NER task by pattern matching according to the belonging list in the dictionary. Friedman et al. [14] developed a clinical document processor that recognized medical information in the medical record and mapped this information into a structured representation containing medical terms. Fukuda et al. [15] proposed a method to identify the names of substances such as proteins from biological papers, using the characteristics of proper noun descriptions in the professional field, which eliminates the need to prepare a professional term dictionary in advance. Names can be extracted with precision, whether they are known or newly defined or are single or compound words.
The completeness and accuracy of the dictionary and the accuracy of the matching algorithm can determine the accuracy of such methods. Therefore, dictionary-based methods are more suitable for fields where proper nouns are fixed and updated infrequently. In the biomedical field, there are problems such as the fast updating of proper nouns and different expressions of the same entity name. Experts need to spend much time and effort writing rules, and the cost is high. In addition, different rules are needed for different systems. They are of poor portability and are hard to reuse quickly.
2.2. Clinical Named Entity Recognition Based on Machine Learning
In the past, traditional machine learning based on CNER has been widely used, including HMM, CRF, Support Vector Machine (SVM) [16], Naive Bayesian Model (NBM) [17], etc. Settles [18] used combined feature sets with CRF in biomedical NER tasks. Tang [19] developed an SVM-based NER system for medical entities in the medical record. Roberts et al. [20] utilized SVM with a manually constructed dictionary to classify. Liu [21] evaluated the contribution of different features in the CRF-based CNER task.
Compared with the methods analyzed in Section 2.1, the method in Section 2.2 does not require the experimenter to master much language knowledge, thus saving time and effort. However, this type of method requires a lot of energy to design features. The effect of the model depends on the designed features. With deep learning modeling, the feature extraction problem in traditional machine learning can be addressed.
2.3. Deep-Learning-Based Clinical Named Entity Recognition
Recently, we have witnessed the great success of deep learning in the field of NLP, such as NER and event extraction tasks. Commonly used network models include Convolutional Neural Networks (CNN) [22], Recurrent Neural Networks (RNN) [23], and LSTM. Ma et al. [24] proposed the Bi-directional LSTM-CNNs-CRF model, character-level representations are extracted using CNN, Bi-directional LSTM (BiLSTM) is responsible for modeling the contextual information of each word. Xu et al. [25] combined bidirectional LSTM and CRF based, BiLSTM-CRF model can learn the information features of a given dataset and achieved a score of 0.8022 at NCBI, outperforming many widely used baseline methods. Yin et al. [26] used convolutional neural nets for Chinese character radical feature extraction and captured the correlation between characters using self-attentiveness. Kong et al. [27] proposed a Chinese medical named entity recognition based on a multi-layer CNN and attention mechanism, constructing a multi-layer CNN to extract short-term and long-term memories and using an attention mechanism to capture global information. However, the above deep neural network-based CNER methods cannot model the ambiguity of Chinese.
The BERT-BiLSTM-CRF model was proposed by Jiang et al. [28] to be applied to CNER. The semantic representation of words was enhanced with a BERT pre-trained language model, and the BiLSTM was to learn contextual information. Qin et al. [29] proposed a BERT-BiGRU-CRF model in the field of Chinese electronic medical records, which uses BERT to convert the electronic medical record text into low-dimensional vectors and BiGRU to obtain contextual features. Wu et al. [30] used a bi-directional LSTM model to learn a medical entity’s partial head information using Roberta to learn medical features. Wang et al. [31] used information from medical encyclopedias as additional information to enhance the recognition of Chinese electronic medical record entities. However, these models do not fully consider the characteristics of medical domain data, and it is not very effective in medical entity extraction.
2.4. Research Status of Word Vector Representation Methods
If you want to reflect a word in a text and perform mathematical calculations, it must be done through word embedding. The bag-of-words model simply represents words without any semantic features. As the number of words increases, so does the dimension. Researchers propose a way to solve this problem using a pre-trained language model for word representation. Pre-training refers to obtaining a training model independent of subsequent tasks from a large-scale corpus using self-supervised learning. The model can be transferred to other tasks, thereby reducing the training burden of subsequent tasks. The Word2Vec model was proposed by Mikolov et al. to obtain vectors. The GloVe algorithm was proposed by Pennington et al. [32]. In recent years, pre-trained models have received increasing attention. Since this type of model is a context-independent word vector trained by static pre-training technology, it cannot accurately model the polysemy of a word. Therefore, Peters et al. [33] proposed the ElMo algorithm. The bidirectional LSTM network structure was used for context encoding, which could effectively capture context information.
2.4.1. Models for BERT and Its Variants
Devlin et al. [34] proposed Bidirectional Encoder (BERT). The emergence of Bert opened a new era of research in the field of NLP. Then some improved pre-training models based on BERT, mainly including ERNIE [35], BERT-WWM [36], RoBerta [37], and XLNet [38]. The ERNIE model is pre-trained using massive corpora in multiple fields, including encyclopedias, news, forums, etc. BERT-WWM’s improvement over BERT is to replace a complete word with a Mask label instead of a subword. The RoBerta model uses a dynamic mask mechanism for pre-training, cancels the NSP task, and expands the batch size. As an auto-regressive model, the XLNet model can expand the language model and increase the prediction of bidirectional words, the above predicting the next word and the following predicting the previous words.
2.4.2. Research on Chinese Characters
The structure of Chinese characters is different from that of English. Chinese characters are pictographs, and their glyphs also contain rich meanings. Therefore, many scholars have carried out characterization studies on the glyph features of Chinese characters. Sun [39] proposed to learn the radical features of Chinese. Wang et al. [40] proposed a Chinese character root and stroke-enhanced embedding method for learning Chinese character roots from the internal information of semantics and form. Wei [41] proposed a visual embedding method for semantic association among visual words, segmented the glyph, spliced the average embedding vectors corresponding to each sub-region, and converted it into a fixed-length vector for keyword detection. Su [42] used convolutional autoencoders to learn glyph features from images of traditional Chinese characters and introduced glyph features during training using the corpus. Meng [6] proposed the Glyce model. It tried to extract the semantics of Chinese characters from various ancient and modern Chinese characters and various writing styles, and the performance was improved.
These are the characteristics of Chinese, which improve CNER tasks. However, the current mainstream CNER methods cannot integrate the pre-trained model with the Chinese glyph information.
3. Proposed Method
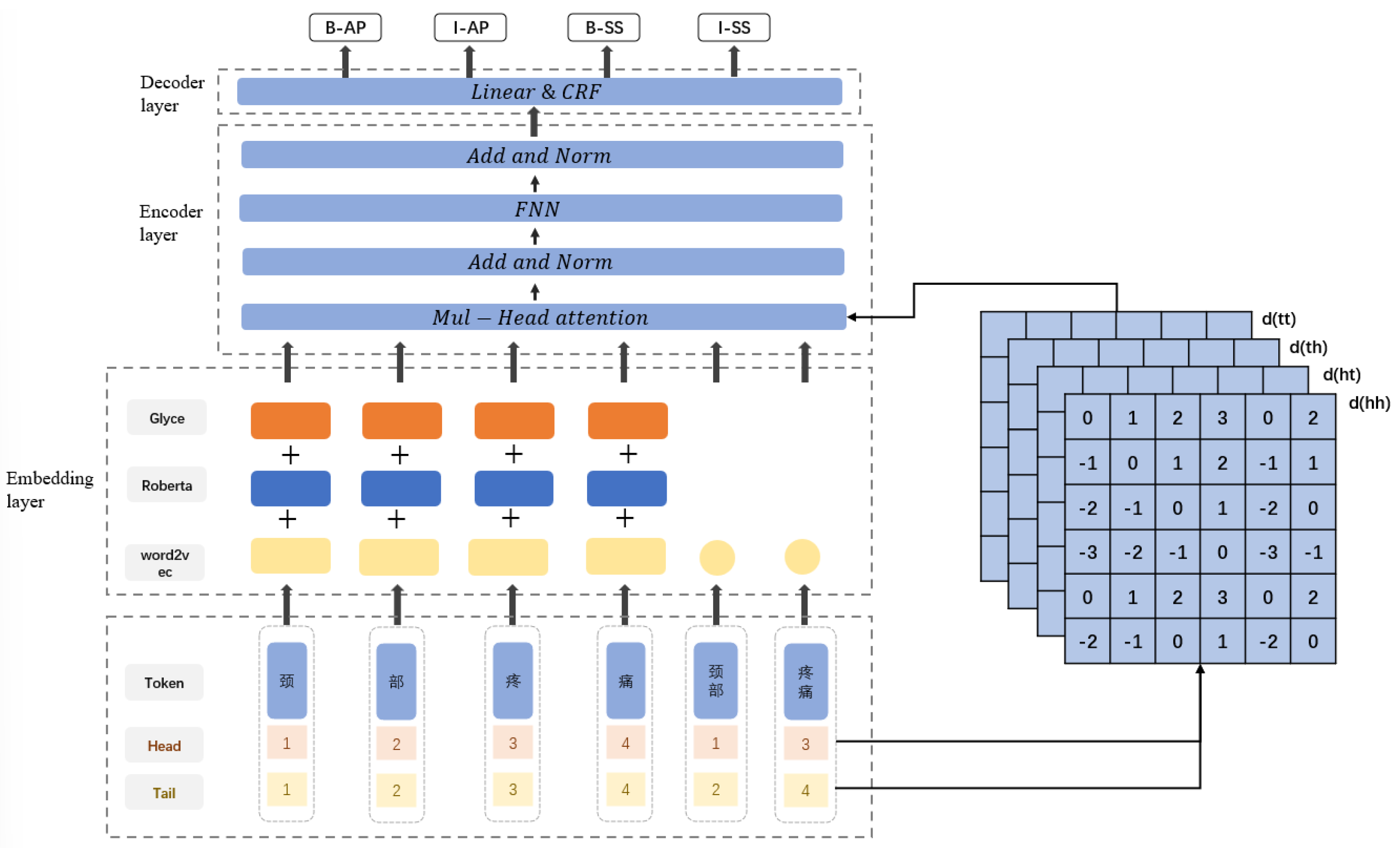
In the NER task, the character sequence of the input text is represented by . The labels of the input text are represented by . The goal of a NER system is to predict the correct sequence Y of labels for the text given the known sequence of characters X of the text. The RG-FLAT-CRF model proposed in this chapter consists of three parts; the embedding layer, the encoding layer, and the decoding layer. The overall structure is shown in Figure 1.
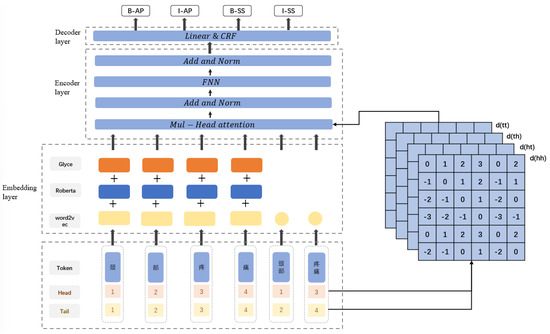
Figure 1.
Model structure diagram of RG-FLAT-CRF.
The model first matches the latent words related to the character in the input text and splices the character information and words information into the embedding layer. The embedding layer consists of three parts, and the character vector is spliced after processing by RoBerta, Glyce, and Word2vec. The word vector is obtained using Word2vec, head and tail position encoding are constructed for each character and word, and the relative position encoding is calculated. The concatenation of word vectors and the corresponding position encoding are input into the encoding layer, consisting of a Transformer neural network that captures deep features and encodes the input sequence. Finally, the output of the encoding layer is input to the decoding layer, which predicts the final label sequence.
This study uses NER to perform entity recognition on Chinese EMR. Specific steps are as follows:
- (1)
- Electronic medical record data preprocessing, that is, the original electronic medical record text data set is processed, and the electronic medical record text set is represented as , where the i-th electronic medical record text is represented as . The predefined entity category , is divided and annotated according to the character level, and the characters and predefined categories are separated by spaces when annotating.
- (2)
- Establish a Chinese EMR text training dataset.
- (3)
- Model training, that is, training the RGT-CRF model. Take the Chinese EMR test text set as input and take the entity and its corresponding category pair as output: . The entity mi represents the entity that appears in the document, and and ei represent the start and end positions of mi, respectively. There is no need to overlap between entities; that is, . represents the predefined category of entity mi, calculates the F1 score according to the precision and recall rate, and uses the F1 score as the comprehensive evaluation index of the model.
3.1. Embedding Layer
The embedding layer consists of three parts: RoBerta layer, Glyce layer, and Word2vec layer:
- (1)
- RoBerta layer: the model adopts the better pre-training model RoBerta to capture the characteristics of medical text and converts each word of medical text into a low-dimensional vector form through RoBerta.
- (2)
- Glyce layer: scan each word in the sentence to obtain the glyph vector corresponding to each word, and enhance the representation of the word.
- (3)
- Word2vec layer: Using Word2vec, the vector representation of each word in the medical text and the vector representation of the latent words can be obtained to enrich the semantic representation.
The character vectors processed by RoBerta, Glyce, and Word2vec are spliced to obtain multi-feature word vectors, and then the character vectors and word vectors processed by Word2vec are spliced together.
3.1.1. RoBerta
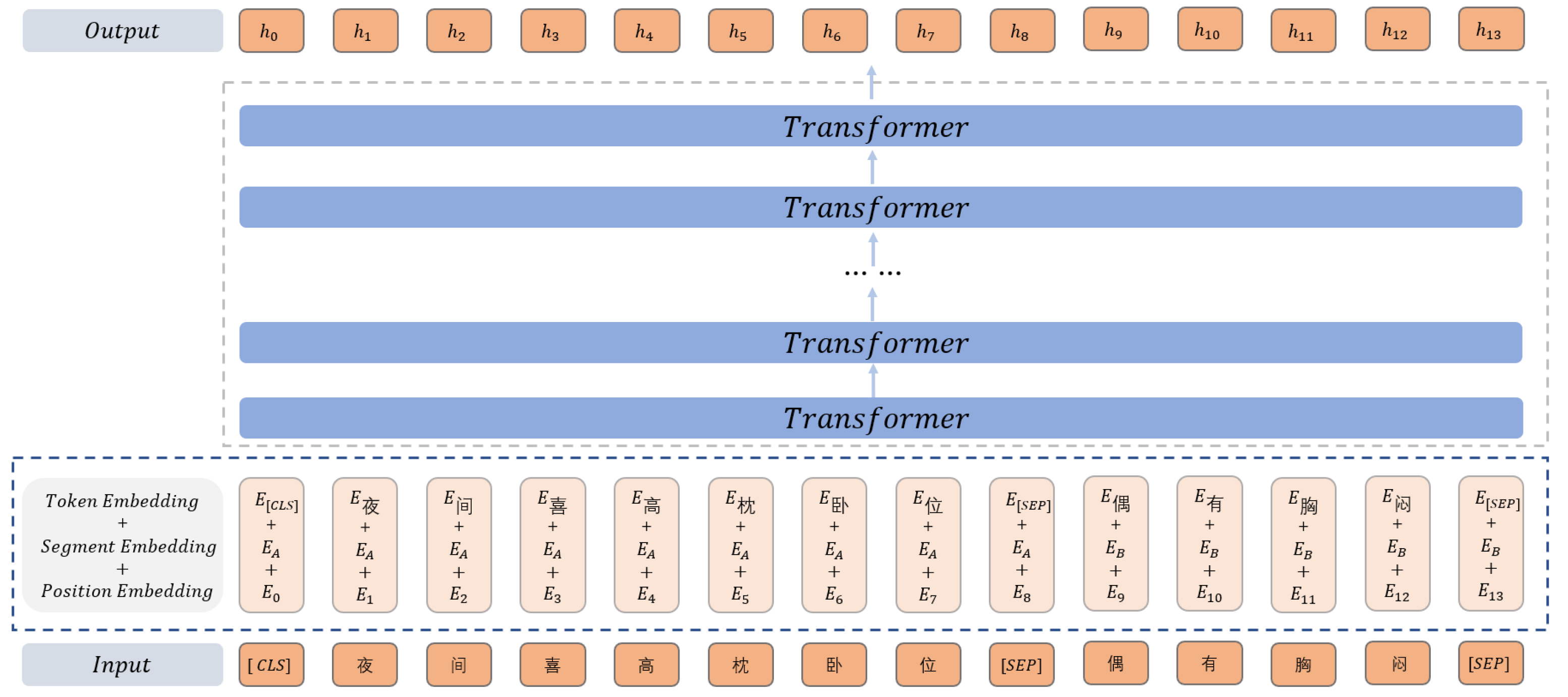
Pretrained language models are often used in NER tasks to generate richer semantic representations. BERT and its variant RoBerta are widely used in research. We use RoBerta for text encoding instead of BERT. Compared with BERT, the model structure of RoBerta has not changed. They are all composed of 12 stacked transformers. Each layer has a hidden state of 768 dimensions. Each Transformer uses a 12-head self-attention mechanism. The only thing that has changed is the pre-training method. Dynamic masks and text encoding are adopted to remove the NSP task and use more data to train the model.
The vector is obtained through the RoBerta. The RoBerta structure is shown in Figure 2. The input text is . First, the sequence is vectorized. This part consists of token embedding, clause embedding, and position embedding. These three embedding layers are essentially equivalent to the static embedding layers, and the table lookup is performed by the embedding matrix. For the x-th token in the processed token sequence, the vector calculation is as follows:
where , , are the token embedding matrix, the clause embedding, and the position matrix.
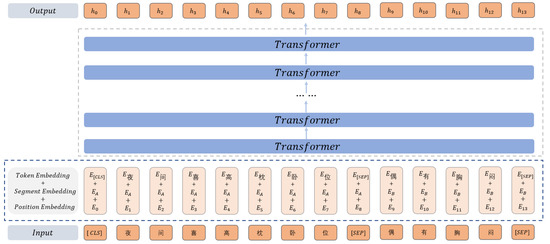
Figure 2.
Structure diagram of RoBerta.
Token Embeddings represent the Embedding vector of each word. Segment Embeddings are used to distinguish different sentences before and after punctuation marks. Position Embeddings represent the embeddings of a word’s position. The input feature of RoBerta is the sum of the above 3 embeddings. “[CLS]” is used as the starting symbol of the input, indicating that the feature can be used in the classification model. “[SEP]” indicates the clause symbol, which is used to cut off the clauses in the sentence.
The obtained vector is input into the stacked Transformer to extract features. The final output is the result of encoding the input sentence text. Finally, we obtained the sentence representation vector with the dependency information among words and words in the sentence text. The calculation is as follows:
where represents the stacked Transformer, outputting the text encoding of the entire sentence through the last layer 𝐻, which can be expressed as . Here is the text representation vector to the xth token.
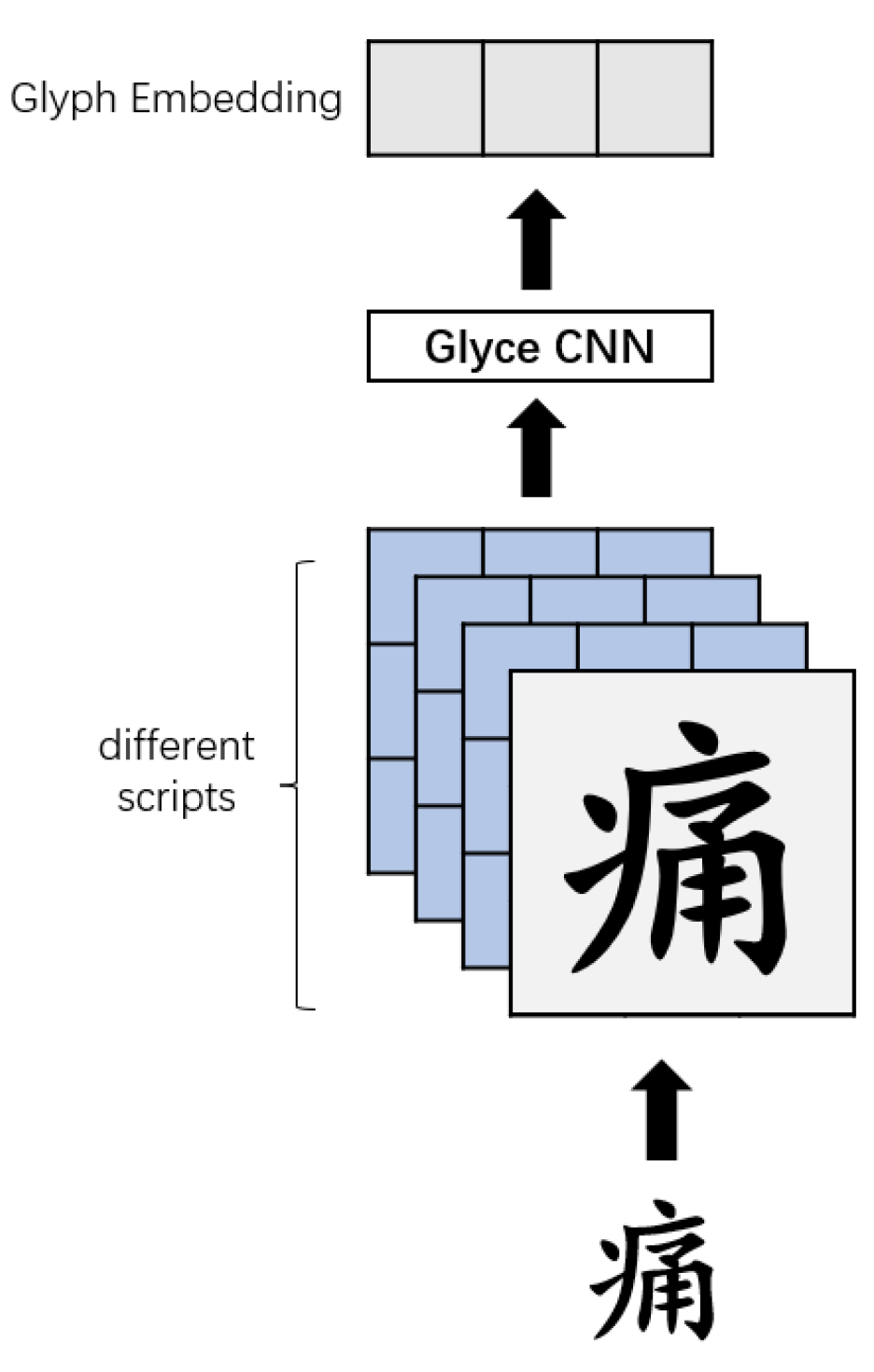
3.1.2. Glyce
Chinese characters are pictographs, and most Chinese characters are evolved from graphics. Chinese characters contain rich semantic information, especially in the medical field. Most of the words for diseases have the same parts. Therefore, we believe that adding glyph information to word vectors can enhance the representation of characters.
Glyce used different versions of the writing method, as well as different writing to enhance the representation of the characters.
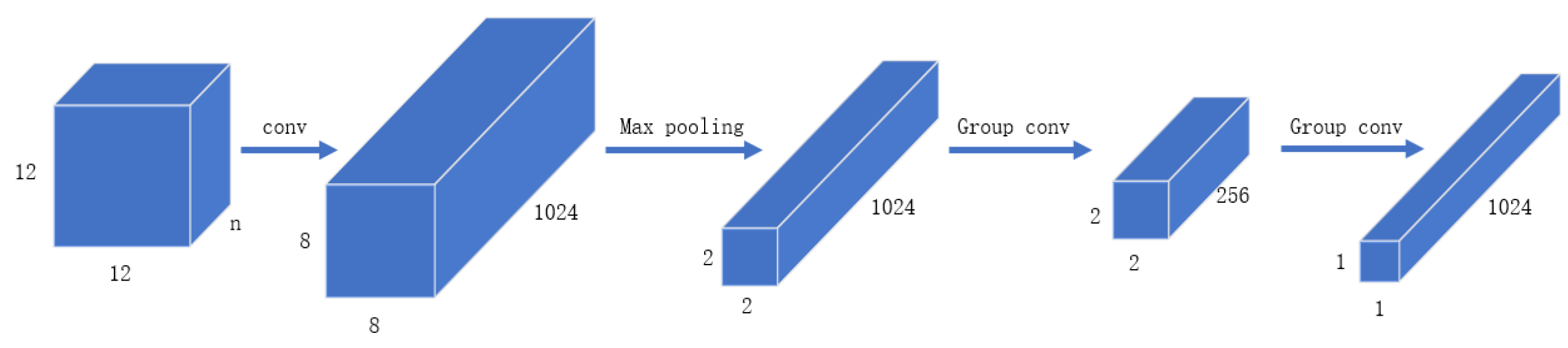
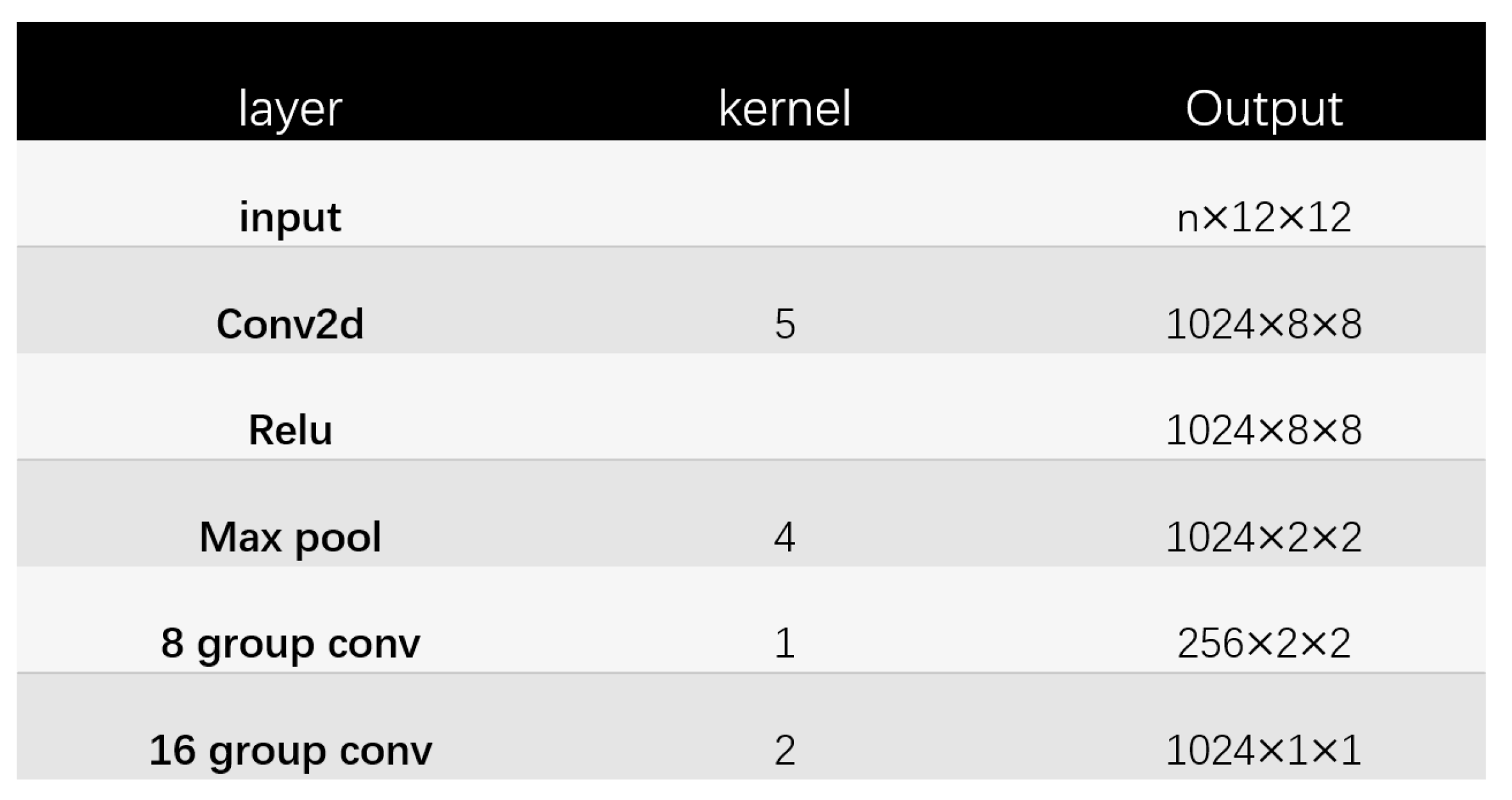
Glyce is different from traditional CNN. There are about 100,000 Chinese characters, but only a few thousand are commonly used. Compared with classification on the ImageNet dataset. There’re few training examples for Chinese characters. Compared with the size of Imagenet images, Chinese images are usually smaller, with a size of 12 × 12. Thus according to the Chinese writing habits, a 2 × 2 Tianzi lattice structure is used. As shown in Figure 3, this structure can reflect the glyph information of Chinese, including components such as radicals, which is suitable for the extraction of glyph information.
Figure 3.
Schematic diagram of the Tianzi lattice.
The structure of Glyce Tianzi lattice-CNN is shown in Figure 4. The processing process is shown in Figure 5. To capture lower-level graph features, the input image approximation firstly passes through a convolutional layer with kernel size 5. In addition, the convolutional layer has to increase the number of feature channels to 1024. Then we apply a max-pooling layer with a pooling kernel of 4 × 4 to perform feature downsampling. After this, the resolution is reduced from 8 × 8 to 2 × 2. This 2 × 2 Tianzi lattice structure shows the glyph features of Chinese characters, and finally, we apply the group convolution operation to map the Tianzi lattice to the final output.
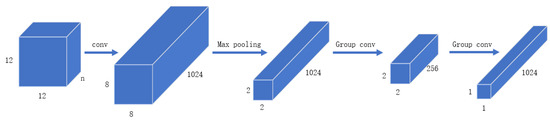
Figure 4.
CNN structure diagram in Glyce.
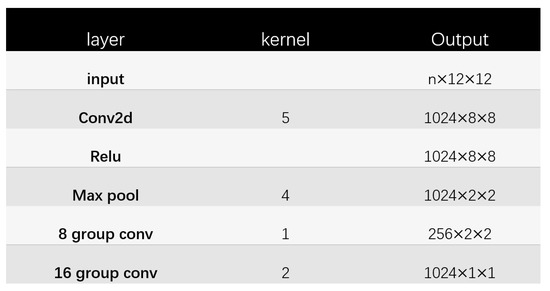
Figure 5.
The Tianzi lattice—CNN structure.
For the input text , the glyph vector obtained by Glyce is as shown in Figure 6.
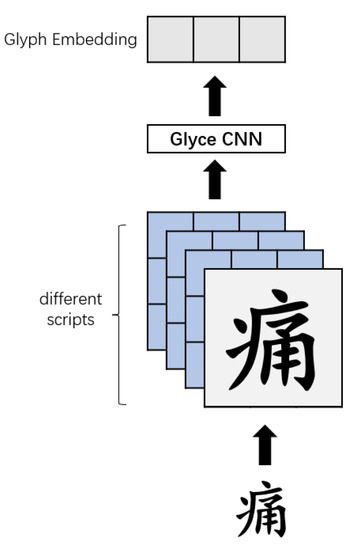
Figure 6.
Glyce character embedding.
3.1.3. Word2vec
We use Word2vec to get word vectors, a typical representative of distributed representation. Compared with one-hot, Word2vec takes into account the relationships among words. In addition, Word2vec also optimizes the training efficiency of the model, so it is used more frequently.
3.2. Position Encoder
Chinese NER tasks are often considered sequence labeling tasks. By calculating the probability of each character corresponding to each entity type label, The label with the highest probability is used as the final identification result. There are usually two vectorization methods to vectorize Chinese characters into the model calculation: methods based on word vectors and methods based on character vectors.
The first task of the word vector-based model is to segment the text into the form of words. The improvement effect of word vectors on entities is significant. The word contains more semantic information, but if there is a false classification, it will affect the results of NER.
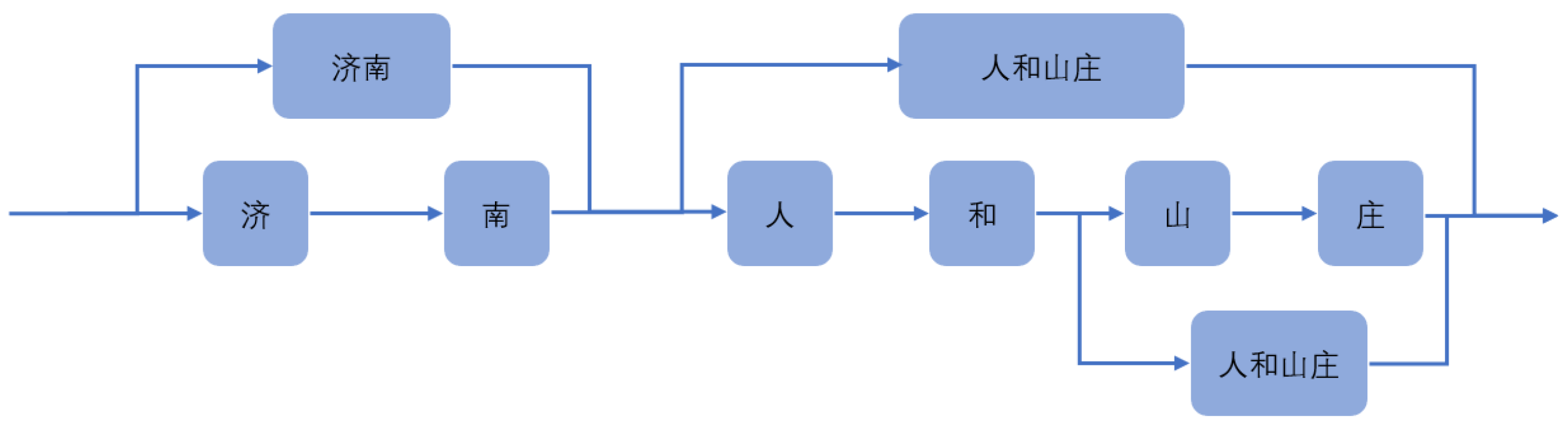
For instance, in Figure 7, this sentence can be divided into ‘济南人(Jinan People)’, ‘和(and)’, ‘山庄(Mountain Villa)’, and can also be divided into ‘济南(Jinan People)’, ‘人和山庄(Renhe Mountain Villa)’. These two-word segmentation methods have a great impact on recognition.
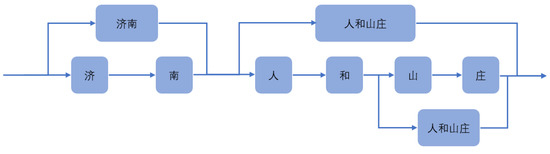
Figure 7.
Structure diagram of Lattice.
Using character vector-based models avoids word segmentation error information but lacks lexical information. For example, ‘感冒(cold)’, separate the word ‘感(feel)’ and ‘冒(emit)’ represent different semantic information. ‘感(feel)’ means feeling, and ‘冒(emit)’ means to penetrate outward or rise upward. It is difficult to express the information of the word ‘感冒(cold)’ in medicine after ‘感(feel)’ and ‘冒(emit)’ are separated, which is especially obvious in the medical field.
To address the above problems, we adopted the FLAT-lattice structure, shown in Figure 8. This structure uses both character vectors and word vectors. Based on character vectors, the latent vocabulary of each character is matched, and the word vectors are added to the model. This method utilizes the semantic relationship of words and avoids the phenomenon of word segmentation errors.
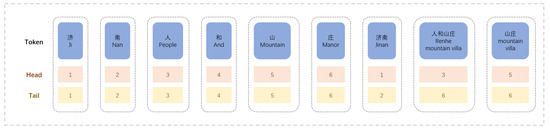
Figure 8.
Structure diagram of Flat-lattice.
After using the dictionary to obtain lattice information from the string, it is flattened, and the structure is shown in Figure 8.
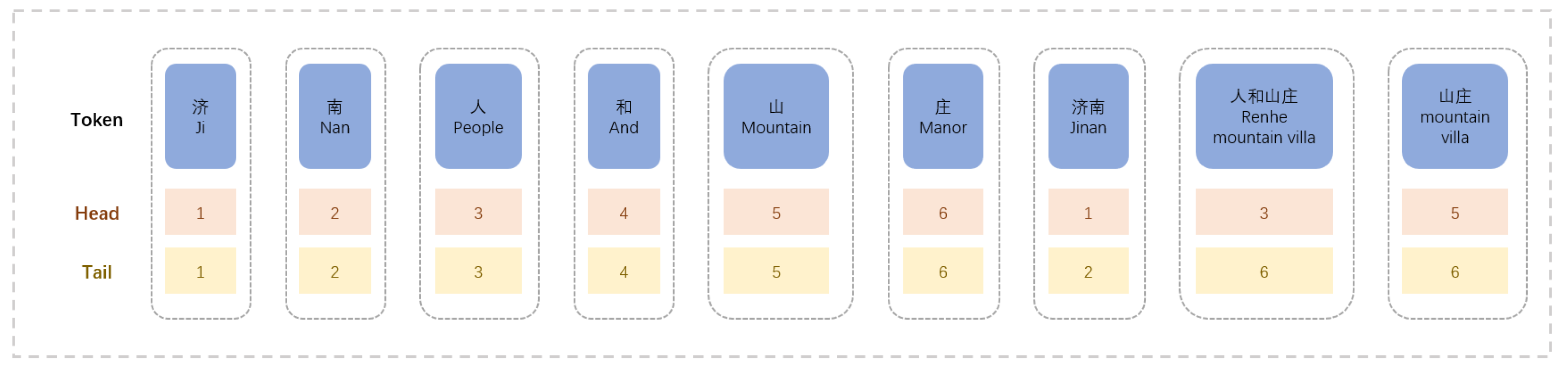
These flat lattices can also be defined as spans. A span comprises a token, a head, and a tail. A token is a word or character, and the head represents the starting position of the token in the original sequence, and the tail represents the ending position of the token in the original sequence. For characters, the head and tail are the same. For the matched words, head indicates the start position of the word in the sequence, and tail indicates the end position of the word in the sequence. The flat lattice can preserve the original structure of the lattice and, at the same time, preserve the word order information of the original sentence.
According to the Flat-lattice structure, there are three interrelationships, intersection, involvement, and separation. We use relative position encoding to encode the positional relationship among each span. Relative position encoding does not directly model the interaction relationship but obtains a dense vector by computing a set of head and tail changes. Not only the interrelationships among spans can be represented, but more detailed sequence relationships can be shown, such as the distance among words and characters. Let and , and denote the head and tail positions of and , respectively. Four kinds of relative distances can be used to represent the relative relationship between and . Their calculation formulas are as follows:
where stands for the distance from the head of to the head of , is the distance from the head of to the tail of , represents the distance from the tail of to the head of , is the distance from the tail of to the tail of . The final relative position encoding is a nonlinear transformation of the four distances, which can be calculated like:
among them, is a learnable parameter, ⨁ represents the connection operator, and the calculation method of refers to the calculation method of the transformer. The calculation is as shown in the equation:
3.3. Encoder
The encoding layer consists of Transformers, which aim to extract semantic and temporal features from the context automatically.
Before the transformer appeared, most NER used BiLSTM as the model’s encoder. However, BiLSTM has some problems: (1) The sequential nature of the recurrent neural network represented by LSTM hinders the parallelization of training samples; (2) The problem of long-term dependence cannot be completely solved.
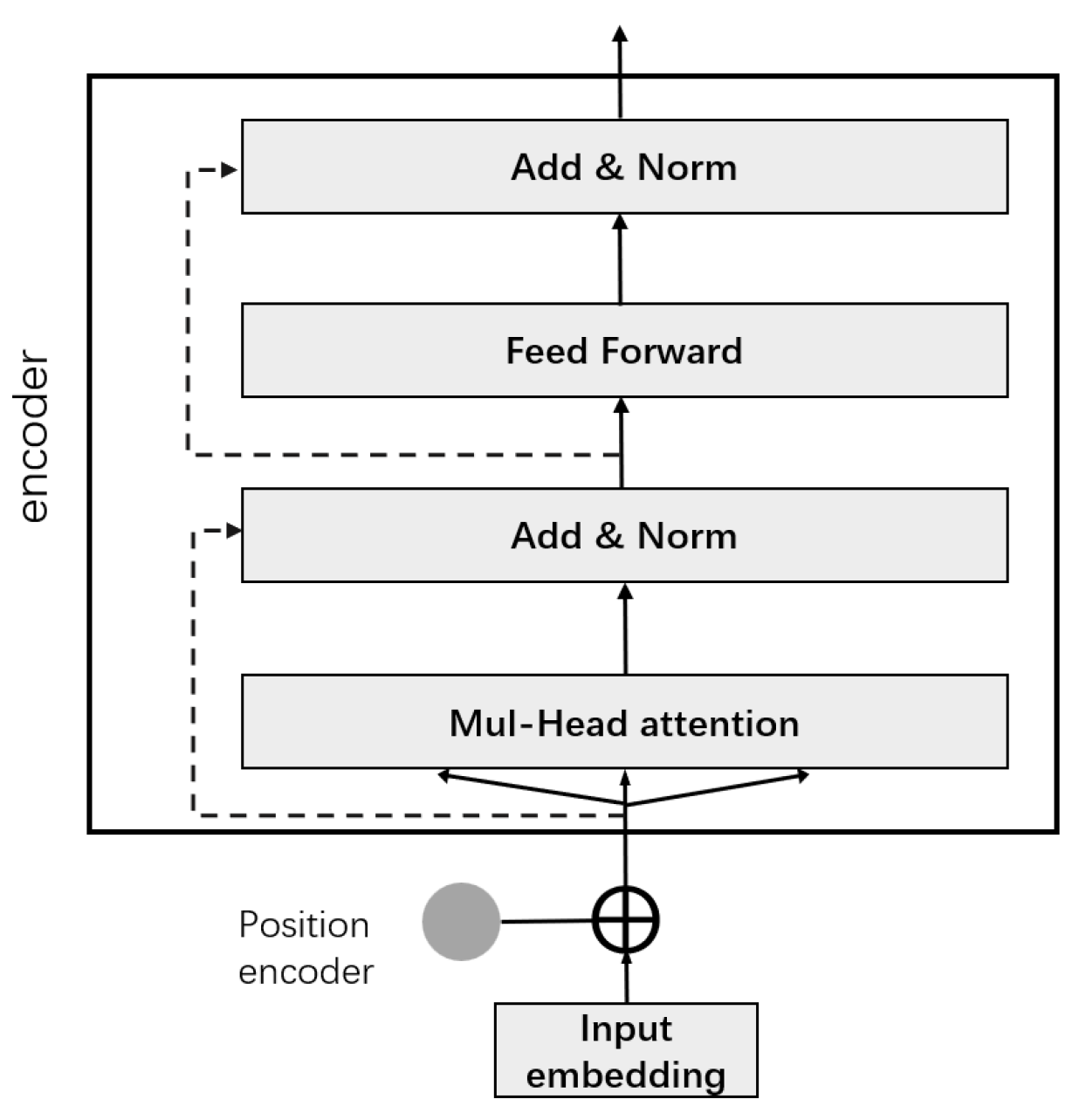
Transformer avoids recurrent model structure and uses attention mechanism for modeling. The structure is shown in Figure 9. We used its encoding part, which consists of two parts, a feedforward network and a multi-head self-attention layer, both of which have a residual network. Multi-head self-attention consists of stacked self-attentions, all accompanied by a “layer normalization” step.
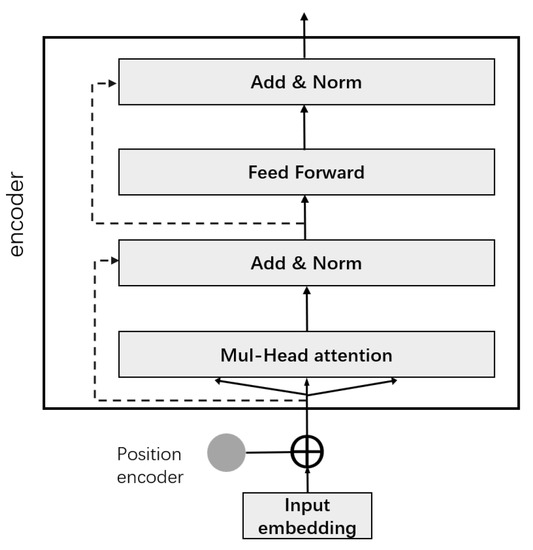
Figure 9.
Structure diagram of Transformer.
When the encoder encodes this word, the self-attention mechanism can take other words in this sentence into consideration.
First, we send the vector output of the embedding layer and the corresponding relative position encoding to the encoding layer, using the encoding layer of the transformer. A Query vector, a Key vector, and a Value vector are created for each word by this self-attention mechanism. They are obtained through the vector multiplication by the three matrices we trained. Their calculation formula is as follows:
The second step is to calculate the score, which will make the gradient more stable, and then it is divided by . The traditional Transformer model can capture contextual semantics by adding position information to the input, but there is a problem of sentence errors in the face of text segmentation input. Therefore, extra position information is added to the Transformer structure of the Transformer-XL model, and the absolute vector is converted into a relative vector. Solve the modeling of long text, capture ultra-long distance dependencies, and calculate the attention score vector among input vectors by the formula:
where are learnable parameters, are the embedded representations of and
Then pass the result through softmax, which normalizes the scores for all words. For the weighted value vector, the output of the self-attention layer at that position is obtained, and the following is its formula:
The multi-head attention mechanism consists of multiple self-attentions. Define multiple groups of different Q, K, and V, and let them focus on different contexts, respectively. The process of calculating Q, K, V is still the same, except that the matrix of linear transformation has changed from one set of to multiple sets of .
For the input matrix X, each group of Q, K, V can get an output matrix Z. Concatenate the different matrices together and multiply with an additional matrix .
The multi-head attention mechanism enhances the attention layer’s performance in two aspects:
- (1)
- It empowers the model with a closer focus on different locations.
- (2)
- Multiple “representation subspaces” are given to the attention layer, and multi-head attention allows us to possess multiple sets of Q, K, and V matrices. After training, each group projects the output into a different representation subspace. The calculation formula is as (15):
The resulting output is subjected to layer normalization and residual connections. The specific formula is as follows:
After the operation of Feedforward, the formulas are shown in equations:
3.4. Decoder
The decoding layer consists of CRFs, whose purpose is to resolve the correlation between the output labels to obtain the globally optimal annotation sequence for the text.
For the input sequence , its predicted label is . The score matrix P output by the encoding layer is n×k in size, n is the length of the input sequence, and q is the different types of labels defined. represents the score of the ith character in the sentence on the label. A state transition score matrix A represents the probability score of transition among different labels. represents the transition score from label to label . , represent the start tag and the end tag, respectively. Under the condition of the given sequence, the score of the corresponding sequence tag is obtained. The functions can be described as follows:
The predicted probability is . The calculation formula is shown in (22):
The loss function, as shown in the formula:
In the last, we adopted the Viterbi algorithm to get the optimal path, that is, a more reasonable predicted label of the input sequence. The calculation formula is as follows (24):
3.5. Time Complexity Analysis
We discuss the time complexity of the model.
where is the sequence length and d is the dimension of embedding. is the number of convolutional kernels the neural network has; is the lth convolutional layer of the neural network; is the number of output channels of the lth convolutional layer of the neural network; and for the lth convolutional layer, the number of input channels is the number of output channels of the -1st convolutional layer. is the number of labels as
4. Experiment Design
This section presents the following aspects: the dataset used for the experiments, the labeling rules, the evaluation metrics, and an introduction to the comparative experimental model.
4.1. Dataset
Our proposed RG-FLAT-CRF model is validated with real datasets of three clinical NER tasks.
These three datasets are all from the CCKS competition dataset. The following is the introduction to these datasets.
CCKS-2017 data is adopted for the experiment. Since we did not participate in the competition, we only found some open-source data. The CCKS-CNER2017 dataset. Provides 300 electronic clinical record texts with 29,865 annotated instances (7816 sentences). It is annotated with five entity types: symptoms and signs, diseases and diagnosis, body parts, examinations and tests, and treatment. Table 1 lists its detailed statistics. The proportion of each part of the data is shown in Figure 10.

Table 1.
Entity statistics of the three datasets.
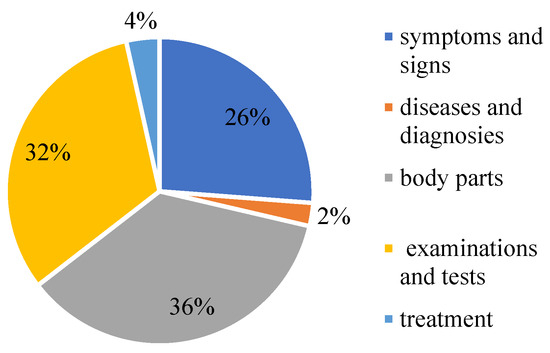
Figure 10.
The proportion of medical entities on CCKS2017.
CCKS-2019 contains 23,384 annotated instances (10,179 sentences). They are annotated with six entity types, namely diseases and diagnosis, examinations, tests, surgery, drugs, and anatomical parts. The elaborated statistics are shown in Table 1. The proportion of each part of the data is shown in Figure 11.
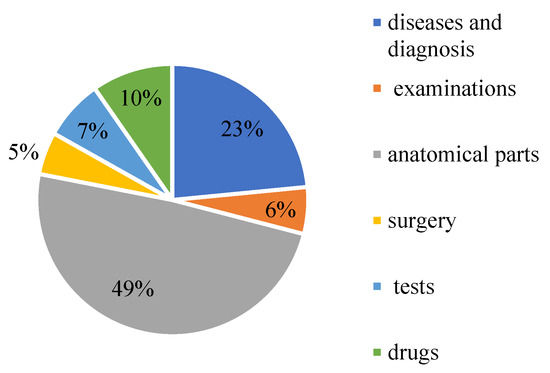
Figure 11.
The proportion of medical entities on CCKS2019.
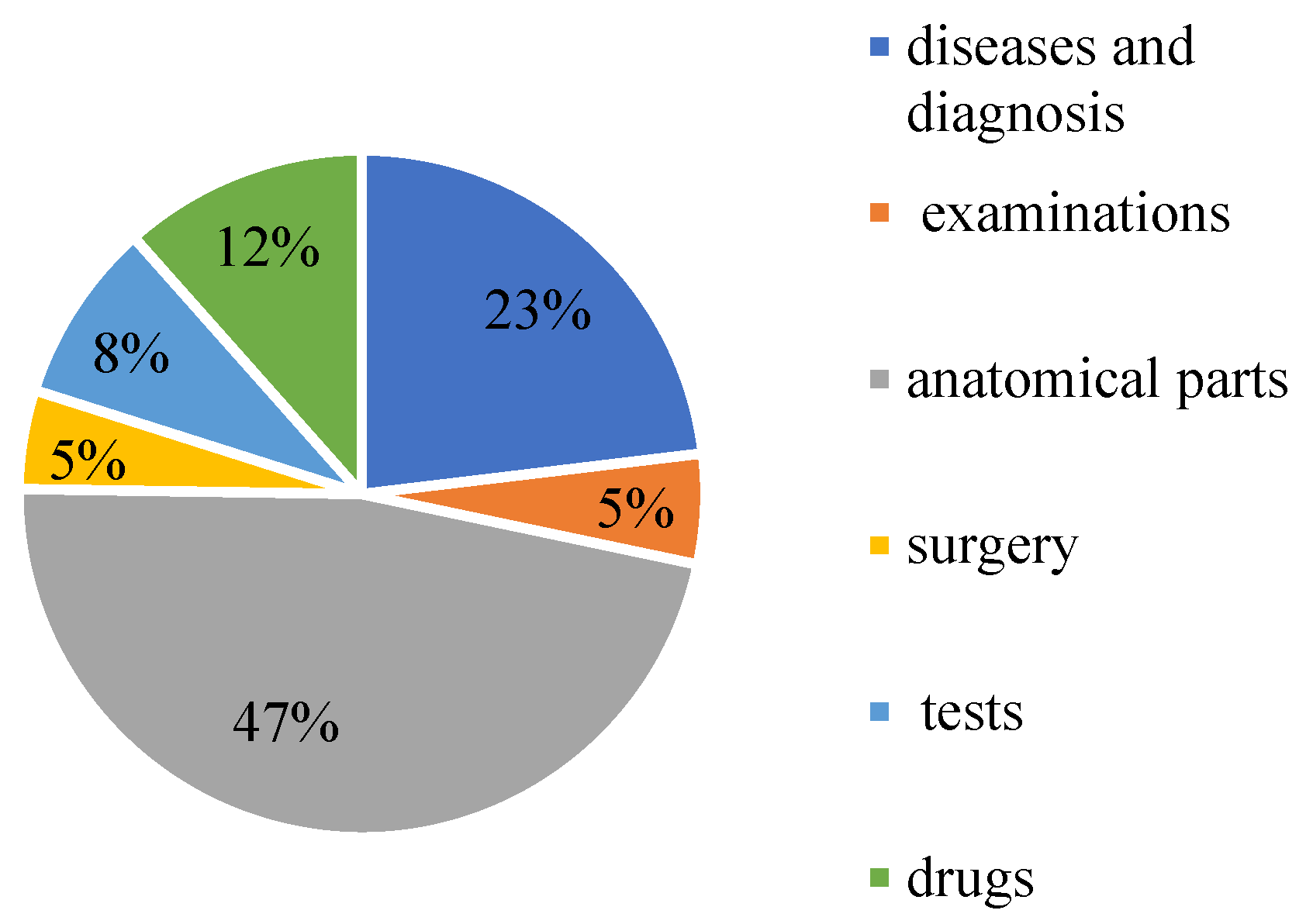
CCKS-2020 contains 24,341 annotated instances (13,308 sentences) with six entity types: diseases and diagnosis, examinations, tests, surgery, drugs, and anatomical parts. Table 1 shows the specific statistics. The proportion of each part of the data is shown in Figure 12.
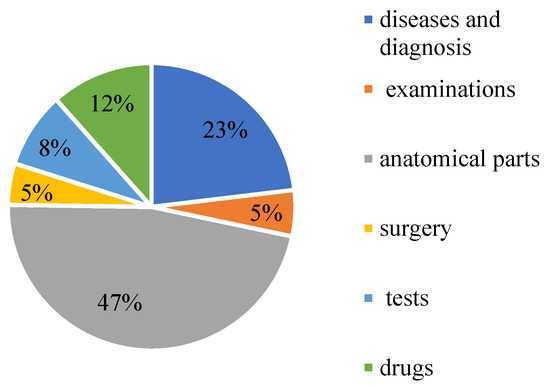
Figure 12.
The proportion of medical entities on CCKS2020.
4.2. Labeling Rules
We adopt the BOI rule, where the entity’s beginning is represented by B, I is the interior, and O stands for the other categories.
Annotation methods of five entity categories in CCKS2017: SS for symptoms and signs, DD for disease and diagnosis, AP for body parts, EE for inspection and examination, TM for treatment.
Annotation methods of six entity types in CCKS2019 and 2020: DD for disease and diagnosis, GEXA for examination, AP for the anatomical site, SU for surgery, EEXA for the test, and DR for the drug.
4.3. Evaluation Indicators
This paper uses the most common evaluation metrics in the NER field Precision, Recall, and F1 scores are used as the evaluation indicators of the model to evaluate the performance of the evaluation model comprehensively. TP is the number of positive samples predicted as positive samples, FN is the number of positive samples predicted as negative samples, and FP is the number of negative samples predicted as positive samples. They are widely used to evaluate classification and sequence annotation tasks [43].
Precision: The ratio of the number of recognized entities to the number of recognized entities is recorded as Precision, abbreviated as P. The calculation formula is Equation (25).
Recall: The percentage of correctly identified entities out of the number of entities in the sample. The calculation formula is Equation (26).
Both take values between 0 and 1, and the closer the value is to 1, the higher the precision or recall. Precision and recall are sometimes contradictory; a weighted harmonic mean that needs to be considered, and the F1-score is a combination of the two. The higher the F1 score, the more robust the classification model is. The calculation formula is Equation (27).
4.4. Experimental Parameters
The parameters of the RG-FLAT-CRF were tuned by Adam, and a hierarchical lr mechanism introduced. For the pre-trained RoBerta model, a learning rate of 3 × 10−5 is used, and for the other parts a learning rate of 2 × 10−4 is used. For the RG-FLAT-CRF model, the batch size used is 12. Details are shown in Table 2.

Table 2.
Parameter settings.
5. Results and Analysis
This part is divided into two parts: performance comparison with existing models, and ablation research.
5.1. Performance Comparison with Existing Models
To verify the effect of the RG-FLAT-CRF-model, the RGT-CRF model is compared to the existing state-of-the-art models. Evaluated on CCKS2017, CCKS2019, and CCKS2020 datasets, respectively. The comparison model is as follows:
- (1)
- RoBerta: Liu et al. [37] improved the BERT model and proposed the RoBerta model. RoBerta performed better than BERT on NLP downstream tasks, and used RoBerta to enhance semantic representation and complete NER tasks.
- (2)
- RoBerta-BiLSTM-CRF: Xu et al. [25] combined the bi-directional LSTM and CRF, which has become a classic model, and combined the RoBerta model with BiLSTM-CRF on this basis. Use RoBerta trained vectors and then use the BiLSTM-CRF model to extract entities.
- (3)
- RoBerta-BiGRU-CRF: Qin et al. [29] proposed a BERT-BiGRU-CRF model in the field of Chinese electronic medical records, where the pre-trained model was replaced with an improved RoBerta.
- (4)
- Ra-RC: Wu et al. [30] used RoBerta to obtain medical semantic features while using a bidirectional long short-term memory network to learn the radical features of Chinese characters.
- (5)
- AR-CCNER: Yin et al. [26] used a convolutional neural network to extract radical features while using a self-attention mechanism to capture the dependencies between characters.
- (6)
- ACNN: Kong et al. [27] used a multi-layer CNN structure to capture short-term and long-term contextual relations. CNN can also solve the problem that LSTM is difficult to exploit GPU parallelism, and the model uses an attention mechanism that can obtain global information.
- (7)
- BE-Bi-CRF-JN: Wang et al. [31] cite additional medical knowledge information to correlate the original text in the named entity recognition task with its encyclopedic knowledge and enhance the ability of entity recognition by building a connection network.
Table 3, Table 4 and Table 5 show the precision, recall, and F1 results detailing various medical entities and all medical entities. From the comparison results of Table 6, the performance of the RGT-CRF model proposed in this chapter has achieved the best results on the three datasets, and the improvement on CCKS2017 is about 2~5%. The improvement is about 0.3~8% on CCKS2019 and about 3~9% on CCKS2020.

Table 3.
Results of different models on CCKS2017.

Table 4.
Results of different models on CCKS2019.

Table 5.
Results of different models on CCKS2020.

Table 6.
Comparison of the results of different F1 of each model on different datasets.
The effect of ACNN is unstable in CCKS2017 and CCKS2019. Compared with other models, ACNN does not use BERT or an improved model based on BERT to enhance semantic representation, but multi-layer CNN and attention mechanisms play a certain positive role. From the three datasets, most of the models use BERT or an improved pre-training model based on BERT to enhance semantic representation and have achieved good experimental results. RoBerta-BiLSTM-CRF performs better than RoBerta-BiGRU-CRF on the three datasets. Although BiGRU has a simpler structure than BiLSTM, it is clear that BiLSTM is more suitable for Chinese electronic medical record NER. At the same time, these two models perform moderately well on the three datasets, as the feature extraction networks of the two models are variations of recurrent neural networks and cannot solve the long-range dependency problem. AR-CCNER and Ra-RC performed better on the CCKS2017 and CCKS2019 datasets overall. Although AR-CCNER did not use a BERT-based pre-training model to enhance semantic representation, both AR-CCNER and Ra-RC were based on the characteristics of Chinese. BiLSTM and CNN are used to extract and use radical features, respectively, which utilize the glyph information of Chinese characters to a certain extent, but do not consider the information of learning the overall glyph structure of Chinese characters, and the model also lacks medical vocabulary information. BE-Bi-CRF-JN also achieved good results, proving that the use of external corpus in Chinese electronic medical records NER is effective. The above analysis shows that the RGT-CRF model is more suitable for Chinese electronic medical record named entity recognition electronic medical record recognition. This is mainly because the model adds glyph information while introducing lexical information based on words.
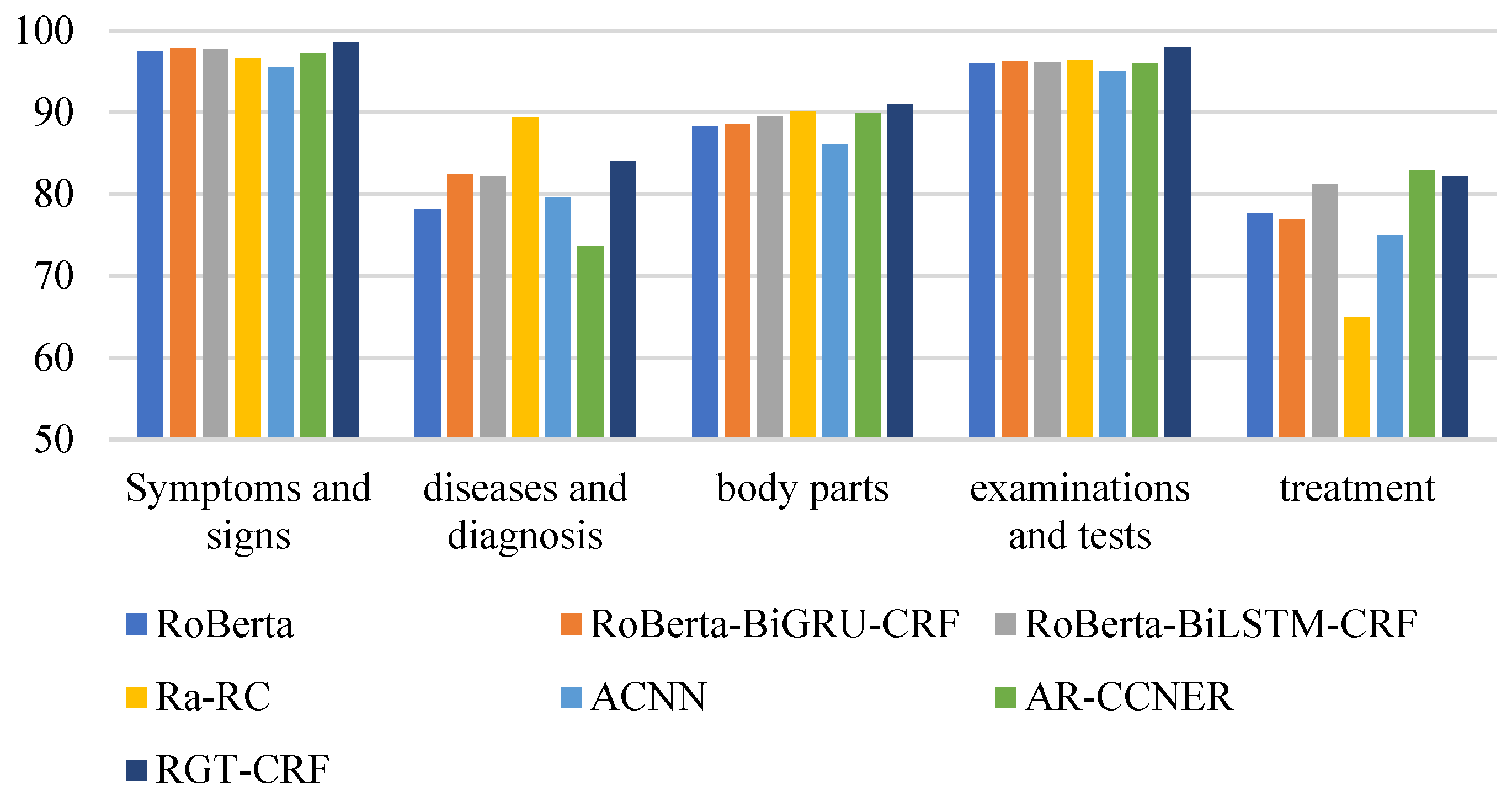
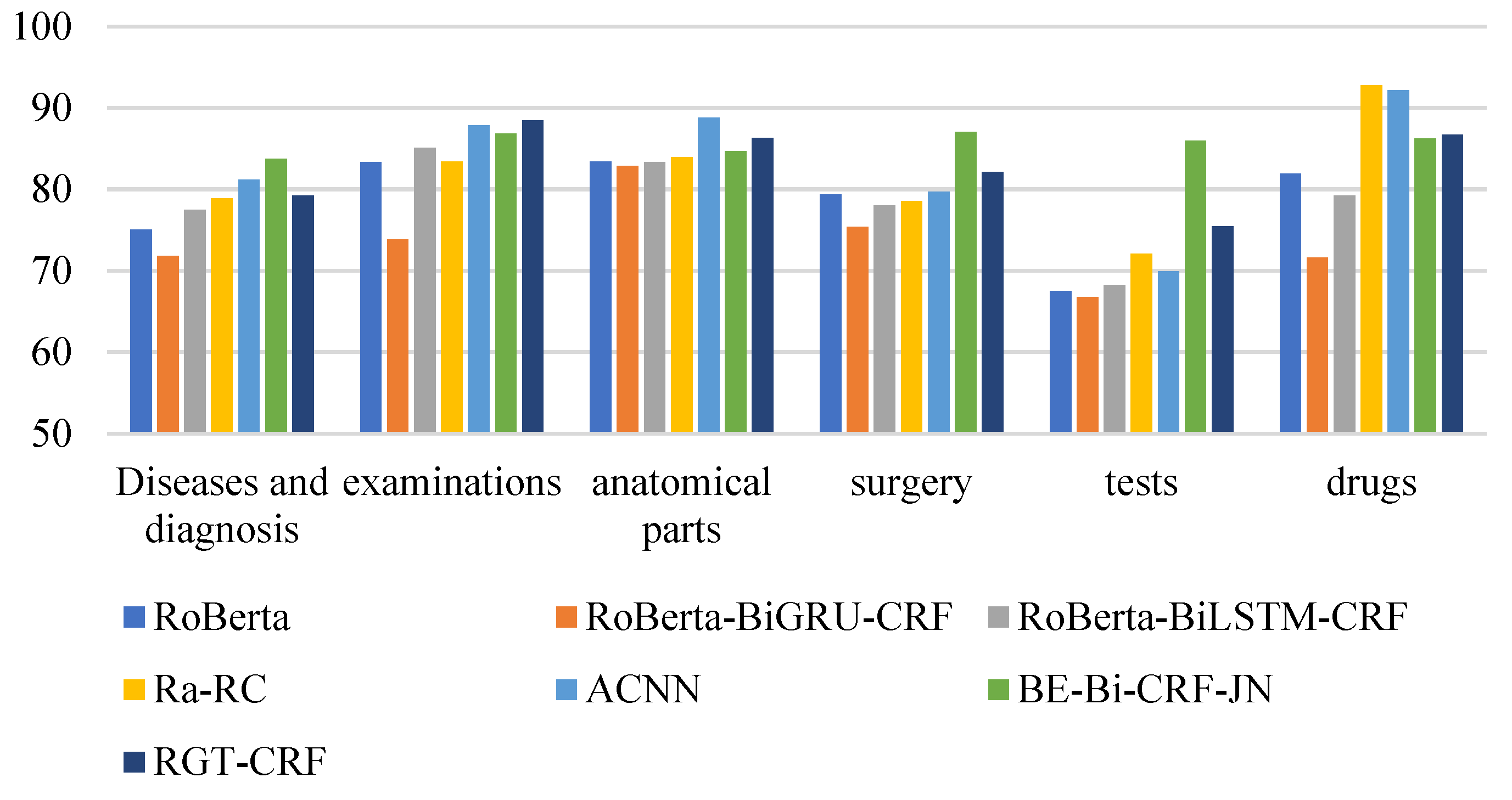
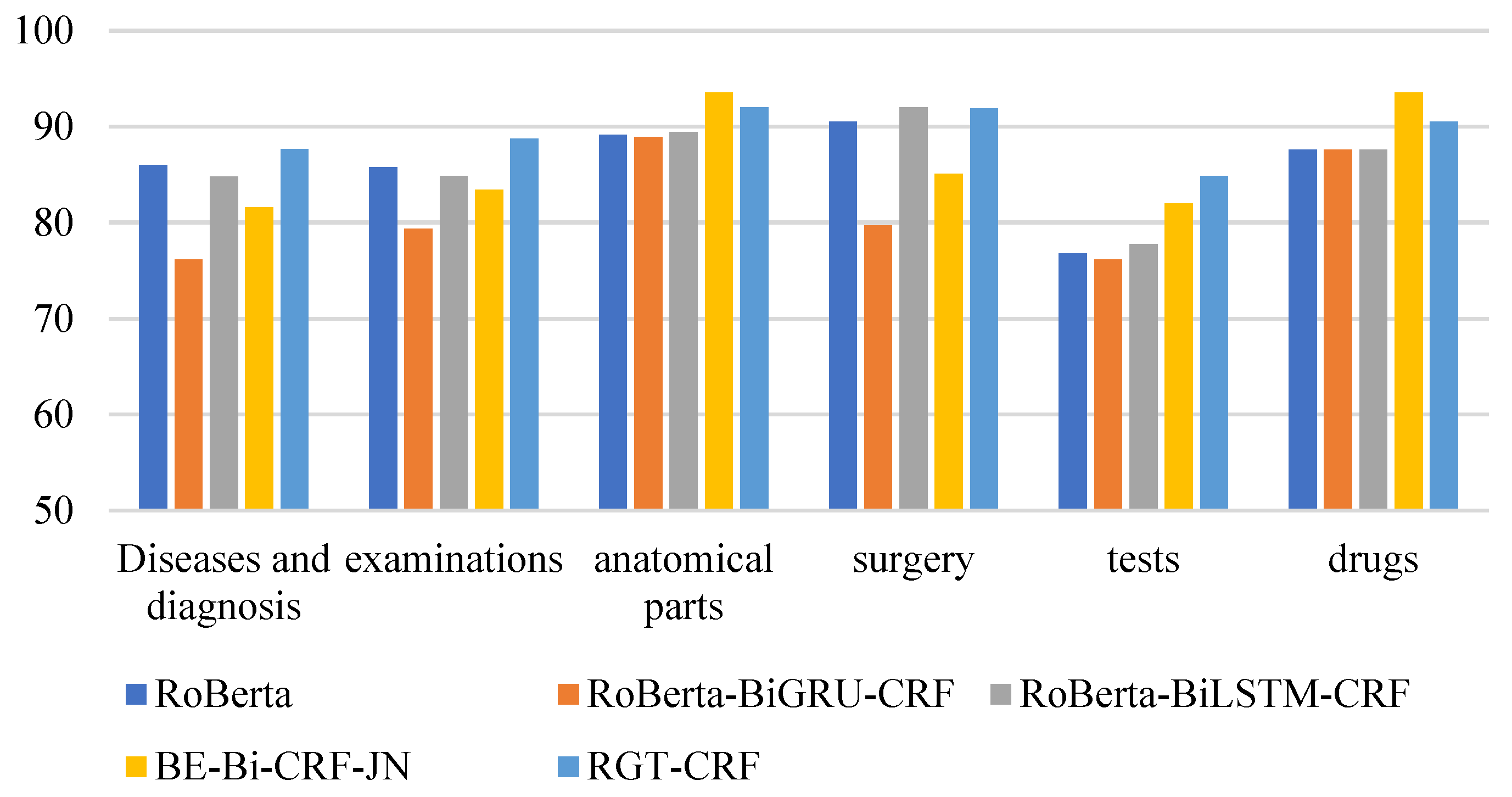
From the perspective of entity type, the overall recognition effect of different medical entities is compared longitudinally. From Figure 13, Figure 14 and Figure 15, it can be seen that the recognition results of different models on CCKS2017 show disease and diagnosis. Poor, because there are many long entities like ‘右股骨颈骨折髋关节股骨头表面置换术(Right femoral neck fracture hip femoral head resurfacing)’ in the two types of entities in the CCKS2017 dataset, and the boundaries of each entity cannot be clearly identified. The recognition results of different models on CCKS2019 and CCKS2020 show disease and diagnosis. The recognition results of these two types of entities are poor because the two types of entities in the CCKS2019 dataset and CCKS2020 dataset are similar to ‘CA125′, ‘CEA’. Many entities coexist with English and numbers, such as ‘CA199′, which will also cause the model to fail to identify the boundaries of each entity.
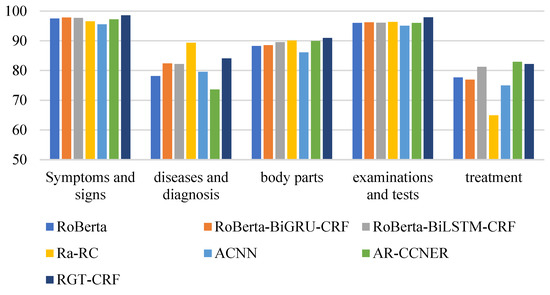
Figure 13.
F1 values of different entities on CCKS2017 for different models.
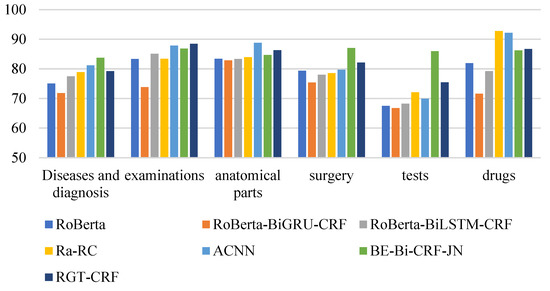
Figure 14.
F1 values of different entities on CCKS2018 for different models.
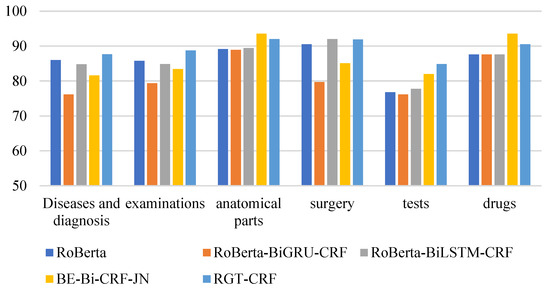
Figure 15.
F1 values of different entities on CCKS2020 for different models.
To make the comparative results more convincing, a further hypothesis test was performed by calculating p-values using the t-test method, and p-values smaller than the significance level (usually 0.05) were considered statistically significant. Table 7 shows the statistical comparison of the proposed method with other methods. Most of the results are significant.

Table 7.
Comparison results with different models on different datasets.
5.2. Ablation Research
We design a set of ablation experiments to verify the contribution of each part to the model, where RGT-CRF-NG indicates that the model does not add glyph information. RGT-CRF-NF shows that the model does not add lexical information and its corresponding positional encoding. Finally, it is compared with RoBerta-BiLSTM-CRF and RGT-CRF on three datasets, and the results are shown in Table 8.

Table 8.
Performance of different variants on three datasets.
The experimental results of RGT-CRF-NF and RGT-CRF-NG are better than the RoBerta-BiLSTM-CRF model regarding the three datasets, indicating that the glyph information and the use of lattice structure to add lexical information are effective for Chinese electronic medical record named entity recognition. The result of RGT-CRF-NG is slightly worse than that of RGT-CRF-NF, indicating that adding medical glyph information to the Chinese electronic medical record NER task is more effective than word information. This comparison can also be found in the above experiments using glyph information. Similarly, the final model with radical information is better than the model without radical information. This is because many Chinese characters in medical entities have the same glyph structure, so their meanings are also similar.
For example, ’疼(pain) ’, ’痛(pain)’, ’病(sick)’, ’腹(belly)’, ’腰(waist)’, ’肝(liver)’, ’脾(spleen)’, ’呕(vomit)’, ’吐(threw up)’, ’咳(cough)’, ’嗽(cough)’, ’胰(pancreatic)’, ’肠(intestinal)’, ’肿(swell)’, ’胀(swell)’. And this is very common in medical entities.
6. Conclusions
In this paper, an RG-FLAT-CRF model is proposed for Chinese CNER, which can learn the glyph features of medical fonts, and at the same time introduces word information to enhance word boundaries, and finally achieves good performance on three datasets. The RG-FLAT-CRF model obtains character vectors through RoBerta, Glyce, word2vec, and word vectors through word2vec. The word information is fused using the Flat-lattice structure and then encoded by the transformer network. In line with the output of the encoding layer, the label of each input character is predicted by the CRF layer. It addresses problems like word segmentation errors and lack of lexical information, given the characteristics of Chinese medical characters and the vector of multi-feature fusion. The final experimental results demonstrate that our proposed model outperformed the baseline models.
Several issues require further research. At this stage, deep learning requires a large amount of annotated data to train the model, as does our proposed model, but large-scale annotated data in the Chinese electronic medical record domain requires medical experts to annotate, which can be time-consuming. Therefore, our next research investigates how to perform named entity recognition on medical record texts with sparse data.
Author Contributions
J.L.: Conceptualization, Methodology, Software, Writing—original draft. Y.W.: Supervision, Project administration. R.L., C.C., and X.S.: Investigation, Writing—review & editing. S.Z.: Data curation, Resources. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key R&D Program (Demonstration of R&D and Application of Integrated Science and Technology Service Platform for Central Plains Urban Agglomeration), grant number 2018YFB1404500.
Data Availability Statement
We used the CCKS open-source Chinese electronic medical record named entity recognition dataset and cite it in the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chowdhury, S.; Dong, X.; Qian, L.; Li, X.; Guan, Y.; Yang, J.; Yu, Q. A multitask bi-directional RNN model for named entity recognition on Chinese electronic medical records. BMC Bioinform. 2018, 19, 75–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Zhou, Y.; Ruan, T.; Gao, D.; Xia, Y.; He, P. Incorporating dictionaries into deep neural networks for the Chinese clinical named entity recognition. J. Biomed. Inform. 2019, 92, 103133. [Google Scholar] [CrossRef] [PubMed]
- Shaukat, K.; Shaukat, U. Comment extraction using declarative crowdsourcing (CoEx Deco). In Proceedings of the 2016 International Conference on Computing, Electronic and Electrical Engineering (ICE Cube), Quetta, Pakistan, 11–12 April 2016; pp. 74–78. [Google Scholar]
- Li, J.; Sun, A.; Han, J.; Li, C. A survey on deep learning for named entity recognition. IEEE Trans. Knowl. Data Eng. 2022, 34, 50–70. [Google Scholar] [CrossRef] [Green Version]
- Alam, T.M.; Shaukat, K.; Hameed, I.A.; Khan, W.A.; Sarwar, M.U.; Iqbal, F.; Luo, S. A novel framework for prognostic factors identification of malignant mesothelioma through association rule mining. Biomed. Signal Processing Control 2021, 68, 102726. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Advances in Neural Information Processing Systems 30; Curran Associates Inc.: Red Hook, NY, USA, 2017; pp. 5998–6008. [Google Scholar]
- Zhang, Y.; Yang, J. Chinese NER using lattice LSTM. arXiv 2018, arXiv:1805.02023. [Google Scholar]
- Li, X.; Yan, H.; Qiu, X.; Huang, X. FLAT: Chinese NER using flat-lattice transformer. arXiv 2020, arXiv:2004.11795. [Google Scholar]
- Dai, Z.; Yang, Z.; Yang, Y.; Carbonell, J.; Le, Q.V.; Salakhutdinov, R. Transformer-xl: Attentive language models beyond a fixed-length context. arXiv 2019, arXiv:1901.02860. [Google Scholar]
- Meng, Y.; Wu, W.; Wang, F.; Li, X.; Nie, P.; Yin, F.; Li, M.; Han, Q.; Sun, X.; Li, J. Glyce: Glyph-vectors for chinese character representations. arXiv 2019, arXiv:1901.10125. [Google Scholar]
- Mikolov, T.; Sutskever, I.; Chen, K.; Corrado, G.S.; Dean, J. Distributed representations of words and phrases and their compositionality. In Advances in Neural Information Processing Systems 26; Curran Associates Inc.: Red Hook, NY, USA, 2013; pp. 3111–3119. [Google Scholar]
- Shaukat, K.; Luo, S.; Varadharajan, V.; Hameed, I.A.; Xu, M. A survey on machine learning techniques for cyber security in the last decade. IEEE Access 2020, 8, 222310–222354. [Google Scholar] [CrossRef]
- Shaukat, K.; Luo, S.; Varadharajan, V.; Hameed, I.A.; Chen, S.; Liu, D.; Li, J. Performance comparison and current challenges of using machine learning techniques in cybersecurity. Energies 2020, 13, 2509. [Google Scholar] [CrossRef]
- Friedman, C.; Alderson, P.O.; Austin, J.H.; Cimino, J.J.; Johnson, S.B. A general natural-language text processor for clinical radiology. J. Am. Med. Inform. Assoc. 1994, 1, 161–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukuda, K.; Tamura, A.; Tsunoda, T.; Takagi, T. Toward information extraction: Identifying protein names from biological papers. Pac. Symp. Biocomput. 1998, 707, 707–718. [Google Scholar]
- McCallum, A.; Freitag, D.; Pereira, F.C. Maximum entropy Markov models for information extraction and segmentation. ICML 2000, 17, 591–598. [Google Scholar]
- Možina, M.; Demšar, J.; Kattan, M.; Zupan, B. Nomograms for visualization of naïve Bayesian classifier. In European Conference on Principles of Data Mining and Knowledge Discovery; Springer: Berlin/Heidelberg, Germany, 2004; pp. 337–348. [Google Scholar]
- Settles, B. Biomedical named entity recognition using conditional random fields and rich feature sets. In Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and Its Applications (NLPBA/BioNLP), Geneva, Switzerland, 28–29 August 2004; pp. 107–110. [Google Scholar]
- Tang, B.; Cao, H.; Wu, Y.; Jiang, M.; Xu, H. Recognizing clinical entities in hospital discharge summaries using Structural Support Vector Machines with word representation features. BMC Med. Inform. Decis. Mak. 2013, 13, S1. [Google Scholar] [CrossRef] [Green Version]
- Roberts, K.; Shooshan, S.E.; Rodriguez, L.; Abhyankar, S.; Kilicoglu, H.; Demner-Fushman, D. The role of fine-grained annotations in supervised recognition of risk factors for heart disease from EHRs. J. Biomed. Inform. 2015, 58, S111–S119. [Google Scholar] [CrossRef]
- Liu, K.; Hu, Q.; Liu, J.; Xing, C. Named entity recognition in Chinese electronic medical records based on CRF. In Proceedings of the 2017 14th Web Information Systems and Applications Conference (WISA), Liuzhou, China, 11–12 November 2017; pp. 105–110. [Google Scholar]
- LeCun, Y.; Boser, B.; Denker, J.S.; Henderson, D.; Howard, R.E.; Hubbard, W.; Jackel, L.D. Backpropagation applied to handwritten zip code recognition. Neural Comput. 1989, 1, 541–551. [Google Scholar] [CrossRef]
- Mikolov, T.; Karafiát, M.; Burget, L.; Cernocky, J.; Khudanpur, S. Recurrent neural network based language model. Interspeech. Makuhari 2010, 2, 1045–1048. [Google Scholar]
- Ma, X.; Hovy, E. End-to-end sequence labeling via bi-directional lstm-cnns-crf. arXiv 2016, arXiv:1603.01354. [Google Scholar]
- Xu, K.; Zhou, Z.; Hao, T.; Liu, W. A bidirectional LSTM and conditional random fields approach to medical named entity recognition. In Proceedings of the International Conference on Advanced Intelligent Systems and Informatics 2017; Springer: Cham, Swizerland, 2017; pp. 355–365. [Google Scholar]
- Yin, M.; Mou, C.; Xiong, K.; Ren, J. Chinese clinical named entity recognition with radical-level feature and self-attention mechanism. J. Biomed. Inform. 2019, 98, 103289. [Google Scholar] [CrossRef]
- Kong, J.; Zhang, L.; Jiang, M.; Liu, T. Incorporating multi-level CNN and attention mechanism for Chinese clinical named entity recognition. J. Biomed. Inform. 2021, 116, 103737. [Google Scholar] [CrossRef]
- Zhang, W.; Jiang, S.; Zhao, S.; Hou, K.; Liu, Y.; Zhang, L. A BERT-BiLSTM-CRF model for Chinese electronic medical records named entity recognition. In Proceedings of the 2019 12th International Conference on Intelligent Computation Technology and Automation (ICICTA), Xiangtan, China, 26–27 October 2019; pp. 166–169. [Google Scholar]
- Qin, Q.; Zhao, S.; Liu, C. A BERT-BiGRU-CRF Model for Entity Recognition of Chinese Electronic Medical Records. Complexity 2021, 2021, 6631837. [Google Scholar] [CrossRef]
- Wu, Y.; Huang, J.; Xu, C.; Zheng, H.; Zhang, L.; Wan, J. Research on Named Entity Recognition of Electronic Medical Records Based on RoBERTa and Radical-Level Feature. Wirel. Commun. Mob. Comput. 2021, 2021, 2489754. [Google Scholar] [CrossRef]
- Wang, Q.; Haihong, E. Bi-directional Joint Embedding of Encyclopedic Knowledge and Original Text for Chinese Medical Named Entity Recognition. In Proceedings of the 2021 2nd International Conference on Electronics, Communications and Information Technology (CECIT), Sanya, China, 27–29 December 2021; pp. 304–309. [Google Scholar]
- Pennington, J.; Socher, R.; Manning, C.D. Glove: Global vectors for word representation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; pp. 1532–1543. [Google Scholar]
- Peters, M.E.; Neumann, M.; Iyyer, M.; Gardner, M.; Clark, C.; Lee, K.; Zettlemoyer, L. Deep contextualized word representations. arXiv 2018, arXiv:1802.05365. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Sun, Y.; Wang, S.; Li, Y.; Feng, S.; Chen, X.; Zhang, H.; Tian, X.; Zhu, D.; Tian, H.; Wu, H. Ernie: Enhanced representation through knowledge integration. arXiv 2019, arXiv:1904.09223. [Google Scholar]
- Cui, Y.; Che, W.; Liu, T.; Qin, B.; Yang, Z. Pre-training with whole word masking for chinese bert. IEEE ACM Trans. Audio Speech Lang. Process. 2021, 29, 3504–3514. [Google Scholar] [CrossRef]
- Liu, Y.; Ott, M.; Goyal, N.; Du, J.; Joshi, M.; Chen, D.; Levy, O.; Lewis, M.; Zettlemoyer, L.; Stoyanov, V. Roberta: A robustly optimized bert pretraining approach. arXiv 2019, arXiv:1907.11692. [Google Scholar]
- Yang, Z.; Dai, Z.; Yang, Y.; Carbonell, J.; Salakhutdinov, R.R.; Le, Q.V. Xlnet: Generalized autoregressive pretraining for language understanding. In Advances in Neural Information Processing Systems 32; Curran Associates Inc.: Red Hook, NY, USA, 2019; p. 32. [Google Scholar]
- Sun, Y.; Lin, L.; Yang, N.; Ji, Z.; Wang, X. Radical-enhanced chinese character embedding. In International Conference on Neural Information Processing; Springer: Cham, Switzerland, 2014; pp. 279–286. [Google Scholar]
- Wang, S.; Zhou, W.; Zhou, Q. Radical and Stroke-Enhanced Chinese Word Embeddings Based on Neural Networks. Neural Process. Lett. 2020, 52, 1109–1121. [Google Scholar] [CrossRef]
- Wei, H.; Zhang, H.; Gao, G. Word image representation based on visual embeddings and spatial constraints for keyword spotting on historical documents. In Proceedings of the 2018 24th International Conference on Pattern Recognition (ICPR), Beijing, China, 20–24 August 2018; pp. 3616–3621. [Google Scholar]
- Su, T.R.; Lee, H.Y. Learning chinese word representations from glyphs of characters. arXiv 2017, arXiv:1708.04755. [Google Scholar]
- Shaukat, K.; Luo, S.; Chen, S.; Liu, D. Cyber threat detection using machine learning techniques: A performance evaluation perspective. In Proceedings of the 2020 International Conference on Cyber Warfare and Security (ICCWS), Islamabad, Pakistan, 20–21 October 2020; pp. 1–6. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).