1. Introduction
Each month, during human female ovulation, the left or right ovary produces and releases an egg for fertilization. Early development of an embryo starts with the fertilization of an egg by a sperm at the fallopian tube. The fertilized egg (embryo), where the egg and the sperm unite, develops and goes through several stages, reaching the blastocyst stage before implanting in the wall of the uterus. The implantation of the embryo is a critical point during pregnancy for embryologists, who aim to accurately identify the quality of an embryo at this point [
1]. Due to the lack of consistent noninvasive methods to determine the quality of embryos, the process of identifying the quality of an embryo is a challenging task [
2]. As bovine and human females share various characteristics in terms of folliculogenesis, the cow has been used as a model to investigate folliculogenesis events in humans [
3,
4]. Bó et al. [
5] described the principles of evaluating bovine embryos, which are important for deciding whether the embryo is worth transferring or frozen for a given recipient and whether the embryo is eligible to be exported for human use. The conventional evaluation method of the quality of bovine embryos is subjective visual evaluation of embryo images by embryologists using stereomicroscopy, followed by a scoring system issued by the International Embryo Technology Society (IETS). The scores produced determine whether the quality of the bovine embryo is excellent, fair, or good.
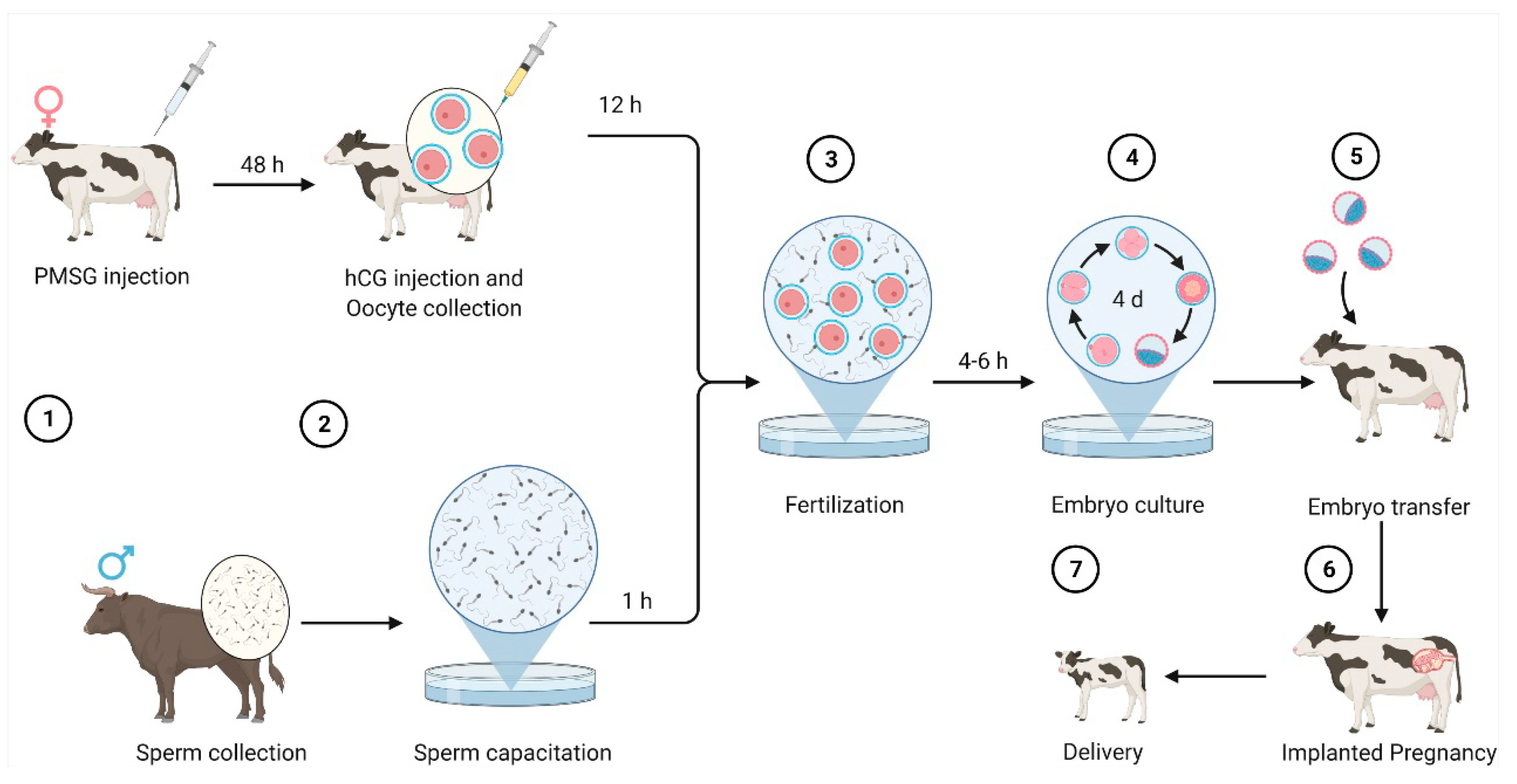
Figure 1 shows the process of bovine embryo production through in vitro fertilization [
6,
7,
8].
Deep Learning (DL) techniques have been successfully applied to solve many real biological problems, including various medical imaging problems [
9,
10,
11,
12,
13,
14,
15]. Specifically, DL techniques aim to improve the prediction performance in a given task via learning complex concepts from simpler ones [
16]. DL methods provide promising solutions in artificial intelligence when applied to medical imaging and could improve the performance of several medical imaging problems, including predicting the quality of embryos, in the coming years.
Predicting the quality of embryos helps embryologists to (1) improve the embryo transfer process, which leads to better genetics and viable embryos [
17]; (2) reduce the time spent searching for viable embryos, which is important for clients in the cattle industry who need to freeze or transfer the embryos to recipients [
18,
19]; and (3) avoid abortion at later stages of pregnancy caused by poor quality of embryos [
20]. Due to the importance of predicting the quality of embryos, researchers from different domains, including developmental biology and machine learning, are involved in this work. For example, Rocha et al. [
21] proposed a machine learning approach to predict the quality of bovine embryos that works as follows. A preprocessing step aims to construct feature vectors via extracting 36 features from images of in vitro bovine images. Then, the constructed features are provided as input to a neural network, to build a model, which is then used to classify unseen images. Manna et al. [
22] proposed an ensemble approach utilizing features extracted from the foreground image of the embryo to construct feature vectors, which are then provided to an ensemble of neural networks to train and predict the quality of unseen embryos. Filho et al. [
23] proposed a machine learning approach to predict the grading of human embryos using extracted image features, provided as input to support vector machines (SVM), to yield a model. The resulting model is then used to evaluate the model using images of unseen human embryos obtained via two independent experts. Other machine learning and deep learning approaches have been developed for other prediction tasks related to embryos [
24,
25]. For example, Miyagi et al. [
26] developed a deep learning approach utilizing a convolutional neural network consisting of two convolutional layers for feature extraction, two pooling layers for dimensionality reduction, and fully connected layers for classification. The developed approach is used to predict the number of live births from images of blastocysts. Experimental results showed that the deep learning approach performs significantly better than the baseline method.
The work described here differs from [
5,
21,
22,
23,
26] in three ways. (1) We present three deep learning approaches, where the first deep learning approach (DL) exploits a convolutional neural network consisting of a convolutional layer for feature extraction, an average pooling layer for dimensionality reduction and fully connected, and output layers for the classification. The second deep learning approach (B1DL) extends the DL approach and is coupled with a boosting technique to improve the prediction performance by selecting an ensemble with the highest area under the curve during the training, which is then used to perform predictions on unseen embryo images. The third deep learning approach (B2DL) is different from the B1DL approach in terms of utilizing a different boosting technique. (2) The proposed deep learning approaches are compared against other deep learning approaches under a supervised learning setting, using only images of in vitro bovine embryos. (3) We conduct statistical tests and evaluate the performance of all approaches using several performance measures.
Conventional approaches have relied on the experience and expertise of humans, following evaluation principles to determine the quality of bovine embryos [
5]. To automate the intelligence of the expert, ML approaches require extracting image features to construct a dataset, which are then provided to a ML algorithm (artificial neural networks) to create a model that automates human intelligence for predicting the quality of unseen bovine embryos [
21]. However, these approaches are (1) not accurate enough as they rely on manually hand crafted features; (2) based on the subjective evaluation of experts who spend much time examining embryo images and can mislabel some due to oversight or fatigue; and (3) based on a set of subjective features that could affect the generalizability of models [
25]. In contrast to previous approaches that rely on a set of specified features, sophisticated deep learning approaches can be used to improve prediction performance without specifying a set of hand-crafted features [
27,
28]. However, these deep learning approaches require extensive computational resources and the adopting of concepts that improve the learning process [
29,
30].
Contributions. Our contributions in this paper are summarized as follows.
- -
We present new deep learning approaches for developmental biology, enabling deep learning algorithms to achieve high-performance results from images of in vitro bovine embryos.
- -
The proposed approaches adopt modified versions of deep convolutional neural networks and boosting techniques, and are the first, to the best of our knowledge, to be applied in the developmental biology discipline pertaining to predicting the quality of embryos.
- -
In contrast to previous works, we assess our approaches by employing several classification performance measures against baselines, including existing deep learning algorithms such as LeNet and AlexNet, based only on in vitro data.
- -
We perform an experimental study to report the predictive performance of all deep learning approaches. The results demonstrate that our approaches significantly outperformed the baseline approaches.
Organization. The rest of this paper is organized as follows. We review existing research related to this paper in
Section 2.
Section 3 and
Section 4 present our proposed deep learning approaches and report performance results of the proposed approaches against the baselines. We provide a discussion of the results in
Section 5. Lastly, we conclude the paper and point out directions for future work in
Section 6.
5. Discussion
Our first deep learning approach (DL) aims to learn a model for classifying in vitro bovine embryos using a CNN variant, which includes a convolutional layer and pooling layer for the feature extraction and dimensionality reduction, fully connected layers, and an output layer for the classification. The resulting model is used to classify test examples of in vitro bovine embryo images. The second deep learning approach (B1DL) utilizes the CNN used in the DL approach, coupled with a boosting technique, to yield an ensemble of models, which are then used to classify test examples. The third deep learning approach (B2DL) behaves like the B1DL approach. However, B2DL has a different boosting technique in terms of the weight update mechanism.
Predicting the quality of bovine embryos is a challenging task. Making accurate predictions of the quality of bovine embryos could be specifically adapted to optimize the cryopreservation and avoid embryo selection in countries where it is not permitted, which in turn could help embryologists to enhance laboratory protocols, leading to successful pregnancies [
20,
22,
61]. In this study, we aim to improve the prediction of the quality of bovine embryos via leveraging deep learning techniques that consist of a variant of CNN with or without boosting techniques. Experimental results show that our approaches perform better than baseline approaches.
For the optimization process during the training of LeNet and AlexNet, we used the Adam optimizer in this study with the default parameters (as suggested in [
62]) to minimize the categorical cross-entropy loss function as in [
63]. For our approaches, categorical cross-entropy is used as a loss function and the optimization is performed via RMSProp optimizer [
64,
65], with default parameters given in [
49].
In this study, we ran the cross-validation 10 times to assess the stability of prediction algorithms. The reported p-values obtained from a Friedman test show that the prediction algorithms utilizing our approaches perform better than the baseline methods that utilize LeNet and AlexNet architectures.
The deep learning training phase requires additional time depending on the number of processing layers. The more layers a network has, the more computational running time is needed during the training. Therefore, we utilized deep learning consisting of fewer layers, because the proposed approaches work in an iterative manner, where DL is trained in each iteration.
While our boosted approaches require the invoking of CNN several times, our computational experiments show that the B2DL prediction algorithm is efficient as it requires less running time than the baseline methods. This is due to B2DL including just four layers compared to other baseline methods (i.e., LeNet and AlexNet) consisting of more than five. AlexNet was the slowest prediction algorithm.
6. Conclusions and Future Work
We propose three deep learning approaches to accurately predict the quality of in vitro bovine embryos. The first approach DL utilizes a convolutional neural network variant consisting of four layers, excluding the input and output layers. The DL approach aims to learn a model from a training set of images pertaining to in vitro bovine embryos. The resulting model is then applied to perform predictions on unseen test examples. The second approach, B1DL, is an extension of the DL approach. The only difference is that B1DL utilizes a variant of the boosting technique to select the best ensemble in terms of the maximum area under the curve during the training, from a set of several ensemble models. Then, the best ensemble is used to perform predictions on unseen text examples. The third approach, B2DL, is similar to B1DL. The only difference is that B2DL utilizes a boosting technique with a different weight update mechanism. These deep learning approaches are designed to improve the predictive performance. Experimental results on images pertaining to in vitro bovine embryos demonstrate that our approaches perform better than existing baseline methods and achieve statistically significant results.
In future work, we plan to (1) adopt the proposed approaches under the transfer learning setting; (2) incorporate the proposed approaches with generative adversarial networks to improve the prediction performance under the transfer learning setting [
66,
67,
68]; and (3) evaluate the proposed deep learning approaches against others in the context of different medical imaging problems [
69,
70].