Abstract
In this study, a knowledge graph of Chinese medical record data was constructed based on graph database technology. An entity extraction method based on natural language processing, disambiguation, and reorganization for Chinese medical records is proposed, and dictionaries of drugs and treatment plans are constructed. Examples of applications of the knowledge graph in diagnosis and treatment prediction are given. Experimentally, it is found that the knowledge graph based on the graph database is 116.7% faster than the traditional database in complex relational queries.
1. Introduction
Medical treatment is a part of Chinese nationals’ health monitoring. At present, medical data are diverse and complex [1]. Although China has a well-established system of recording medical records, in most cases, patients’ symptoms, medications, and therapies are recorded in natural language and are highly fragmented. This makes it difficult for researchers in the medical field to retrieve and organize medical records. Knowledge graphs based on graph database technology can effectively solve these problems. They can transform large-scale medical record data into a form that conforms to the graph database model through data analysis, processing, and normalization operations, which facilitates the subsequent construction and querying of the knowledge graph. Through the construction of entity extraction and dictionaries of drugs and treatment plans based on the method of participle reorganization, the key information in Chinese medical records can be effectively extracted and indexed to provide the basis for the subsequent knowledge graph modeling.
Getting the information we need from unstructured natural language medical records requires entity extraction. Many scholars have made some efforts in entity extraction research in different fields in recent years. Jinfeng Yang [2] attempted to construct a corpus of named entities and entity relationships in Chinese electronic medical records. Hongyang Chang [3], based on cardiovascular disease (CVD) electronic medical record texts, presented an annotation scheme for named entities and entity relationships in the Chinese electronic medical record (CEMR). Sune Pletscher-Frankild [4] proposed a text mining and data integration method for disease–gene associations. George Hripcsak [5] proposed management and utilization methods for electronic health records (EHR). Jianqin Liang [6] proposed a new model for naming entity recognition of crop pests and diseases in China, aiming to solve the problems of uneven distribution of entities, incomplete recognition of complex terms, and unclear entity boundaries. Koichi Takeuchi [7] used support vector machines (SVMs) to identify and semantically annotate terms in the field of molecular biology. Yan Gao [8] proposed a character and word attention enhancement (CWAE) neural network based on Chinese resident admission notes as a medical entity recognition model. Chenyuan Hu [9] used bi-directional long- and short-term memory (Bi-LSTM) and text-based convolutional neural networks (TextCNN) to differentiate the syndromes of Traditional Chinese Medicine (TCM).
Knowledge graphs connect entities through relationships and form a network knowledge structure [10]. In recent years, the establishment of knowledge graphs has become a visualization method chosen by many scholars. Scholars have used this method to achieve certain research results in different fields. Jingchi Jiang [11] proposed a recursive neural knowledge network (RNKN) to process Chinese electronic medical records. Xuqing Chai [12] uses a knowledge graph to train the bidirectional long short-term memory network (BLSTM) disease diagnosis model. Jiajing Hu [13] has developed a website called DGLinker based on knowledge graphs to predict new genetic factors associated with diseases. Zhenfeng Lei [14] proposed a data-driven framework called d-DC, which uses knowledge graphs to classify diseases. Zhiqing Li [15] constructed a Neo4j-based knowledge graph by preprocessing the collected clinical data and analyzing statistical rules, and relied on it to predict the risk of diabetic macular edema. Linfeng Li [16] proposed a new quadratic structure to denote medical knowledge in the construction of knowledge graphs, and proposed a novel ranking function of relevant entities considering probability, specificity, and reliability (PSR).
Machine learning methods are also widely used in many areas of medicine, like cancer research [17,18,19], toxinology [20], and wearable sensors [21]. The current research trend is to use neural networks for entity extraction of medical records and importing them into graph databases. The aim is to analyze the natural language in medical records using the extremely strong fitting and classification capabilities of neural networks to obtain words that may be useful entities. However, training a neural network model requires highly capable dedicated computing equipment, which is difficult to achieve in many healthcare organizations [22,23]. Moreover, the approach of classifying and extracting only words does not make good use of the natural language structure in medical records written by healthcare workers, where there is less research on dedicated and efficient methods for entity extraction for medical records [24].
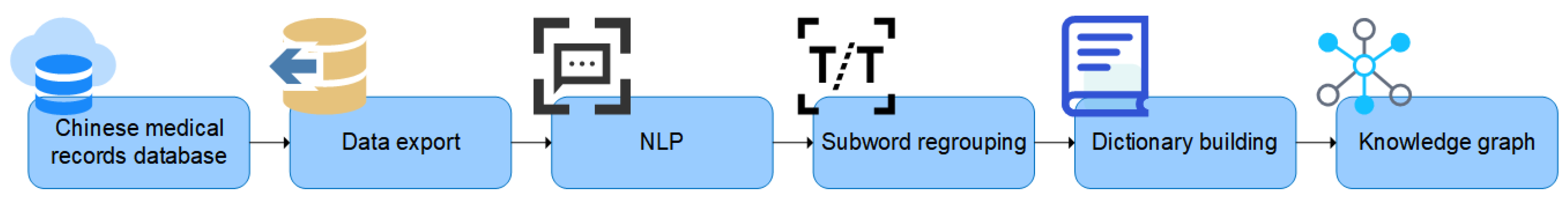
This study proposes an entity extraction method based on natural language processing and disambiguation reorganization with the help of the medical record database of a cooperative medical institution, in order to address the problem of excessive training overhead of existing neural network models. First, the sentences are subjected to natural language processing and word splitting, after which the words are combined and categorized into three categories: drug name, dosage, and connectives, based on the contextual information of the words. Drugs and dosage are extracted along with other structured data to construct a knowledge graph. The method of using a knowledge graph for disease prediction and diagnosis is proposed, and some specific examples are given. The specific process is shown in Figure 1.
Figure 1.
Study framework.
The main contribution of this paper is to propose an entity extraction method based on word segmentation and recombination using linguistic features of Chinese medical records and contextual relationships between words, unlike commonly used neural network methods, which alleviates the performance overhead. A knowledge graph model is established and some concrete examples of applications are given.
Establishing a knowledge graph in medicine can structure non-isomorphic knowledge in the medical domain and build associations between data items [25]. It mainly solves the problems of scattered, diverse, complex data in the medical field, and low value of single data. A map that naturally displays and relates the knowledge in the field of medicine explicitly was built [26]. We can realize data mining and analysis based on the characteristics of the native graph. At the same time, this promotes the development of smart medical care [27], realizes various functions such as auxiliary diagnosis and treatment, and saves manpower and material resources. If the knowledge map of all diseases is integrated, then in medicine, the study of diseases will not become isolated, and the research of various diseases will become closely related.
2. Entity Extraction of Chinese Medical Records
2.1. Preprocessing of Diagnosis and Treatment Data
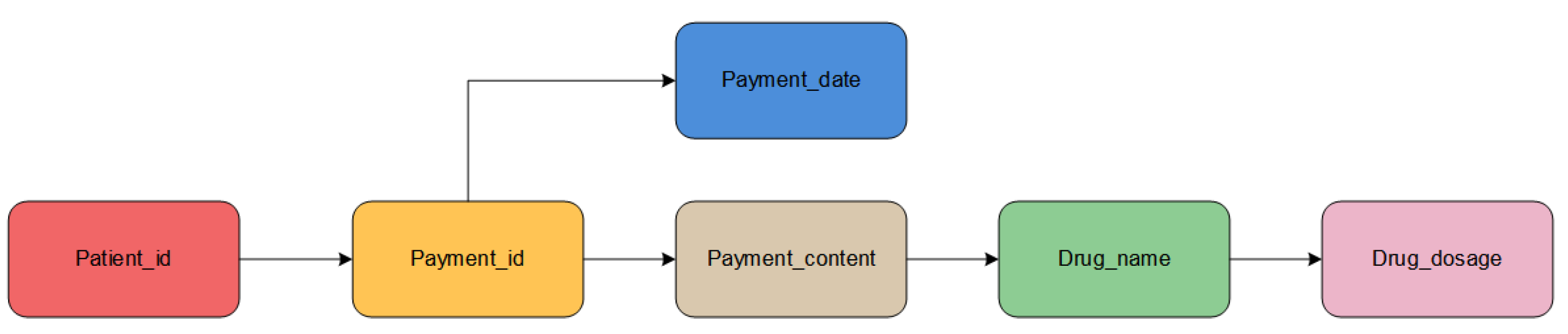
In order to ensure the accuracy of relevant data, we have obtained a detailed database including examination results, payment orders, and surgical conditions through communication with cooperative medical institutions. It covers patients who went to the medical facility in 2016. In order to protect the personal privacy of patients, all data have been desensitized. For simplicity, we will take the payment order data as an example to carry out the analysis and processing and the construction of the knowledge graph in the following text. Some fields and their meanings included in the payment order database are shown in Table 1.

Table 1.
Payment order database field information, where “力扑素” means “Paclitaxel Liposom” and “赫赛丁” means “Trastuzumab”.
In Table 1, the payment number, payment date, and patient ID are all structured data, which can be directly processed in the next step. The payment content consists of unstructured data, which is close to natural language, and there is a large amount of redundant data, which requires entity extraction of useful information that can be obtained from it.
In addition, there are some missing values in the payment order date column and payment content column in the data, which will affect subsequent processing and require filling in. According to the characteristics of the data, for the payment date, we use the approximate average value to fill. For payment content, because it is unstructured data, which is difficult to fill in a general way, we use the decision tree classification algorithm to directly fill in the drug information after entity extraction, and then generate payment content from the drug information.
The processed drug information is in the form of an array. When filling in missing values, it can be understood as multiple 0–1 classification tasks, which determine whether various drugs exist in the array. Among various classification algorithms, we have chosen the decision tree method with relatively low performance and cost to train various drugs as labels.
Common decision tree algorithms include ID3, C4.5, CART, etc. [28]. Due to the large number of feature classifications in the samples, in order to prevent the excessive number of labels from affecting the classification effect, we chose the C4.5 algorithm with a modified number of labels for training and prediction. The decision feature selection formula of the C4.5 algorithm is as follows:
where
is the information gain when selecting each feature for classification, which describes the change of sample purity before and after classification; IV (a) reflects the number of classifications, and dividing two numbers can reduce the impact of classifications on information gain. After training and testing on complete data and some manually labeled data, the accuracy rate can reach more than 76.8%. This completes the filling in of missing values.
2.2. Entity Extraction Based on Word Segmentation and Recombination Method
Entity extraction refers to the process of processing and recognizing natural language to obtain entities. In this study, entity extraction is mainly applied to the payment content field.
We downloaded the Chinese and foreign drug name thesaurus [29] from the public vocabulary as the initial dictionary. For some cases of missing drug names, this article adopts a word segmentation reorganization scheme to extract some of the missing entities.
By observing the recording structure of payment content, it can be found that although payment content is recorded in natural language, there are still certain rules to follow. For the vast majority of records, the drug name is in Chinese. The dosage information is usually composed of numbers and English units after the drug name. Half- or full-width symbols are used as separators between drug information. For example:
where “力扑素” means “Paclitaxel Liposom” and “卡铂” means “Carboplatin”. Specifically,
Therefore, we can use the word segmentation tool to split the payment content string according to the characteristics of the words to obtain the components in the sentence. And then, based on their positional relationships in the sentence, we can obtain the three types of entities A, B, and C, as mentioned earlier, as well as their relationships. For the word , obtained after splitting, we first need to determine its components:
- , if and only if is completely composed of Chinese characters.
- , if and only if is completely composed of English letters and Arabic numerals.
- , if and only if is completely composed of non-Chinese characters, English letters, and Arabic numerals, and the length is 1.
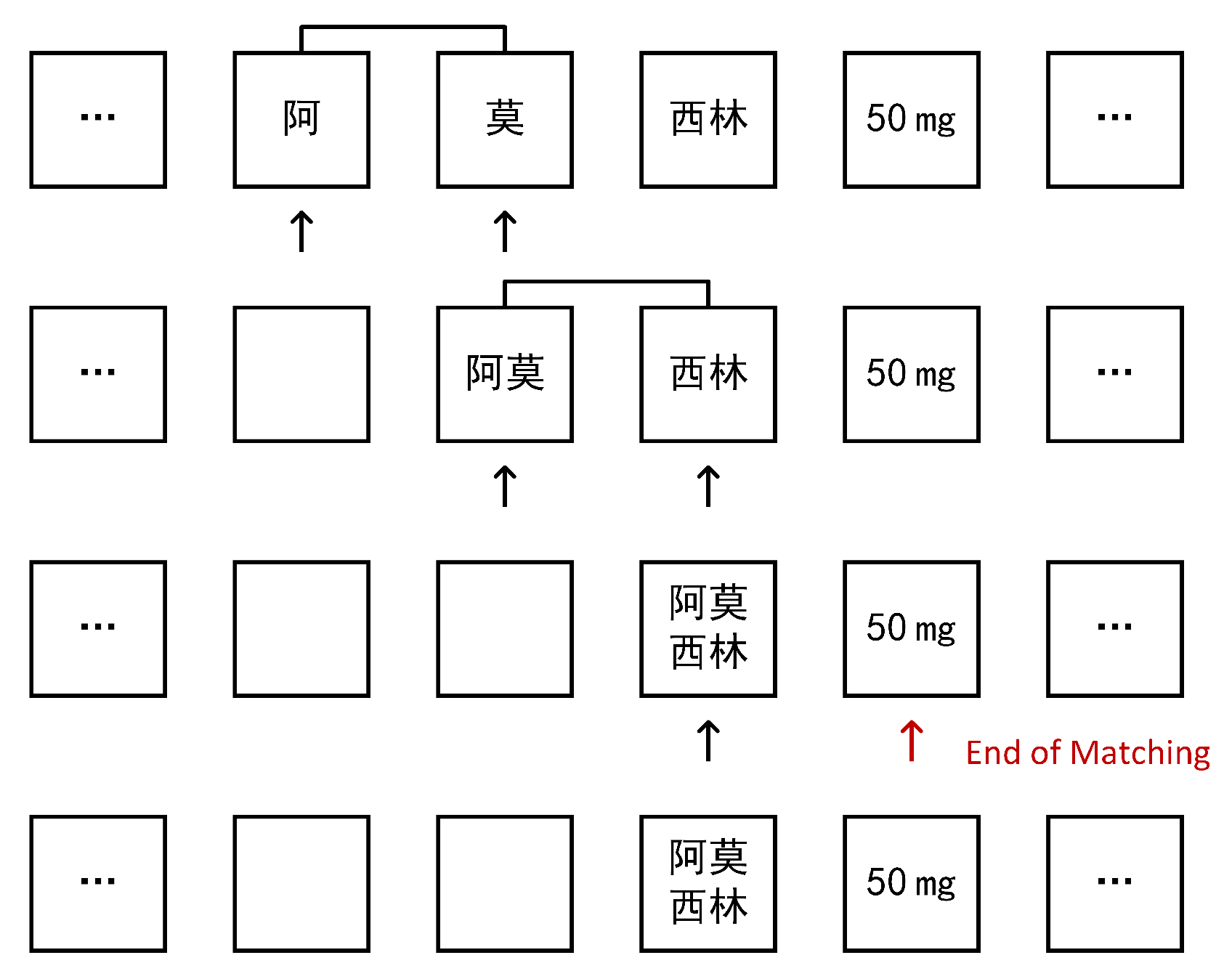
Furthermore, we process drugs that were not included in the initial dictionary. Due to the rarity of drug names, they are generally not fully preserved during word segmentation, but rather split into single or short words. Therefore, we can use word recombination for heuristic extraction. We assume that the drug name has not been included in the initial dictionary, and in word segmentation is split into sub-words , we scan the word-segmented string array, if appears, and replace with back into the original array and continue to scan until the above conditions are not met, so we reorganize , namely into is . Then, we update the dictionary and add into the original dictionary. In this way, the next time the drug name is encountered, the word breaker will automatically extract the complete word, eliminating the need for recombination and improving processing efficiency. The specific process of the algorithm is shown in Figure 2.
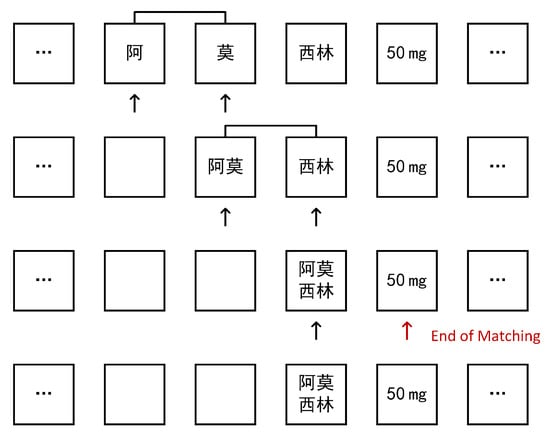
Figure 2.
Split word restructuring algorithm process, where “阿莫西林” means “Amoxicillin”.
3. Implementation of Knowledge Graph
3.1. Entity Relationship Construction
In the payment order database used in this article, we extracted five relationships, as follows:
By this processing, we can create relational data tables for the construction of knowledge graphs. Most of the content of the relational data table can be obtained directly from the original database. For the relationship and relationship that cannot be obtained directly, the former can be obtained from the inclusion relationship between the word and the sentence , and the latter can be constructed based on the word position relationship mentioned in Section 2.2. Tuples . Where , and and . If exists, it means that there is such a relationship between and .
Through the above processing, we can obtain a relational table, which consists of fields shown in Table 2.

Table 2.
Payment order database field information.
It is noticed that the at field is not a field commonly found in relational tables. This is because, in this study, different patients may use the same medication, but the dosage of the medication varies from person to person, and the dosage of a certain patient may also vary depending on the course of treatment and the condition. If no distinction is made, the structure of the knowledge graphs will be confused, and the required information cannot be found correctly. Therefore, we have added the field to save the primary key of the record where the relationship is located in the original database (i.e., the payment ID) to distinguish it.
3.2. Construction of Knowledge Graph Based on Neo4j
Knowledge Graph is a structured semantic knowledge base used to describe concepts and their interrelationships in the physical world in symbolic form. Its basic unit of composition is the “entity-relationship-entity” triplet, as well as entities and their related attribute value pairs. Entities are interconnected through relationships, forming a network of knowledge arranges. Here:
- Entity: corresponds to the semantic ontology of the real world.
- Relationship: corresponds to the relationship between ontologies, connecting different types of entities.
- Attributes: describes the characteristics of an entity.
In the task of building a knowledge graph, a graph-based database, Neo4j [30], is used. Neo4j mainly connects user-defined nodes through specific relationships in the form of graphs, so that we can start from the explored nodes and find out the connection between two nodes through the user-defined relationship between nodes.
The knowledge graph entity design includes five items, namely , , , , , and . A total of more than 120,000 nodes have been established.
After importing the CSV file into the Neo4j graph database, the created knowledge graph data will be visualized. Each entity category is represented by a circle of a different color, and the relationship between entities is marked on the connection line between each entity. The entities are connected as shown in Figure 3.
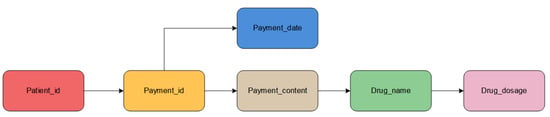
Figure 3.
Entity connection diagram.
4. Application of Knowledge Graph
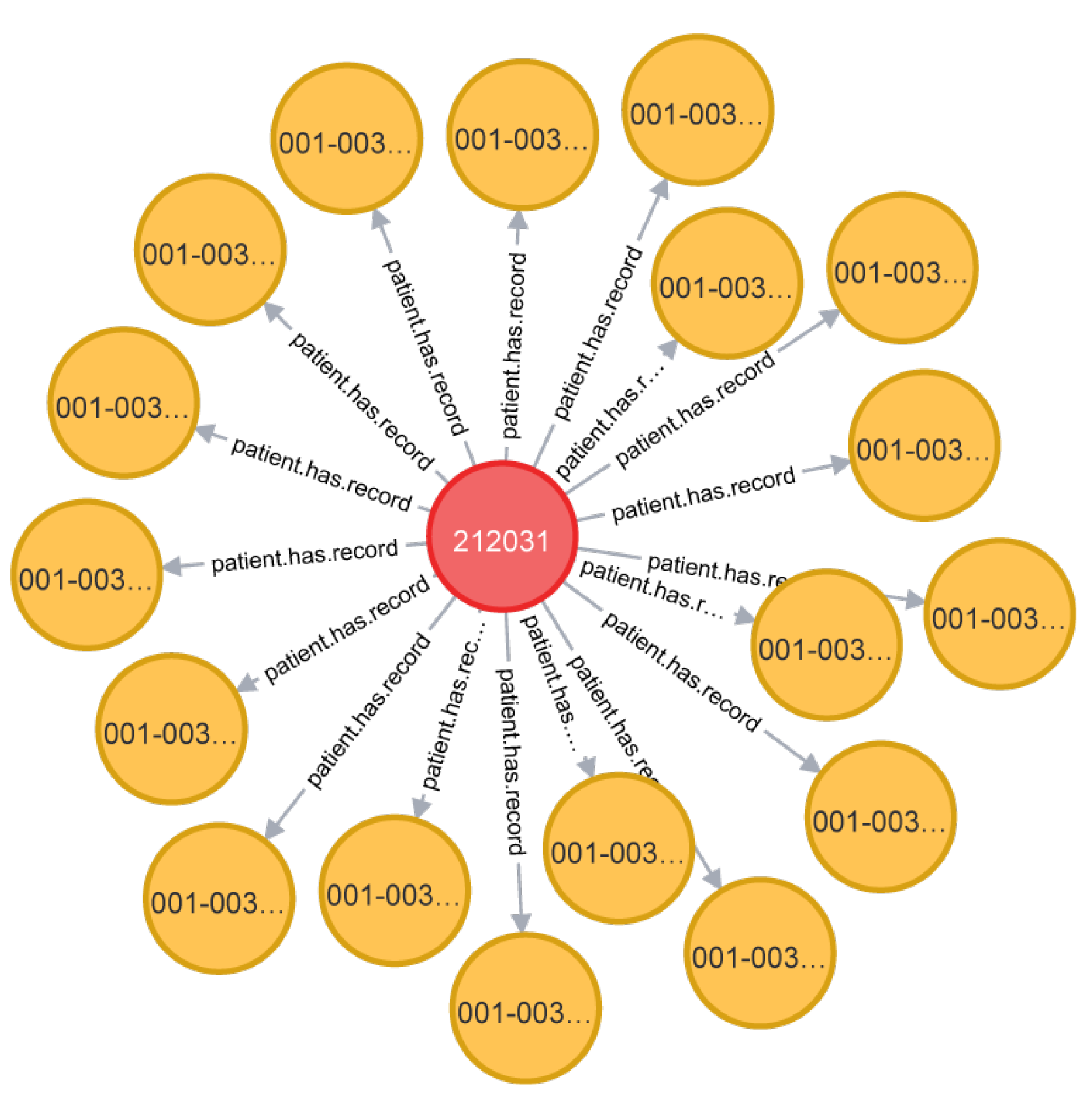
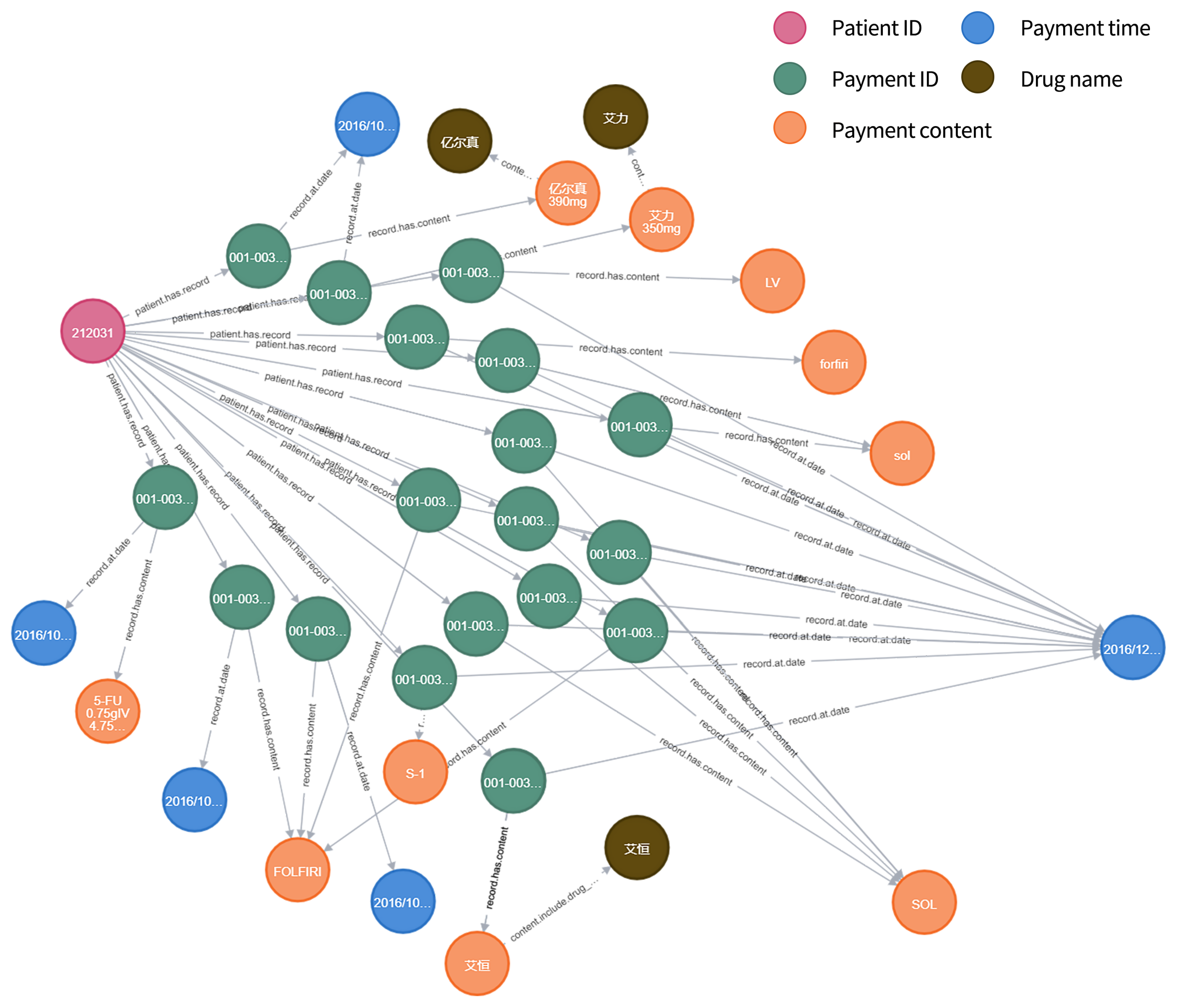
Take the relationship “patient has a record number” as an example. Due to the privacy of information, patients are identified by patient number in the data table. Stored in a graph database, a graph about the custom relationship can be obtained. As shown in Figure 4:
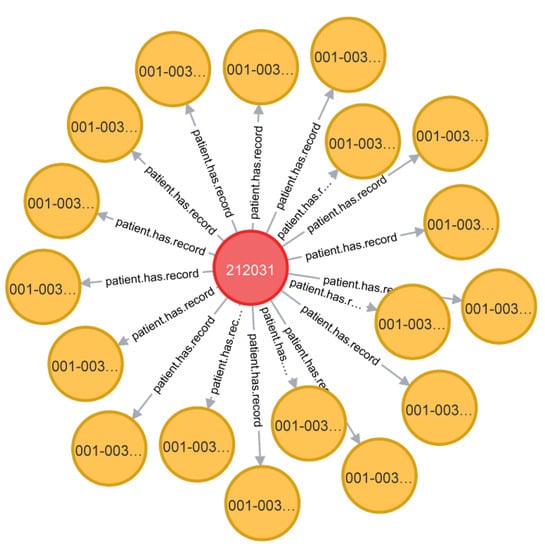
Figure 4.
Customized relationship diagram.
When all the nodes and custom relationships in the data table are stored, a knowledge graph about the chemotherapy schedule can be obtained, as shown in Figure 5.
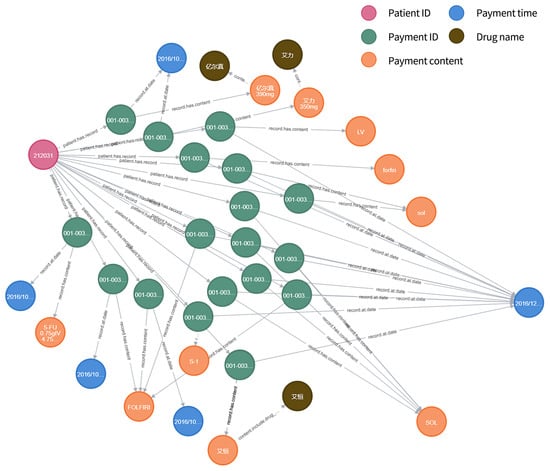
Figure 5.
Payment detail data sheet knowledge graph (partial), where “亿尔真” means “Calcium Levofolinate for Injection”, “艾力” means “Irinotecan Hydrochloride for Injection” and “艾恒” means “Oxaliplatin for Injection”.
To query a single object, such as patient number, drug regimen, and drug name, you only need to click the corresponding icon in the graph database or use the Cypher query language to query. The method of querying custom relationships is similar to the above method.
However, the purpose of building a knowledge graph is not just to query these single objects, but mainly to study the connection between two objects in the data table, that is, two nodes in the graph database that are associated with other nodes to achieve the purpose of convenient analysis. For example, we can obtain the treatment records of the patient through the patient number, and the drug regimen from the treatment records used in this treatment, and then we can find out the patients through the drug regimen who use the same drug regimen, which is conducive to the management of treatment. It is also convenient to explore the similarities and differences between patients using the same drug regimen, which also has a certain significance for medical research.
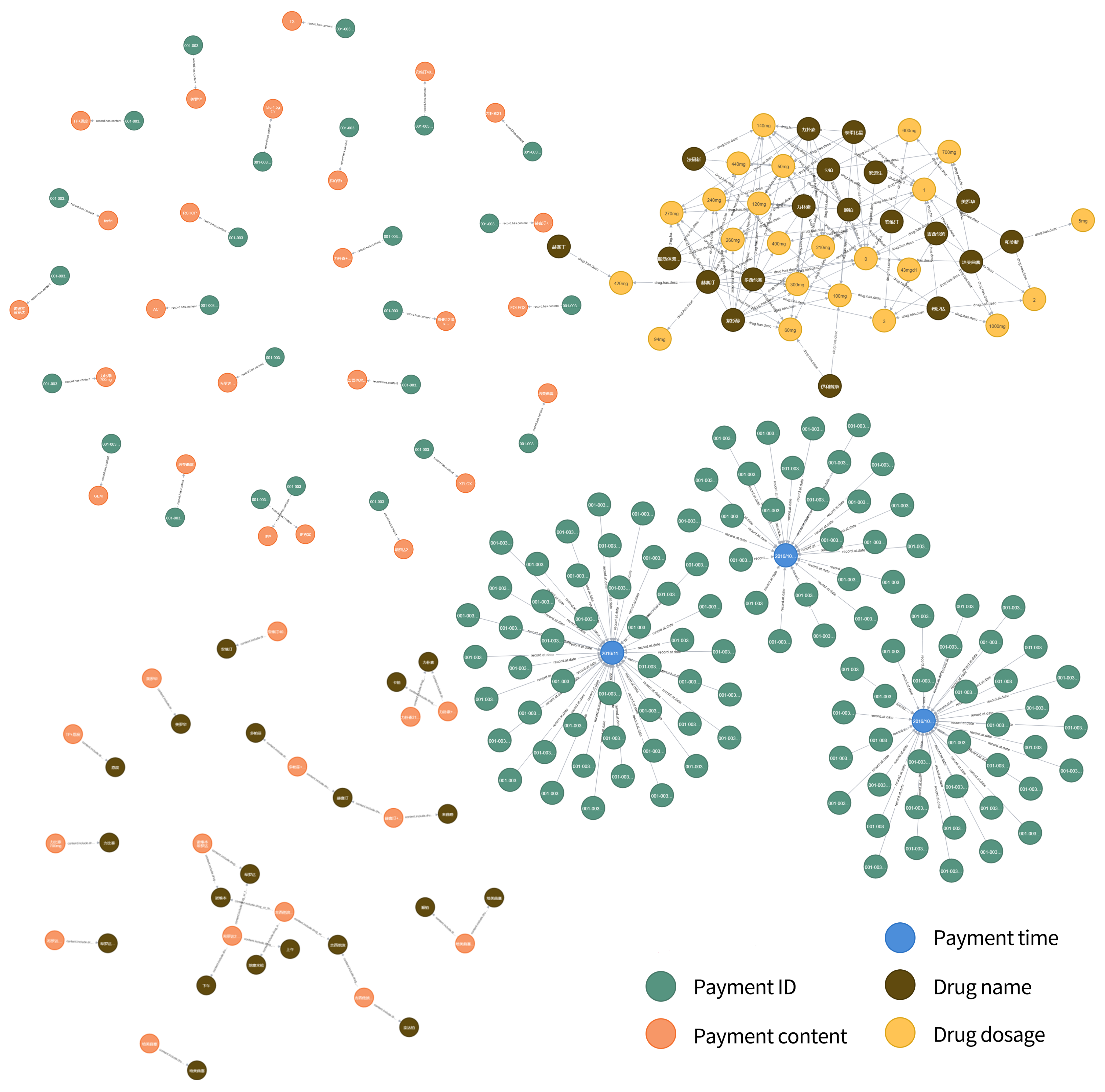
When all datasets (not just the payment order database mentioned earlier as an example) are all stored in a graph database, the resulting knowledge graph will be huge (like Figure 6), which is also of great significance for disease research. When patients undergo examinations in the hospital, the test situation is similar to that of some patients, so the knowledge graph has the function of disease prediction. When patients have confirmed their illness, the treatment plan of patients with similar conditions in previous examinations can be referred to for auxiliary diagnosis and treatment cost prediction during the examination.
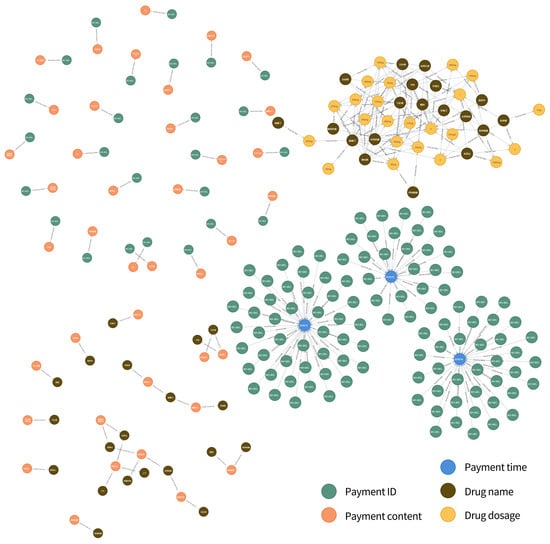
Figure 6.
Connection diagram of several relationships.
For example, in the basic search, when we want to obtain the patient’s basic information, such as gender, age, blood type, place of origin, etc., we need to use the Cypher query language to query the patient ID. From patient ID, we can find other entities linked to it. Entities correspond to multiple attributes, including gender, age, blood type, place of origin, etc. Similarly, we can also obtain all the record IDs of patients entered into the database, including multiple body part examination records, various index parameter test records, pathology report records, surgical records, hospitalization records, expense records, etc., We only need to know the type of relationship between the patient number and the object to be queried. For example, to find the expense records for a patient, we can find relations like:
This can be easily performed with the Cypher language. However, the purpose of building a knowledge graph is not only to query these objects, but the patient information data level is also diverse and complex. The graph database is more suitable for querying datasets with higher correlation, and we mainly study objects with multiple correlations or multi-level relationships between objects. For the database demanders, we mainly divide it into three parts: patients and their families, medical staff, and hospital managers.
For the first group, they may be more concerned with the cost of treatment and the length of treatment for the disease. For example, when they have an initial understanding of their condition, they can predict the cost and duration of treatment by referring to other anonymous patients of similar age and severity, so that they can roughly estimate the cost and duration of treatment they need. Then the database creator can set the “severity” attribute to the object of “symptom” when establishing the database, and this attribute can of course also contain multiple indicators, including the severity of the specific examination site, such as “nasopharynx is serious, nasal cavity is serious, oral cavity is normal” and other general attributes. Then when querying, we find relations like:
Through similar methods, patients can even try to check the cost and length of treatment for patients with other diseases that have the same aspects as themselves, because other diseases can also cause different degrees of damage to the patient’s body.
For the second group of people, data analysis is more important. The graph database can first compare the data of medical staff’s examination of the patient’s body parts and index parameter test with the database data to predict the type of disease. Taking nasopharyngeal carcinoma as an example:
Entity z found in this query is the treatment we want. In the process of treatment, the patient’s condition can also be judged according to the indicator parameter data of the disease patients at different stages recorded in the database, and reminding the doctor that the next stage of diagnosis and treatment or the patient may have some emergencies is also the direction of the database’s future efforts. Of course, the graph database can also obtain past drug use records according to treatment options, and explore whether different treatment regimens use the same drug and dosage, which is conducive to the management of treatment. This approach also facilitates the exploration of similarities and differences between patients who use the same drug regimen, which also has some implications for medical research.
For the last option, the graph data searches for surgical records, hospitalization records, drug regimens, and treatment costs of patients with the same disease and multiple body parts examined and with similar parameters. Hospital managers can reasonably allocate medical resources according to the search results, avoid the problem of an unbalanced distribution of medical resources, and also play a great role in the construction and development of hospitals.
5. Performance Analysis
The insertion and query performance of the graph database is compared with that of the traditional database. The extracted entity information is imported into graph database Neo4j and traditional database MySQL, respectively, on Intel(R) Core(TM) i7-10750H CPU @ 2.60 GHz, and the time taken is shown in the Table 3:

Table 3.
Insert performance comparison (Unit: second).
It is found that the overall insertion speed of the traditional database is 46% faster than that of the graph database, which is because the traditional database only inserts records when inserting information and does not establish relationships between entities, and does less processing of data; while the graph database requires graph construction when inserting information, connects data while inserting data, and constructs a knowledge graph.
Comparison of query performance. A query is performed for the drugs used by a patient. The time spent is shown in Table 4:

Table 4.
Query performance comparison (Unit: second).
The graph database is 116.7% faster than the traditional database, which is due to the fact that various types of data in the graph database are stored in the same graph and can find the path quickly with the help of its own connection relationship, while the traditional database needs to connect multiple tables in each query and make correlation queries, which greatly slows down the query time.
In summary, although the insertion speed of the traditional database is faster than the graph database, the query data are much slower than the graph database. In practical applications, the insertion operation to the database is usually discrete and only inserted once when a new record is available for the patient, and the perception of the insertion time is not obvious, while the query operation is continuous and often requires batch export of patient data for analysis, which is more sensitive to export time. Therefore, graph databases are superior to traditional databases in the process of analyzing medical data.
6. Discussion
In this paper, we propose an entity extraction method based on natural language processing and disambiguation reorganization with the help of the medical record database of a cooperative medical institution, in order to solve the problem of excessive training overhead of existing neural network models. First, the sentences are subjected to natural language processing and disambiguation, followed by merging and categorizing the words into three categories: drug names, dosages, and connectives based on their contextual information. A knowledge graph is constructed by extracting structured data such as drug and dose. Meanwhile, this paper also proposes a method for disease prediction and diagnosis using a knowledge graph, and gives some specific examples.
The approach presented in this paper has the following implications for the management of medical records, the practice, the academy, the company, and the economy:
- Reducing neural network model training overhead: traditional neural network models require a large amount of labeled data and computational resources in the training phase, which increases the difficulty and cost of training. The method proposed in this paper avoids the dependence on large-scale labeled data, alleviates the training overhead, and improves efficiency by utilizing the medical record database of cooperative medical institutions for entity extraction.
- Constructing knowledge graphs to assist medical decision-making: constructing knowledge graphs by extracting structured data such as drugs and dosages from medical records can provide more comprehensive information support for medical practice. Knowledge graphs are able to model the relationships between different entities and provide the functionality to assist medical decision-making through reasoning and querying. Doctors can utilize knowledge graphs for disease prediction and diagnosis, thereby improving medical outcomes and accuracy.
- Enhancement of medical record management and information retrieval: the method in this paper can transform textual information in medical records into structured data and establish connections between entities through entity extraction and knowledge graph construction. This enables the medical record management system to search for and retrieve the text more flexibly, improves the efficiency of organizing and utilizing the medical record information, and provides more convenient and accurate support for medical work.
- This study can strengthen the connection between the academy, the company, and the hospital. The hospital provides data and real cases and realistic scene construction to make the research more real and reliable, after the company and the academy jointly research and develop the needs, the academy opens up new ideas and provides theoretical support, and then the company puts it into production, turns the theory into reality, and finally puts the product into the hospital, and the hospital gives feedback to improve the research.
- The construction of the knowledge graph based on the graph database can also give the company certain inspiration, lead to data penetrating into all aspects of life, increase the research on data, and innovate the research methods of data, whether it is the development of enterprise transformation, optimizing the enterprise structure to promote economic development, research or innovation, and promoting the development of science and technology [31]. The proposal of new research can also provide jobs, ease the employment pressure of the country or region, and at the same time, the comprehensive development of new research from its inception to its implementation canot only control the growth of high medical costs, but also avoid or alleviate disease. It has important social value and scientific significance and can create value and stimulate domestic demand.
This research paper has some limitations and directions that can be followed up in the future. For example, compared to the neural network model, although the entity extraction method proposed in this paper can alleviate the computational overhead, there may still be some challenges in processing long text and complex contexts. Future research can further optimize the performance of the entity extraction model and improve the understanding and processing of complex contexts to enhance the stability and reliability of the method. Modern generative models such as BERT and GPT can partly address problems such as generating keywords or a brief summary from text. However, this approach incurs a greater performance overhead than ever before [32].
The method proposed in this paper has an impact on medical record management and medical practice, which can alleviate the training overhead of neural network models, construct knowledge graphs to assist medical decision-making, and enhance medical record management and information retrieval. All of these contribute to the improvement of medical efficiency, accuracy, and quality, and have positive implications for promoting the development of the medical field and improving the patient experience. For patients, a user-friendly query interface is desirable.
7. Conclusions
In this paper, we propose a specialized entity extraction model for Chinese medical records based on natural language processing and word segmentation and reorganization, and use the extracted entities to construct a knowledge graph of medical data, with examples of the application of the knowledge graph in the process of disease diagnosis and treatment. The results are as follows: on the same hardware, the knowledge graph model built in this paper is 116.7% faster than traditional databases in processing multi-hop complex relationships. Knowledge graphs can assist medical decision-making, improve the efficiency of medical record management and information retrieval, and have a positive impact on healthcare and patient experience improvement. However, the method still faces the challenge of dealing with more complex contexts and long texts, and a user-friendly interface is also needed for non-specialists, which requires further optimization and research.
Author Contributions
Conceptualization, J.W. and Y.J.; Data curation, J.W., Z.Q. and Q.L.; Formal analysis, Y.H.; Funding acquisition, Y.J.; Investigation, Y.J.; Methodology, J.W.; Project administration, J.W. and Y.J.; Resources, Y.J.; Software, J.W. and Z.Q.; Supervision, Y.J.; Validation, Y.H., Q.L. and J.Y.; Visualization, Z.Q.; Writing—original draft, J.W., Z.Q., Y.H., Q.L. and J.Y.; Writing—review & editing, J.W. and Y.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Ministry of Education, Science and Technology Development Center grant number 2018A03031.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available because the research data are confidential.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhaoxia, L.; Yong, Y.; Yin, X. Big Data in Health Care: Theory and Practice; Beijing Book Company Co., Inc.: Beijing, China, 2017. [Google Scholar]
- Yang, J.; Guan, Y.; He, B.; Qu, C.; Yu, Q.; Liu, Y.; Zhao, Y. Corpus construction for named entities and entity relations on Chinese electronic medical records. J. Softw. 2016, 27, 2725–2746. [Google Scholar]
- Chang, H.; Zan, H.; Zhang, S.; Zhao, B.; Zhang, K. Corpus Construction for Named-Entity and Entity Relations for Electronic Medical Records of Cardiovascular Disease. In Proceedings of the China Health Information Processing Conference, Hangzhou, China, 21–23 October 2022; Tang, B., Chen, Q., Lin, H., Wu, F., Liu, L., Hao, T., Wang, Y., Wang, H., Eds.; Springer: Singapore, 2023; pp. 3–18. [Google Scholar]
- Pletscher-Frankild, S.; Pallejà, A.; Tsafou, K.; Binder, J.X.; Jensen, L.J. DISEASES: Text mining and data integration of disease–gene associations. Methods 2015, 74, 83–89. [Google Scholar] [CrossRef]
- Hripcsak, G.; Albers, D.J. Next-generation phenotyping of electronic health records. J. Am. Med. Inform. Assoc. 2012, 20, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Li, D.; Lin, Y.; Wu, S.; Huang, Z. Named Entity Recognition of Chinese Crop Diseases and Pests Based on RoBERTa-wwm with Adversarial Training. Agronomy 2023, 13, 941. [Google Scholar] [CrossRef]
- Takeuchi, K.; Collier, N. Bio-medical entity extraction using support vector machines. Artif. Intell. Med. 2005, 33, 125–137. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, Y.; Wang, P.; Gu, L. Medical Named Entity Extraction from Chinese Resident Admit Notes Using Character and Word Attention-Enhanced Neural Network. Int. J. Environ. Res. Public Health 2020, 17, 1614. [Google Scholar] [CrossRef]
- Hu, C.; Zhang, S.; Gu, T.; Yan, Z.; Jiang, J. Multi-Task Joint Learning Model for Chinese Word Segmentation and Syndrome Differentiation in Traditional Chinese Medicine. Int. J. Environ. Res. Public Health 2022, 19, 5601. [Google Scholar] [CrossRef]
- Qu, J. A Review on the Application of Knowledge Graph Technology in the Medical Field. Sci. Program. 2022, 2022, 3212370. [Google Scholar] [CrossRef]
- Jiang, J.; Wang, H.; Xie, J.; Guo, X.; Guan, Y.; Yu, Q. Medical knowledge embedding based on recursive neural network for multi-disease diagnosis. Artif. Intell. Med. 2020, 103, 101772. [Google Scholar] [CrossRef] [PubMed]
- Chai, X. Diagnosis Method of Thyroid Disease Combining Knowledge Graph and Deep Learning. IEEE Access 2020, 8, 149787–149795. [Google Scholar] [CrossRef]
- Hu, J.; Lepore, R.; Dobson, R.J.B.; Al-Chalabi, A.; Bean, D.M.; Iacoangeli, A. DGLinker: Flexible knowledge-graph prediction of disease–gene associations. Nucleic Acids Res. 2021, 49, W153–W161. [Google Scholar] [CrossRef]
- Lei, Z.; Sun, Y.; Nanehkaran, Y.; Yang, S.; Islam, M.S.; Lei, H.; Zhang, D. A novel data-driven robust framework based on machine learning and knowledge graph for disease classification. Future Gener. Comput. Syst. 2020, 102, 534–548. [Google Scholar] [CrossRef]
- Li, Z.Q.; Fu, Z.X.; Li, W.J.; Fan, H.; Li, S.N.; Wang, X.M.; Zhou, P. Prediction of Diabetic Macular Edema Using Knowledge Graph. Diagnostics 2023, 13, 1858. [Google Scholar] [CrossRef]
- Li, L.; Wang, P.; Yan, J.; Wang, Y.; Li, S.; Jiang, J.; Sun, Z.; Tang, B.; Chang, T.H.; Wang, S.; et al. Real-world data medical knowledge graph: Construction and applications. Artif. Intell. Med. 2020, 103, 101817. [Google Scholar] [CrossRef]
- Iqbal, M.J.; Javed, Z.; Sadia, H.; Qureshi, I.A.; Irshad, A.; Ahmed, R.; Malik, K.; Raza, S.; Abbas, A.; Pezzani, R.; et al. Clinical applications of artificial intelligence and machine learning in cancer diagnosis: Looking into the future. Cancer Cell Int. 2021, 21, 270. [Google Scholar] [CrossRef] [PubMed]
- Zaev, R.I.; Romanov, A.Y.; Solovyev, R.A. Segmentation of Prostate Cancer on TRUS Images Using ML. In Proceedings of the 2023 International Russian Smart Industry Conference (SmartIndustryCon), Sochi, Russia, 27–31 March 2023; pp. 460–465. [Google Scholar] [CrossRef]
- Wozniak, J.M.; Jain, R.; Balaprakash, P.; Ozik, J.; Collier, N.T.; Bauer, J.; Xia, F.; Brettin, T.; Stevens, R.; Mohd-Yusof, J.; et al. CANDLE/Supervisor: A workflow framework for machine learning applied to cancer research. BMC Bioinform. 2018, 19, 59–69. [Google Scholar] [CrossRef]
- Jiang, C.; Yang, H.; Di, P.; Li, W.; Tang, Y.; Liu, G. In silico prediction of chemical reproductive toxicity using machine learning. J. Appl. Toxicol. 2019, 39, 844–854. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Lorincz, K.; Hughes, R.; Huggins, N.; Growdon, J.; Standaert, D.; Akay, M.; Dy, J.; Welsh, M.; Bonato, P. Monitoring motor fluctuations in patients with Parkinson’s disease using wearable sensors. IEEE Trans. Inf. Technol. Biomed. 2009, 13, 864–873. [Google Scholar] [CrossRef]
- Sarker, I.H. Deep Learning: A Comprehensive Overview on Techniques, Taxonomy, Applications and Research Directions. SN Comput. Sci. 2021, 2, 420. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, G.; Fang, Y. Exploring the use of information technology in hospital case management work. Electron. Commun. Comput. Sci. 2022, 4, 160–162. [Google Scholar]
- Murali, L.; Gopakumar, G.; Viswanathan, D.M.; Nedungadi, P. Towards electronic health record-based medical knowledge graph construction, completion, and applications: A literature study. J. Biomed. Inform. 2023, 143, 104403. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, B.; Zhao, Y.; Mao, S.; Tang, Y. A knowledge graph method for hazardous chemical management: Ontology design and entity identification. Neurocomputing 2021, 430, 104–111. [Google Scholar] [CrossRef]
- Tan, J.; Qiu, Q.; Guo, W.; Li, T. Research on the Construction of a Knowledge Graph and Knowledge Reasoning Model in the Field of Urban Traffic. Sustainability 2021, 13, 3191. [Google Scholar] [CrossRef]
- Cheng, B.; Zhang, J.; Liu, H.; Cai, M.; Wang, Y. Research on Medical Knowledge Graph for Stroke. J. Healthc. Eng. 2021, 2021, 5531327. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.Y.; Ying, L. Decision tree methods: Applications for classification and prediction. Shanghai Arch. Psychiatry 2015, 27, 130. [Google Scholar]
- SogouIME. Chinese and Foreign Drug Names [Official Recommendation]_Sogou Input Method Thesaurus. 2010. Available online: https://pinyin.sogou.com/dict/detail/index/20666?rf=dictindex (accessed on 27 May 2022).
- Webber, J. A Programmatic Introduction to Neo4j. In Proceedings of the SPLASH ’12: 3rd Annual Conference on Systems, Programming, and Applications: Software for Humanity, Tucson, AZ, USA, 19–26 October 2012; Association for Computing Machinery: New York, NY, USA, 2012; pp. 217–218. [Google Scholar] [CrossRef]
- Xie, Z.d.; Wu, J.c.; Li, Z.m.; Wu, G.h. A Study of Construction and Cultivation of Big Data Capacity of Enterprise. J. Guangdong Univ. Technol. 2017, 34, 110. [Google Scholar] [CrossRef]
- Tang, R.; Lu, Y.; Liu, L.; Mou, L.; Vechtomova, O.; Lin, J. Distilling Task-Specific Knowledge from BERT into Simple Neural Networks. arXiv 2019, arXiv:1903.12136. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).