1. Introduction
Skin disease is a severe global public health problem that affects a large number of people [
1]. The symptoms of skin diseases are diverse, and the changing of the symptoms is a long-term process. It is difficult for ordinary people to determine the type of skin disease with the naked eye, and most people often neglect the changes in their skin symptoms, which can lead to severe consequences such as permanent skin damage and even the risk of skin cancer [
2]. In addition, the early treatment of skin cancer can decrease morbidity and mortality [
3].
In addition, due to the rapid development of deep learning technology, it has rapidly become the preferred method for medical image analysis [
4,
5]. In addition, compared with traditional classification methods, deep learning has a stronger robustness and a better generalization ability [
6]. In the meantime, convolutional neural networks are one of the most well-known and representative deep learning models [
7,
8]. It has been widely used in many aspects of medical image analysis [
9,
10], and great progress has been made in medical image classification. For example, Datta. et al. [
11] combined soft-attention and Inception ResNet-V2 [
12] (IRv2) to construct an IRV2-SA model for dermoscopic image classification. This combination improved the sensitivity score compared to the baseline model, reaching 91.6% on the ISIC2017 [
13] dataset. Apart from that, its accuracy on the HAM10000 [
14] dataset was 93.7%, which was 4.7% higher than the baseline model. Lan. et al. [
15] proposed a capsule network method called FixCaps. It is an improved convolutional neural network model based on CapsNets [
16] with a larger receptive domain. It works by applying a high-performance large kernel with a kernel size of up to 31
31 at the bottom convolutional layer. At the same time, an attention mechanism was introduced to reduce the loss of spatial information caused by convolution and pooling, and it achieved an accuracy of 96.49% and an f1-score of 86.36% on the HAM10000 dataset.
The IRV2-SA model and FixCaps model perform well in terms of classification accuracy. However, they are not impeccable in terms of other classification performance evaluation criteria, and the classification performance is not satisfactory in classifications with a restricted individual sample data. Enhancing their classification accuracy is problematic because of the restricted available image data of skin diseases and the extreme imbalance of lesion samples. In addition, the categories of skin diseases are elaborate, and the symptoms are very analogous in the early stages, which causes the model classification to be more problematic. At the same time, the generalization ability of a single reliable network model qualified with restricted data is weak, and the feature extraction ability is insufficient. Attaining a high classification accuracy is still challenging. The common research strategy to solve the problem of small data samples and class imbalance is data augmentation or enhancing the feature extraction ability of the model.
All in all, the main contributions of this paper can be summarized in the following points:
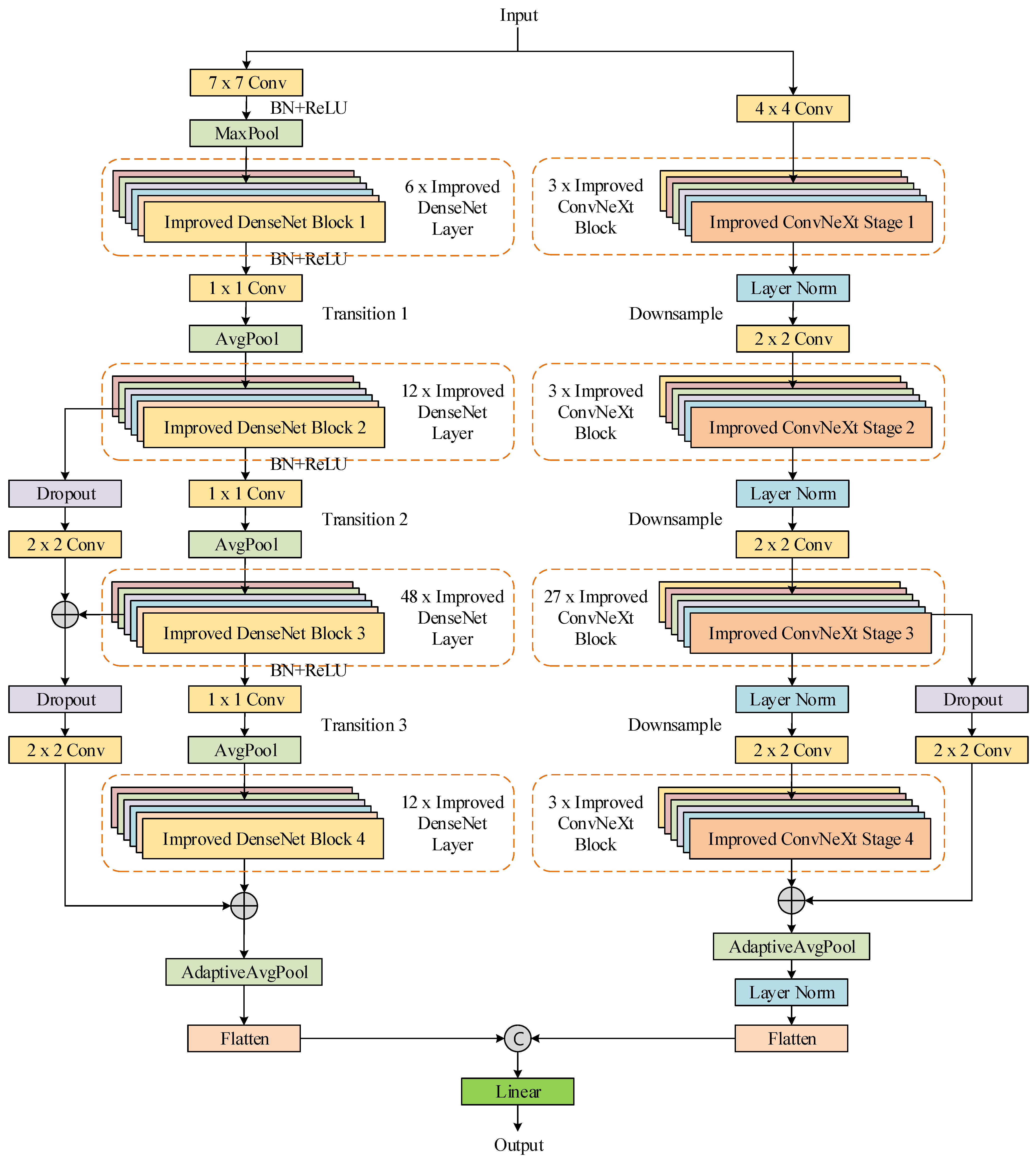
In this work, a convolutional neural network (CNN) model based on model fusion was proposed for skin disease classification. DenseNet201 [
17] and ConvNeXt_L [
18] were selected as the backbone sub-classification models for the model fusion.
To enhance the feature extraction ability of the proposed network model, the Efficient Channel Attention [
19] module and the Gated Channel Transformation [
20] attention module were introduced into the core blocks of DenseNet201 and ConvNeXt_L, respectively.
A parallel strategy was applied to fuse the features of the deep and shallow layers to further enhance the feature-extraction ability of the model.
The classification performance of the model was improved through a series of works such as model pre-training, data augmentation, and parameter fine-tuning.
Extensive experiments were conducted to compare the proposed model with the basic CNN models commonly used in recent years to ensure the validity of this work. The experiments were carried out by the proposed network model on a private dataset dominated by acne-like skin diseases, and training and testing were conducted on the public HAM10000 [
14] (Human-Against-Machine with 10000 training images) dataset with an extreme imbalance in skin diseases, and the proposed model was compared with other state-of-the-art models on the HAM10000 dataset. This verified the generalization capacity and the accuracy of the proposed network model.
2. Related Work
CNN models have been widely explored for skin disease classification, and some of these models have achieved very good classification performances. Below, we summarized the relevant published work of some researchers in the field of skin disease image classification.
Many researchers have proposed reliable multi-class CNN models. Mobiny et al. [
21] proposed an approximate risk-aware deep Bayesian model named Bayesian DenseNet-169, which outputs an estimate of model uncertainty without additional parameters or significant changes to the network architecture. It increased the classification accuracy of the base DenseNet169 [
17] model from 81.35% to 83.59% on the HAM10000 dataset. Wang et al. [
22] propose an interpretability-based CNN model. It is a multi-class classification model that takes skin lesion images and patient metadata as the input for skin lesion diagnosis. It achieved a 95.1% and 83.5% accuracy and sensitivity, respectively, on the HAM10000 dataset. Allugunti et al. [
23] created a multi-class CNN model for diagnosing skin cancer. The proposed model makes a distinction between lesion maligna, superficial spreading, and nodular melanoma. This permits the early diagnosis of the virus and the quick isolation and therapy necessary to stop the further transmission of infection. Anand et al. [
24] modified the Xception [
25] model by adding layers such as a pooling layer, two dense layers, and a dropout layer. A new fully connected (FC) layer changed the original FC layer with seven skin disease classes. It had a classification accuracy of 96.40% on the HAM10000 dataset.
Improving the classification accuracy of the model by using ensemble learning is also an effective method. Thurnhofer-Hemsi et al. [
26] proposed an ensemble composed of improved CNNs combined with a regularly spaced test-time-shifting technique for skin lesion classification. It builds up multiple test input images via a shift technique and passes it to each classifier passed to the ensemble and then combines all the outputs for classification. It had a classification accuracy of 83.6% on the HAM10000 dataset.
Through the introduction of an attention module, the feature extraction ability of a model can be enhanced, thereby improving the classification performance of the model. Karthik et al. [
27] replaced the standard Squeeze-and-Excite [
28] block in the EfficientNetV2 [
29] model with an Efficient Channel Attention [
19] block, and the total number of training parameters dropped significantly. The test accuracy of the model reached 84.70% in four types of skin disease datasets including acne, actinic keratosis, melanoma and psoriasis.
Through image processing techniques such as image conversion, equalization, enhancement and segmentation, the accuracy of image classification can be enhanced. Abayomi-Alli et al. [
30] propose an improved data augmentation model for the effective detection of melanoma skin cancer. The method was based on oversampling data embedded in a nonlinear low-dimensional manifold to create synthetic melanoma images. It achieved a 92.18%, 80.77%, 95.1% and 80.84% accuracy, sensitivity, specificity and f1-score, respectively, on the PH2 [
31] dataset. Hoang et al. [
32] proposed a novel method using a new segmentation approach and wide-ShuffleNet for skin lesion classification. It first separates the lesion from the background by computing an entropy-based weighted sum first-order cumulative moment (EW-FCM) of the skin image. The segmentation results are then input into a new deep learning structure, wide-ShuffleNet, and classified. It achieved a 96.03%, 70.71%, 75.15%, 72.61% and 84.80% specificity, sensitivity, precision, f1-score and accuracy, respectively, on the HAM10000 dataset. Malibari et al. [
33] proposed an Optimal Deep-Neural-Network-Driven Computer-Aided Diagnosis Model for their skin cancer detection and classification model. The model primarily applies a Wiener-filtering-based pre-processing step followed by a U-Net segmentation approach. The model achieved a maximum accuracy of 99.90%. Nawaz et al. [
34] proposed an improved Deep-Learning-based method, namely, the DenseNet77-based UNET model. Their experiments demonstrated the robustness of the model and its ability to accurately identify skin lesions of different colors and sizes. It obtained a 99.21% and 99.51% accuracy on the ISIC2017 [
13] and ISIC2018 [
35] datasets, respectively.
Therefore, by summarizing the related work published by these researchers in the field of skin disease image classification, we proposed a CNN model for skin disease classification based on model fusion. In addition, through a series of work such as model fusion, deep and shallow feature fusion, the introduction of an attention module, model pre-training, data augmentation and parameter fine-tuning, the classification performance of the proposed model was enhanced.
6. Conclusions
In this paper, we proposed a convolutional neural network model for skin disease classification based on model fusion. We chose DenseNet201 and ConvNeXt_L as the backbone sub-classification models of our model fusion. In addition, on the core block of each sub-classification model, an attention module was introduced to assist the network in acquiring a region of interest in order to enhance the ability of the network model to extract image features. In addition, the features extracted by the shallow network could capture more details, and the features extracted by the deep network contained more abstract semantic information. Combining the characteristics of the two, a parallel strategy was adopted to fuse the features of the deep and shallow layers. Finally, through a series of works such as model pre-training, data augmentation and parameter fine-tuning, the classification performance of the proposed model was further improved.
On the private dataset, the proposed model achieved an accuracy of 96.49%, which was 4.42% and 3.66% higher than the two baseline models, respectively. On the public dataset, HAM10000, the accuracy and f1-scores of the proposed model were 95.29% and 89.99%, respectively, which also achieved good results compared to the other state-of-the-art models. It was demonstrated that the proposed model possessed a good classification performance on the datasets with an extreme imbalance or a small number of samples as well as a good generalization ability.