BioEdge: Accelerating Object Detection in Bioimages with Edge-Based Distributed Inference
Abstract
:1. Introduction
- We present BioEdge, a distributed inference framework for the bioimage object detection task. This framework can improve the throughput of the detection process in small hospitals or biomedical laboratories that lack GPU resources. It leverages the collaboration between local devices and edge servers while addressing privacy concerns.
- We employ a computationally lightweight autoencoder to reduce the communication overhead when transferring data between the local device and the edge server. This autoencoder is incorporated at the distribution layer and demonstrates a negligible drop in accuracy.
- We utilize the Scaled-YOLOv4 and YOLOv7 algorithms for object detection, supporting biomedical applications that demand high accuracy. For the evaluation, we utilize the high-precision COVID-19 dataset provided by the Society for Imaging Informatics in Medicine (SIIM).
2. Related Work
2.1. CNN Architecture and Models
2.2. Biomedical Image Preprocessing and Augmentation
2.3. Distributed Inference
3. BioEdge
3.1. Dataset Preparation
3.2. Scaled-YOLOv4
3.3. YOLOv7
3.4. Overall Design of BioEdge
3.5. Autoencoder
- A convolutional (CONV) layer performs a convolution operation on the input data. The convolution operation involves sliding a small filter over the input data and computing the dot product between the filter and the input at each position. The result of the convolution operation is a feature map that captures the presence of certain features in the input data. In the encoder, the convolutional layers are used to extract features from the input data and create a compressed representation of the data.
- Batch normalization involves normalizing the inputs to a layer by subtracting the mean and dividing by the standard deviation. This helps to reduce the internal covariate shift, which is a phenomenon where the distribution of the inputs to a layer changes during training. By reducing the internal covariate shift, batch normalization helps to stabilize the training process and improve the performance of the network. In the encoder, the batch normalization layer is used to normalize the inputs to the convolutional layers and improve the training process.
- A deconvolutional (DECONV) layer, also known as a transposed convolutional layer, performs an inverse convolution operation on the input data. The deconvolution operation involves sliding a small filter over the input data and computing the dot product between the filter and the input at each position. The result of the deconvolution operation is a feature map that captures the presence of certain features in the input data. In the decoder, the deconvolutional layers are used to reconstruct the original data from the compressed representation created by the encoder.
- The Sigmoid function is an activation function that maps any input value to a value between 0 and 1. In the encoder, the Sigmoid function is used in the last layer of the encoder. Because the output values are constrained to the range [0, 1] in the Sigmoid function, the values can be scaled to satisfy the transmitter output power constraint. Namely, the output values can be adjusted to fit within a certain power budget, which is important in resource-constrained mobile devices where power consumption is a critical factor. It also benefits the quantization of data in the digital communication system. Quantization is the process of mapping a continuous range of values to a finite set of discrete values, which is necessary for transmitting data over the network. By constraining the output values to [0, 1], the quantization process can be simplified and made more efficient, further reducing communication overhead and improving the overall performance of the system.
- The ReLU (Rectified Linear Unit) function is an activation function that maps any input value less than 0 to 0, and any input value greater than 0 to the same value. In the decoder, the ReLU function is used in the first layer of the decoder. The ReLU function is well-suited for tasks that involve detecting and extracting features from data. The ReLU function helps to activate the relevant features in the data and suppress the irrelevant features.
4. Results
4.1. Experimental Environment
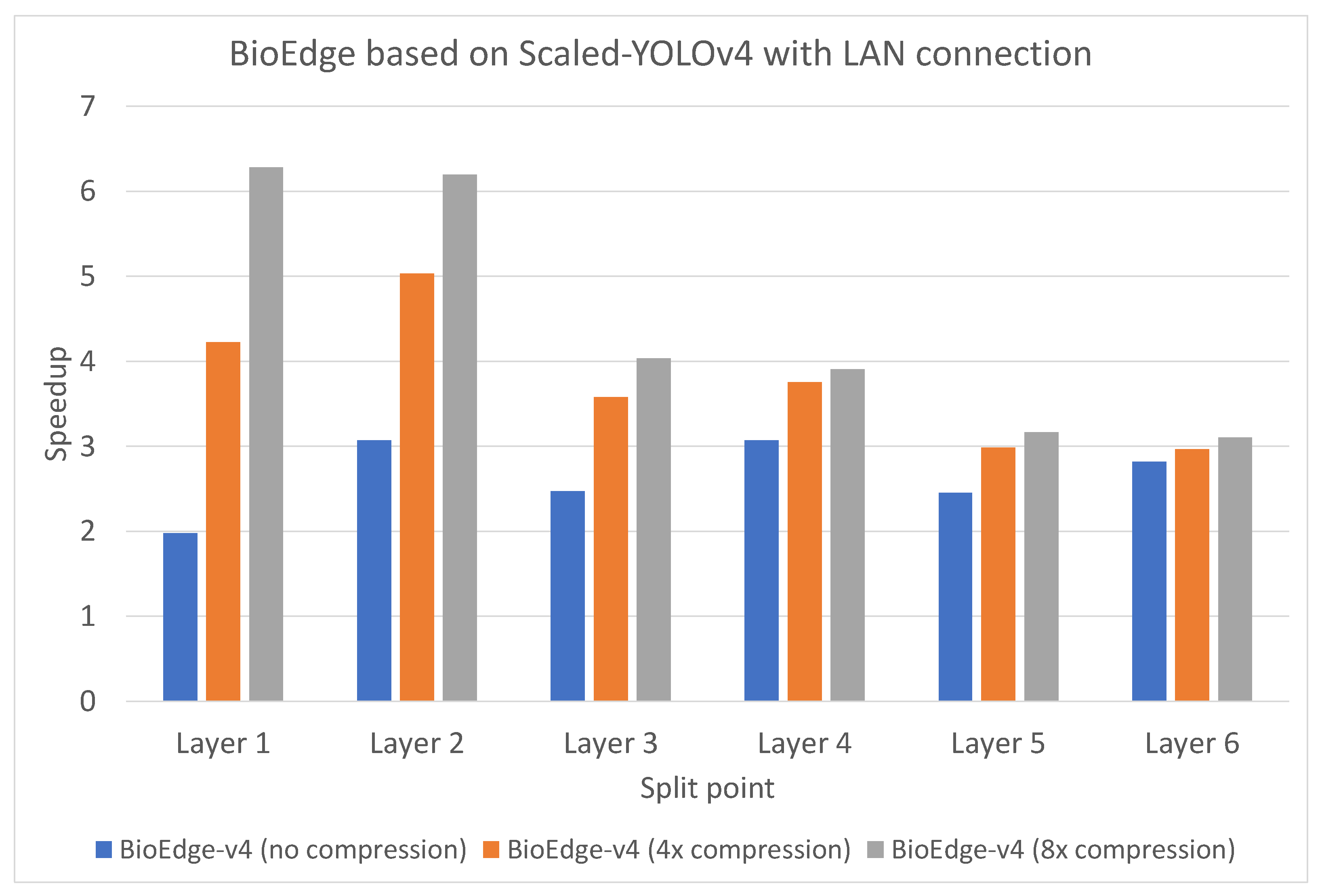
4.2. Inference Latency
4.3. Inference Accuracy
4.4. Memory Consumption
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Min, S.; Lee, B.; Yoon, S. Deep learning in bioinformatics. Brief. Bioinform. 2017, 18, 851–869. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Roa, A.; Gilmore, H.; Basavanhally, A.; Feldman, M.; Ganesan, S.; Shih, N.N.; Tomaszewski, J.; González, F.A.; Madabhushi, A. Accurate and reproducible invasive breast cancer detection in whole-slide images: A deep learning approach for quantifying tumor extent. Sci. Rep. 2017, 7, 46450. [Google Scholar] [CrossRef]
- Kim, Y.; Yi, S.; Ahn, H.; Hong, C.H. Accurate Crack Detection Based on Distributed Deep Learning for IoT Environment. Sensors 2023, 23, 858. [Google Scholar] [CrossRef] [PubMed]
- Robinson, T.; Harkin, J.; Shukla, P. Hardware acceleration of genomics data analysis: Challenges and opportunities. Bioinformatics 2021, 37, 1785–1795. [Google Scholar] [CrossRef]
- Hong, C.H.; Varghese, B. Resource management in fog/edge computing: A survey on architectures, infrastructure, and algorithms. ACM Comput. Surv. (CSUR) 2019, 52, 1–37. [Google Scholar] [CrossRef]
- Wang, C.Y.; Bochkovskiy, A.; Liao, H.Y.M. Scaled-yolov4: Scaling cross stage partial network. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Virtual. 19–25 June 2021; pp. 13029–13038. [Google Scholar]
- Wang, C.Y.; Bochkovskiy, A.; Liao, H.Y.M. YOLOv7: Trainable bag-of-freebies sets new state-of-the-art for real-time object detectors. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 7464–7475. [Google Scholar]
- Ha, Y.J.; Yoo, M.; Lee, G.; Jung, S.; Choi, S.W.; Kim, J.; Yoo, S. Spatio-Temporal Split Learning for Privacy-Preserving Medical Platforms: Case Studies With COVID-19 CT, X-ray, and Cholesterol Data. IEEE Access 2021, 9, 121046–121059. [Google Scholar] [CrossRef]
- Lakhani, P.; Mongan, J.; Singhal, C.; Zhou, Q.; Andriole, K.P.; Auffermann, W.F.; Prasanna, P.; Pham, T.; Peterson, M.; Bergquist, P.J.; et al. The 2021 SIIM-FISABIO-RSNA Machine Learning COVID-19 Challenge: Annotation and Standard Exam Classification of COVID-19 Chest Radiographs. J. Digit. Imaging 2023, 36, 365–372. [Google Scholar] [CrossRef]
- Abumalloh, R.A.; Nilashi, M.; Ismail, M.Y.; Alhargan, A.; Alghamdi, A.; Alzahrani, A.O.; Saraireh, L.; Osman, R.; Asadi, S. Medical image processing and COVID-19: A literature review and bibliometric analysis. J. Infect. Public Health 2022, 15, 75–93. [Google Scholar] [CrossRef]
- Oh, Y.; Park, S.; Ye, J.C. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imaging 2020, 39, 2688–2700. [Google Scholar] [CrossRef]
- Toğaçar, M.; Ergen, B.; Cömert, Z. COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput. Biol. Med. 2020, 121, 103805. [Google Scholar] [CrossRef] [PubMed]
- Akkus, Z.; Galimzianova, A.; Hoogi, A.; Rubin, D.L.; Erickson, B.J. Deep learning for brain MRI segmentation: State of the art and future directions. J. Digit. Imaging 2017, 30, 449–459. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Boser, B.; Denker, J.S.; Henderson, D.; Howard, R.E.; Hubbard, W.; Jackel, L.D. Backpropagation applied to handwritten zip code recognition. Neural Comput. 1989, 1, 541–551. [Google Scholar] [CrossRef]
- Yap, M.H.; Pons, G.; Marti, J.; Ganau, S.; Sentis, M.; Zwiggelaar, R.; Davison, A.K.; Marti, R. Automated breast ultrasound lesions detection using convolutional neural networks. IEEE J. Biomed. Health Inform. 2017, 22, 1218–1226. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1–9. [Google Scholar] [CrossRef]
- Elaziz, M.A.; Hosny, K.M.; Salah, A.; Darwish, M.M.; Lu, S.; Sahlol, A.T. New machine learning method for image-based diagnosis of COVID-19. PLoS ONE 2020, 15, e0235187. [Google Scholar] [CrossRef]
- Karthik, R.; Menaka, R.; Hariharan, M. Learning distinctive filters for COVID-19 detection from chest X-ray using shuffled residual CNN. Appl. Soft Comput. 2021, 99, 106744. [Google Scholar] [CrossRef] [PubMed]
- Heidari, M.; Mirniaharikandehei, S.; Khuzani, A.Z.; Danala, G.; Qiu, Y.; Zheng, B. Improving the performance of CNN to predict the likelihood of COVID-19 using chest X-ray images with preprocessing algorithms. Int. J. Med. Inform. 2020, 144, 104284. [Google Scholar] [CrossRef]
- Turkoglu, M. COVIDetectioNet: COVID-19 diagnosis system based on X-ray images using features selected from pre-learned deep features ensemble. Appl. Intell. 2021, 51, 1213–1226. [Google Scholar] [CrossRef]
- Goel, T.; Murugan, R.; Mirjalili, S.; Chakrabartty, D.K. OptCoNet: An optimized convolutional neural network for an automatic diagnosis of COVID-19. Appl. Intell. 2021, 51, 1351–1366. [Google Scholar] [CrossRef]
- Sahlol, A.T.; Yousri, D.; Ewees, A.A.; Al-Qaness, M.A.; Damasevicius, R.; Elaziz, M.A. COVID-19 image classification using deep features and fractional-order marine predators algorithm. Sci. Rep. 2020, 10, 15364. [Google Scholar] [CrossRef]
- Pathak, Y.; Shukla, P.K.; Tiwari, A.; Stalin, S.; Singh, S. Deep transfer learning based classification model for COVID-19 disease. Irbm 2022, 43, 87–92. [Google Scholar] [CrossRef]
- Duran-Lopez, L.; Dominguez-Morales, J.P.; Corral-Jaime, J.; Vicente-Diaz, S.; Linares-Barranco, A. COVID-XNet: A custom deep learning system to diagnose and locate COVID-19 in chest X-ray images. Appl. Sci. 2020, 10, 5683. [Google Scholar] [CrossRef]
- Xu, X.; Jiang, X.; Ma, C.; Du, P.; Li, X.; Lv, S.; Yu, L.; Ni, Q.; Chen, Y.; Su, J.; et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering 2020, 6, 1122–1129. [Google Scholar] [CrossRef] [PubMed]
- Chandra, T.B.; Verma, K.; Singh, B.K.; Jain, D.; Netam, S.S. Coronavirus disease (COVID-19) detection in chest X-ray images using majority voting based classifier ensemble. Expert Syst. Appl. 2021, 165, 113909. [Google Scholar] [CrossRef] [PubMed]
- Ismael, A.M.; Şengür, A. The investigation of multiresolution approaches for chest X-ray image based COVID-19 detection. Health Inf. Sci. Syst. 2020, 8, 29. [Google Scholar] [CrossRef] [PubMed]
- Ucar, F.; Korkmaz, D. COVIDiagnosis-Net: Deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med. Hypotheses 2020, 140, 109761. [Google Scholar] [CrossRef]
- Tan, C.; Sun, F.; Kong, T.; Zhang, W.; Yang, C.; Liu, C. A survey on deep transfer learning. In Proceedings of the Artificial Neural Networks and Machine Learning–ICANN 2018: 27th International Conference on Artificial Neural Networks, Rhodes, Greece, 4–7 October 2018; Proceedings, Part III 27. Springer: Berlin/Heidelberg, Germany, 2018; pp. 270–279. [Google Scholar]
- Long, M.; Zhu, H.; Wang, J.; Jordan, M.I. Deep transfer learning with joint adaptation networks. In Proceedings of the International Conference on Machine Learning, PMLR, Sydney, Australia, 6–11 August 2017; pp. 2208–2217. [Google Scholar]
- Loey, M.; Smarandache, F.; M. Khalifa, N.E. Within the lack of chest COVID-19 X-ray dataset: A novel detection model based on GAN and deep transfer learning. Symmetry 2020, 12, 651. [Google Scholar] [CrossRef]
- Shazia, A.; Xuan, T.Z.; Chuah, J.H.; Usman, J.; Qian, P.; Lai, K.W. A comparative study of multiple neural network for detection of COVID-19 on chest X-ray. EURASIP J. Adv. Signal Process. 2021, 2021, 50. [Google Scholar] [CrossRef]
- Mohammed, A.; Wang, C.; Zhao, M.; Ullah, M.; Naseem, R.; Wang, H.; Pedersen, M.; Cheikh, F.A. Weakly-supervised network for detection of COVID-19 in chest CT scans. IEEE Access 2020, 8, 155987–156000. [Google Scholar] [CrossRef]
- Razzak, M.I.; Naz, S.; Zaib, A. Deep learning for medical image processing: Overview, challenges and the future. In Classification in BioApps: Automation of Decision Making; Springer: Berlin/Heidelberg, Germany, 2018; pp. 323–350. [Google Scholar]
- Bhattacharya, S.; Maddikunta, P.K.R.; Pham, Q.V.; Gadekallu, T.R.; Chowdhary, C.L.; Alazab, M.; Piran, M.J. Deep learning and medical image processing for coronavirus (COVID-19) pandemic: A survey. Sustain. Cities Soc. 2021, 65, 102589. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Sedik, A.; Iliyasu, A.M.; Abd El-Rahiem, B.; Abdel Samea, M.E.; Abdel-Raheem, A.; Hammad, M.; Peng, J.; Abd El-Samie, F.E.; Abd El-Latif, A.A. Deploying machine and deep learning models for efficient data-augmented detection of COVID-19 infections. Viruses 2020, 12, 769. [Google Scholar] [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Panwar, H.; Gupta, P.; Siddiqui, M.K.; Morales-Menendez, R.; Singh, V. Application of deep learning for fast detection of COVID-19 in X-Rays using nCOVnet. Chaos Solitons Fractals 2020, 138, 109944. [Google Scholar] [CrossRef]
- Öztürk, Ş.; Özkaya, U.; Barstuğan, M. Classification of Coronavirus (COVID-19) from X-ray and CT images using shrunken features. Int. J. Imaging Syst. Technol. 2021, 31, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Hauswald, J.; Gao, C.; Rovinski, A.; Mudge, T.; Mars, J.; Tang, L. Neurosurgeon: Collaborative intelligence between the cloud and mobile edge. ACM Sigarch Comput. Archit. News 2017, 45, 615–629. [Google Scholar] [CrossRef]
- Zhang, S.; Mu, X.; Kou, G.; Zhao, J. Object detection based on efficient multiscale auto-inference in remote sensing images. IEEE Geosci. Remote Sens. Lett. 2020, 18, 1650–1654. [Google Scholar] [CrossRef]
- Li, C.T.; Wu, X.; Özgür, A.; El Gamal, A. Minimax learning for distributed inference. IEEE Trans. Inf. Theory 2020, 66, 7929–7938. [Google Scholar] [CrossRef]
- Shao, J.; Zhang, J. Bottlenet++: An end-to-end approach for feature compression in device-edge co-inference systems. In Proceedings of the 2020 IEEE International Conference on Communications Workshops (ICC Workshops), Dublin, Ireland, 7 June 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–6. [Google Scholar]
- Huang, K.; Jiang, Z.; Li, Y.; Wu, Z.; Wu, X.; Zhu, W.; Chen, M.; Zhang, Y.; Zuo, K.; Li, Y.; et al. The classification of six common skin diseases based on Xiangya-Derm: Development of a Chinese database for artificial intelligence. J. Med. Internet Res. 2021, 23, e26025. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, X.; Liu, F.; He, L.; Zhang, L.; Yang, Y.; Li, W.; Wang, S.; Liu, L.; Liu, Z.; et al. An interpretable and expandable deep learning diagnostic system for multiple ocular diseases: Qualitative study. J. Med. Internet Res. 2018, 20, e11144. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Liao, H.Y.M.; Wu, Y.H.; Chen, P.Y.; Hsieh, J.W.; Yeh, I.H. CSPNet: A new backbone that can enhance learning capability of CNN. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Seattle, WA, USA, 14–19 June 2020; pp. 390–391. [Google Scholar]
- Tiwari, V.; Singhal, A.; Dhankhar, N. Detecting COVID-19 Opacity in X-ray Images Using YOLO and RetinaNet Ensemble. In Proceedings of the 2022 IEEE Delhi Section Conference (DELCON), New Delhi, India, 11–13 February 2022; pp. 1–5. [Google Scholar] [CrossRef]
- He, Y.; Kang, G.; Dong, X.; Fu, Y.; Yang, Y. Soft filter pruning for accelerating deep convolutional neural networks. arXiv 2018, arXiv:1808.06866. [Google Scholar]







| Framework | Accuracy (mAP50) |
|---|---|
| Original Scaled-YOLOv4 | 0.543 |
| BioEdge (no compression) | 0.543 |
| BioEdge (4-fold compression) | 0.541 |
| BioEdge (8-fold compression) | 0.540 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahn, H.; Lee, M.; Seong, S.; Lee, M.; Na, G.-J.; Chun, I.-G.; Kim, Y.; Hong, C.-H. BioEdge: Accelerating Object Detection in Bioimages with Edge-Based Distributed Inference. Electronics 2023, 12, 4544. https://doi.org/10.3390/electronics12214544
Ahn H, Lee M, Seong S, Lee M, Na G-J, Chun I-G, Kim Y, Hong C-H. BioEdge: Accelerating Object Detection in Bioimages with Edge-Based Distributed Inference. Electronics. 2023; 12(21):4544. https://doi.org/10.3390/electronics12214544
Chicago/Turabian StyleAhn, Hyunho, Munkyu Lee, Sihoon Seong, Minhyeok Lee, Gap-Joo Na, In-Geol Chun, Youngpil Kim, and Cheol-Ho Hong. 2023. "BioEdge: Accelerating Object Detection in Bioimages with Edge-Based Distributed Inference" Electronics 12, no. 21: 4544. https://doi.org/10.3390/electronics12214544
APA StyleAhn, H., Lee, M., Seong, S., Lee, M., Na, G.-J., Chun, I.-G., Kim, Y., & Hong, C.-H. (2023). BioEdge: Accelerating Object Detection in Bioimages with Edge-Based Distributed Inference. Electronics, 12(21), 4544. https://doi.org/10.3390/electronics12214544








