A Novel Hybridoma Cell Segmentation Method Based on Multi-Scale Feature Fusion and Dual Attention Network
Abstract
:1. Introduction
- We construct a new hybridoma cell image segmentation dataset. We accurately label the hybridoma cells in the image and design a tool to enhance the original data. Furthermore, we use three data enhancement methods—rotation reflection transformation, contrast transformation, and brightness transformation—to enhance the datasets and improve the generalization ability of the deep learning network.
- In order to further improve the segmentation accuracy, we propose multi-scale feature fusion and dual attention mechanisms for hybridoma cell image segmentation based on non-local and SENet attention mechanisms.
- We improve the traditional image segmentation network, U-Net. Moreover, we use the residual network to optimize the loss function. A focal loss function is used instead of the traditional cross-entropy loss function to overcome the class imbalance problem.
2. Related Works
2.1. Traditional Image Segmentation
2.2. Deep Learning-Based Image Segmentation
2.3. Attention Mechanism-Based Methods
2.4. Feature Pyramid Networks
3. The Proposed Hybridoma Cell Segmentation Method
- (1)
- Improving each module in the encoder and decoder to a residual structure with skip connections. Furthermore, the ability to extract cell features is enhanced through residual learning.
- (2)
- Improving the cross-entropy loss function in the U-Net network by the focal loss function, increase the proportion of cell categories in the total loss, and alleviate the category imbalance problem in the dataset.
- (3)
- Adding a batch normalization layer before the activation function layer so that the training loss can converge faster, and also effectively prevent the network from overfitting. In order to strengthen the extraction of cell features from the network, we perform feature fusion on the feature maps in the encoder and then apply a dual-attention mechanism.
- (4)
- Adjusting the structure of the encoder and using the feature pyramid to fuse the feature maps of the final output of different modules in the encoder to obtain a variety of semantic expression capabilities, and then obtaining the feature map of the corresponding level in the decoder.
- (5)
- In the first module of the decoder, the global attention mechanism (non-local) is injected into the feature map after channel splicing so as to expand the receptive field of the feature map fusion and improve the network’s ability to perceive the spatial information.
- (6)
- Adding channel attention SENet. We adjust the importance of each channel by learning the dependencies between channels, enhance the ability to obtain semantic information about key features, and improve the segmentation effect of the network.
3.1. Feature Fusion Module Design
- Firstly, for the final feature map of each module in the encoder, we use 256 1 × 1 convolution kernels for performing convolution operations to obtain 256-dimensional feature maps , , , .
- Secondly, the obtained feature maps after layer upsampling are then fused by element-wise addition to obtain the four fused feature maps.
- Finally, the four fused feature maps are spliced into the decoder.
3.2. Dual Attention Mechanism Module
3.2.1. Global Attention Mechanism
- Linear mapping: For an input feature matrix X where, T represents the number of samples selected for one training, H and W represent the height and width of the input feature map; respectively, and 768 is the feature fusion map passed into the encoder. After stitching, we convolve the input matrix with a 1 × 1 × 1 convolution kernel, and then linearly map it to three different types of feature spaces, obtaining three new feature matrices, A, B, and C, with a size of T × H × W × 384.
- Autocorrelation calculation: First, we reduce the dimension of the two feature matrices A and B from a four-dimensional matrix to a two-dimensional matrix by the reshape operation. After dimension reduction, the size of the feature matrices A and B is (T × H × W, 384). Second, we transpose the feature matrix B, and perform matrix multiplication on the processed feature matrix A and the transposed feature matrix B to obtain a matrix of size (T × H × W, T × H × W), which represents the input feature matrix. Then, the autocorrelation inside X is calculated, where, the dependence of each pixel on all other pixels in the image is normalized by using softmax. Similarly, the dimension reduction process is also applied to the feature matrix C, and the size of C will be (T × H × W, 384).
- Attention coefficient calculation: The obtained feature matrix after softmax processing and the feature matrix C after dimension reduction are multiplied together to obtain a feature matrix of size (T × H × W, 384), that is, the attention mechanism coefficient is applied to all the feature maps of the channel corresponding to the pixel. Finally, a 1 × 1 × 1 convolution kernel is used to perform the convolution operation. After adding the obtained result to the input feature matrix X point by point, the output feature matrix Z of the non-local module is finally obtained. The size of the output matrix Z is the same as the size of the input feature matrix X.
3.2.2. Channel Attention Mechanism
3.3. Loss Function Optimization
4. Experimental Results and Analysis
4.1. Dataset Preparation
4.2. Experiments Environment
4.3. Evaluation Metrics
4.4. Attention Mechanism Comparison and Verification
- Experiment 1 is conducted without adding any attention mechanisms to the RA-UNet network.
- Experiment 2 employs a non-local attention mechanism for the RA-UNet network.
- Experiment 3 employs a channel attention mechanism (SENet) for the RA-UNet network.
- In experiment 4, both attention mechanisms (non-local and SENet) are added to the RA-UNet network.
4.5. Feature Fusion Module Comparison and Verification
- Experiment 1 is performed without feature fusion on the feature map on the encoder side, but directly performs channel splicing with the feature map of the corresponding resolution in the decoder.
- In experiment 2, the feature map of each module in the encoder is fused. Then, the fused feature map and the feature map of the corresponding resolution on the decoder side are channel-spliced.
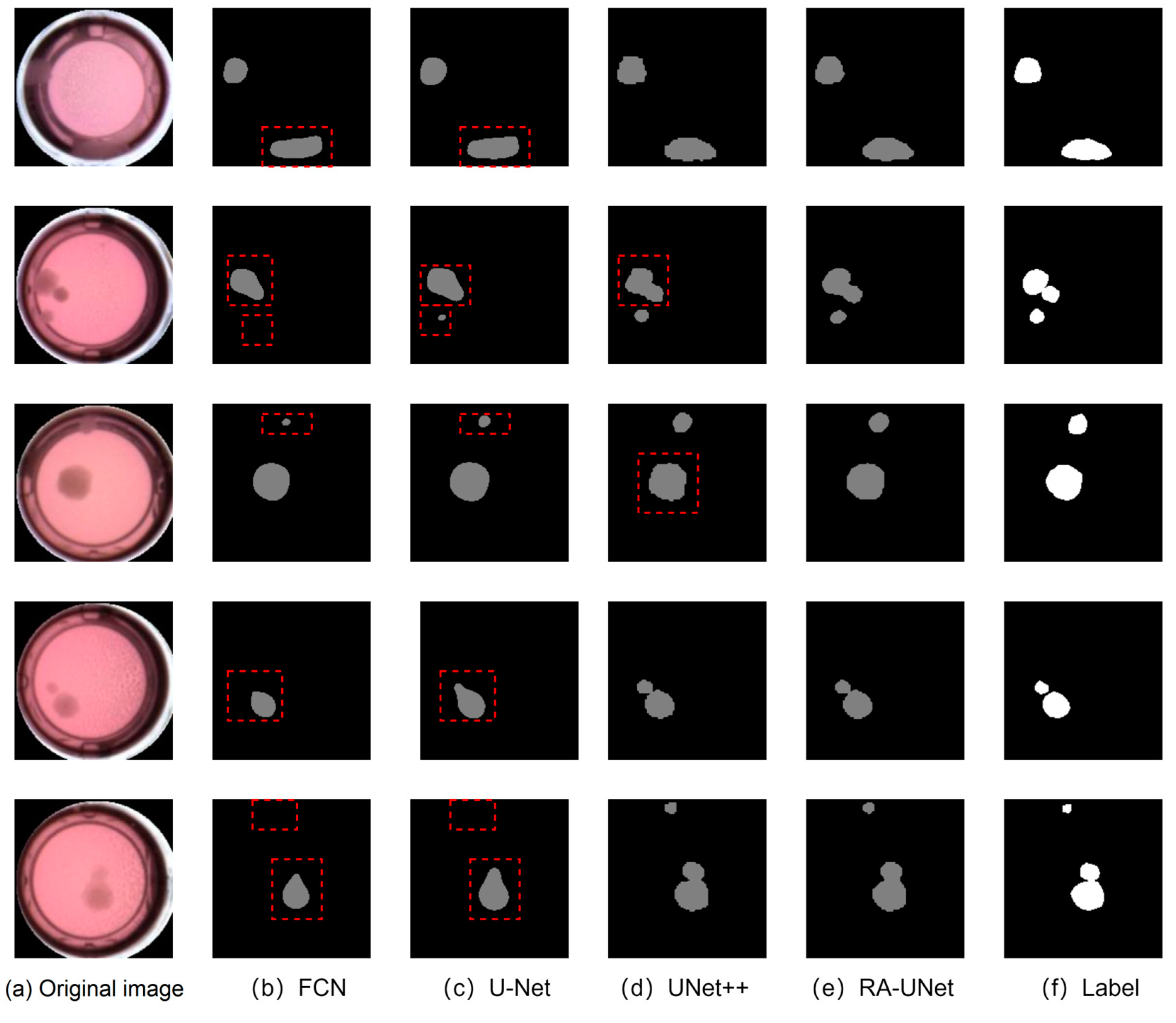
4.6. Segmentation Accuracy Comparison
5. Conclusions and Future Direction
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moraes, J.Z.; Hamaguchi, B.; Braggion, C.; Speciale, E.R.; Cesar, F.B.V.; da Silva Soares, G.D.F.; Osaki, J.H.; Pereira, T.M.; Aguiar, R.B. Hybridoma technology: Is it still useful? Curr. Res. Immunol. 2021, 2, 32–40. [Google Scholar] [CrossRef]
- Minaee, S.; Boykov, Y.; Porikli, F.; Plaza, A.; Kehtarnavaz, N.; Terzopoulos, D. Image Segmentation Using Deep Learning: A Survey. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 44, 3523–3542. [Google Scholar] [CrossRef]
- Malhotra, P.; Gupta, S.; Koundal, D.; Zaguia, A.; Enbeyle, W. Deep neural networks for medical image segmentation. J. Healthc. Eng. 2022, 2022, 9580991. [Google Scholar] [CrossRef]
- Araujo, F.H.; Silva, R.R.; Ushizima, D.M.; Rezende, M.T.; Carneiro, C.M.; Bianchi, A.G.C.; Medeiros, F.N. Deep learning for cell image segmentation and ranking. Comput. Med. Imaging Graph. 2019, 72, 13–21. [Google Scholar] [CrossRef]
- Al-Kofahi, Y.; Zaltsman, A.; Graves, R.; Marshall, W.; Rusu, M. A deep learning-based algorithm for 2-D cell segmentation in microscopy images. BMC Bioinform. 2018, 19, 365. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Zhang, L.; Chen, S.; Ni, D.; Lei, B.; Wang, T. Accurate segmentation of cervical cytoplasm and nuclei based on multiscale convolutional network and graph partitioning. IEEE Trans. Biomed. Eng. 2015, 62, 2421–2433. [Google Scholar] [CrossRef]
- Kothari, S.; Chaudry, Q.; Wang, M.D. Automated cell counting and cluster segmentation using concavity detection and ellipse fitting techniques. In Proceedings of the 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Boston, MA, USA, 28 June–1 July 2009; pp. 795–798. [Google Scholar]
- Gibbs, P.; Buckley, D.L.; Blackband, S.J.; Horsman, A. Tumour volume determination from MR images by morphological segmentation. Phys. Med. Biol. 1996, 41, 2437. [Google Scholar] [CrossRef]
- Kaus, M.R.; Warfield, S.K.; Nabavi, A.; Black, P.M.; Jolesz, F.A.; Kikinis, R. Automated segmentation of MR images of brain tumors. Radiology 2001, 218, 586–591. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Li, C.; Wang, J.; Wei, X.; Li, Y.; Zhu, Y.; Zhang, S. Threshold segmentation algorithm for automatic extraction of cerebral vessels from brain magnetic resonance angiography images. J. Neurosci. Methods 2015, 241, 30–36. [Google Scholar] [CrossRef]
- Wong, D.; Liu, J.; Fengshou, Y.; Tian, Q.; Xiong, W.; Zhou, J.; Qi, Y.; Han, T.; Venkatesh, S.; Wang, S.C. A semi-automated method for liver tumor segmentation based on 2D region growing with knowledge-based constraints. MICCAI Workshop 2008, 41, 159. [Google Scholar]
- Chen, X.; Zheng, C.; Yao, H.; Wang, B. Image segmentation using a unified Markov random field model. IET Image Process. 2017, 11, 860–869. [Google Scholar] [CrossRef]
- Rosenfeld, A. The max Roberts operator is a Hueckel-type edge detector. IEEE Trans. Pattern Anal. Mach. Intell. 1981, 3, 101–103. [Google Scholar] [CrossRef]
- Lang, Y.; Zheng, D. An improved Sobel edge detection operator. In Proceedings of the 2016 6th International Conference on Mechatronics, Computer and Education Informationization (MCEI 2016), Chengdu, China, 9–11 July 2016; pp. 590–593. [Google Scholar]
- Jiang, X.; Zhang, R.; Nie, S. Image Segmentation Based on PDEs Model: A Survey. In Proceedings of the 3rd International Conference on Bioinformatics and Biomedical Engineering, Beijing, China, 11–13 June 2009; pp. 1–4. [Google Scholar] [CrossRef]
- Chan, T.F.; Vese, L.A. Active contours without edges. IEEE Trans. Image Process. 2001, 10, 266–277. [Google Scholar] [CrossRef] [Green Version]
- Celebi, M.E.; Iyatomi, H.; Schaefer, G. Contrast enhancement in dermoscopy images by maximizing a histogram bimodality measure. In Proceedings of the 2009 16th IEEE International Conference on Image Processing (ICIP), Cairo, Egypt, 7–10 November 2009; pp. 2601–2604. [Google Scholar]
- Yuan, X.; Wu, L.; Peng, Q. An improved Otsu method using the weighted object variance for defect detection. Appl. Surf. Sci. 2015, 349, 472–484. [Google Scholar] [CrossRef] [Green Version]
- Vicar, T.; Balvan, J.; Jaros, J.; Jug, F.; Kolar, R.; Masarik, M.; Gumulec, J. Cell segmentation methods for label-free contrast microscopy: Review and comprehensive comparison. BMC Bioinform. 2019, 20, 360. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015, Proceedings of the Part III; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018, Proceedings of the 4; Springer: Berlin/Heidelberg, Germany, 2018; pp. 3–11. [Google Scholar]
- Guan, S.; Khan, A.A.; Sikdar, S.; Chitnis, P.V. Fully dense UNet for 2-D sparse photoacoustic tomography artifact removal. IEEE J. Biomed. Health Inform. 2019, 24, 568–576. [Google Scholar] [CrossRef] [Green Version]
- Khan, N.; Ullah, A.; Haq, I.U.; Menon, V.G.; Baik, S.W. SD-Net: Understanding overcrowded scenes in real-time via an efficient dilated convolutional neural network. J. Real-Time Image Process. 2021, 18, 1729–1743. [Google Scholar] [CrossRef]
- Hao, S.; Lee, D.-H.; Zhao, D. Sequence to sequence learning with attention mechanism for short-term passenger flow prediction in large-scale metro system. Transp. Res. Part C Emerg. Technol. 2019, 107, 287–300. [Google Scholar] [CrossRef]
- Jaderberg, M.; Simonyan, K.; Zisserman, A. Spatial transformer networks. Adv. Neural Inf. Process. Syst. 2015, 28, 1–9. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June2018; pp. 7132–7141. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Girshick, R.; Gupta, A.; He, K. Non-local neural networks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7794–7803. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. CBAM: Convolutional block attention module. In Proceedings of the 15th European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. SSD: Single shot multibox detector. In European Conference on Computer Vision; Springer: Cham, Switzerland, 2016; pp. 21–37. [Google Scholar]
- Xiaoguo, Z.; Ye, G.; Fei, Y.; Qihan, L.; Kaixin, Z. An Approach to Improve SSD through Skip Connection of Multiscale Feature Maps. Comput. Intell. Neurosci. 2020, 2020, 2936920. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2117–2125. [Google Scholar]
- Mnih, V.; Heess, N.; Graves, A. Recurrent models of visual attention. Adv. Neural Inf. Process. Syst. 2014, 27, 2204–2212. [Google Scholar]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal loss for dense object detection. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]












| Experiments Environment | Specific Configuration |
|---|---|
| operating system | Ubuntu 18.04.5 LTS |
| Memory | 32G |
| procesI | IIl(R) Xeon(R) Silver 4210 CPU @ 2.20GHz |
| Graphics card | GeForce RTX 2080 Ti |
| development environment | Visual Studio Code |
| Programming language | Python 3.6.13 |
| Deep Learning Framework | Pytorch 1.7.1 |
| Experiment No. | Attention | Accuracy | ||||
|---|---|---|---|---|---|---|
| Non-Local | SENet | Dice | PA | MPA | MIoU | |
| Experiment1 | - | - | 0.8824 | 0.9917 | 0.9426 | 0.8910 |
| Experiment2 | √ | - | 0.8843 | 0.9918 | 0.9469 | 0.8927 |
| Experiment3 | - | √ | 0.8875 | 0.9923 | 0.9487 | 0.8955 |
| Experiment4 | √ | √ | 0.8937 | 0.9926 | 0.9512 | 0.9007 |
| Model | Accuracy | |||
|---|---|---|---|---|
| Dice | PA | MPA | MIoU | |
| without feature fusion | 0.8820 | 0.9917 | 0.9452 | 0.8901 |
| with feature fusion | 0.8937 | 0.9926 | 0.9512 | 0.9007 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, J.; Ren, H.; Shi, M.; Cui, C.; Zhang, S.; Emam, M.; Li, L. A Novel Hybridoma Cell Segmentation Method Based on Multi-Scale Feature Fusion and Dual Attention Network. Electronics 2023, 12, 979. https://doi.org/10.3390/electronics12040979
Lu J, Ren H, Shi M, Cui C, Zhang S, Emam M, Li L. A Novel Hybridoma Cell Segmentation Method Based on Multi-Scale Feature Fusion and Dual Attention Network. Electronics. 2023; 12(4):979. https://doi.org/10.3390/electronics12040979
Chicago/Turabian StyleLu, Jianfeng, Hangpeng Ren, Mengtao Shi, Chen Cui, Shanqing Zhang, Mahmoud Emam, and Li Li. 2023. "A Novel Hybridoma Cell Segmentation Method Based on Multi-Scale Feature Fusion and Dual Attention Network" Electronics 12, no. 4: 979. https://doi.org/10.3390/electronics12040979







