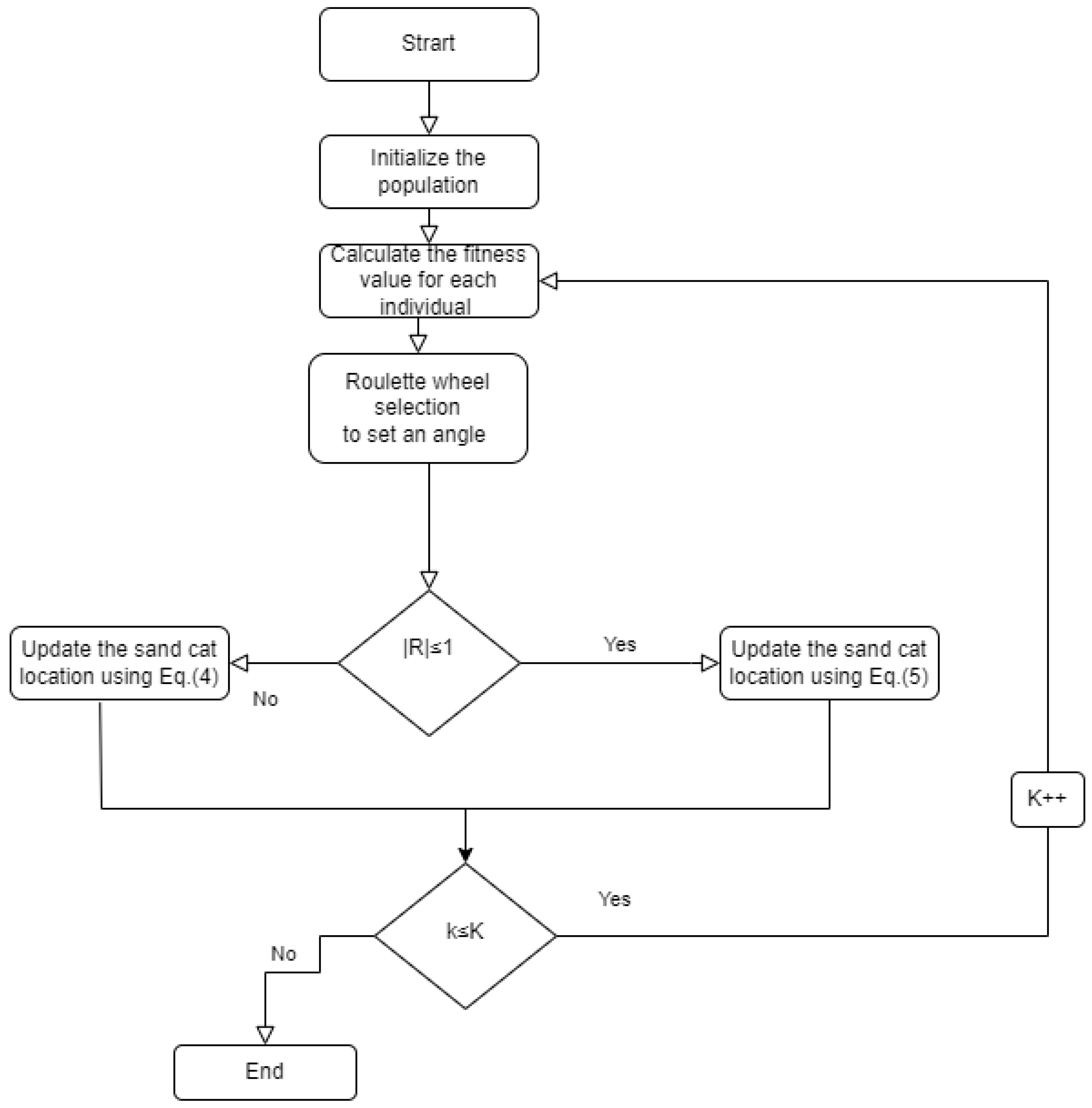
6.3. Performance Evaluation
To thoroughly examine the proposed BMSCSO’s performance, its results on FS problems were compared to those of the standard BSCSO and another seven FS methods that were published in the literature: Binary Biogeography-Based Optimization (BBBO) [
61], Binary Moth-Flame Optimization (BMFO) algorithm [
62], Binary Particle Swarm Optimization (BPSO) [
63], Binary Teaching-Learning-Based Optimization (BTLBO) [
64], Binary Ali Baba and the Forty Thieves (BAFT) algorithm [
10], Binary Honey Badger Algorithm (BHBA) [
65] and Binary Success-History based Adaptive Differential Evolution with Linear population size reduction (BLSHADE) [
66]. In this comparison, not only were the specifics of the experimental findings provided, but also a thorough and meticulous comparison with those comparative optimization techniques was made. In order to fairly compare the proposed BMSCSO with the comparative optimization techniques, the same experimental settings, including population size and number of iterations, were used to all of the competing algorithms. The parameter settings for each of the contending methods are listed in
Table 3.
The experiments were conducted 30 separate runs in order to get results that were statistically significant. The outcomes of the statistical analysis were then obtained based on the general capabilities and conclusions gained during these runs. Each dataset has the same amount of features as it has in terms of dimension. The proposed BMSCSO’s performance was assessed using classification accuracy, sensitivity, specificity, fitness values, and the average amount of selected features during the independent runs. This performance was then compared to the performance of other FS methods using all of the aforementioned rendering criteria. In all comparison tables, the best results are emboldened to give them more prominence over the other results.
The average classification accuracy scores for the standard BSCSO, the proposed BMSCSO, and other competing methods are presented in
Table 4 along with their associated standard derivation findings.
Greater accuracy results indicate superior robustness, while lower SD values indicate algorithmic stability.
Table 4 shows that the proposed BMSCSO obtained the maximum accuracy in a total of 14 datasets and was rated first solely based on having the best accuracy results in 10 datasets. BSCSO was able to acquire the optimal accuracy results in 3 datasets, namely Hepatitis, Leukemia and ProstateGE. BHBA came in second place by exclusively achieving the best results in 5 datasets and realized the best accuracy score in a total of 6 datasets. BBBO ranked third by getting the best accuracy results exclusively in 1 dataset, namely Parkinsons, and also got the best result in the Parkinsons dataset. Outstandingly, both BSCSO and BMSCSO obtained 100% accuracy in the Hepatitis, Leukemia and ProstateGE datasets. When reading
Table 4, one can notice that the proposed BMSCSO was more reliable than its competitors, having the lowest SD values in several datasets and much better than many other rival methods.
Table 5 shows the average fitness value and standard deviation of all competitors in each dataset.
It should be understood that the lowest fitness scores divulge better performance degrees for the optimization techniques. From
Table 5, it can be observed that BHBA and BMSCSO ranked first and second, where each exclusively got the minimum fitness values in an overall of 11 and 4 datasets, respectively. BTLBO ranked third by exclusively obtaining the minimal fitness scores in 4 datasets, namely, Coimbra, Parkinsons, Cleveland and Hepatitis. BBBO exclusively obtained the minimum fitness value in one dataset, namely the Leukemia dataset. BLSHADE and BHBA shared the same minimum fitness value of 0.0144 in the Thyroid dataset. Finally, BAFT, BMFO, BSCSO and BPSO did not disclose any lowest fitness values in any of the datasets investigated in this study. A second reading of the standard derivation findings listed in
Table 5 shows that BHBA’s and BMSCSO’s performance are superior to those of their rivals by achieving the lowest SD results across the majority of the datasets.
Table 6 provides the sensitivity findings of the proposed BMSCSO compared to BSCSO and the other competitors.
Next, we evaluate the proposed BMSCSO with the other competing algorithms in terms of the sensitivity findings that the FS algorithms aim to improve. It should be mentioned that better performance level corresponds to higher sensitivity outcomes.
Table 6 summarizes the outcomes of these opposing algorithms in terms of the average sensitivity findings correlated with their standard derivation values. Regarding the outcomes shown in this table, there is no doubt that BLSHADE has the highest sensitivity values when compared to all of its competitors. In particular, 9 out of 21 datasets, BLSHADE alone, obtained the greatest sensitivity values. By having the greatest sensitivity findings across 6 out of 21 datasets, the proposed BMSCSO clearly came in second place. BMFO has the best exclusive sensitivity results in 4 datasets, namely, Retinopathy, ILPD, Parkinsons and Heart, whereas BHBA placed next despite not achieving the best sensitivity values compared to all other competing algorithms. BBBO came in the next place with the best sensitivity findings only in the ParkinsonC and ProstateGE datasets with values of 0.8829 and 0.8947, respectively. Finally, BAFT, BPSO, and BTLBO did not disclose any distinguished sensitivity results. The proposed BMSCSO’s standard deviation values are low, which shows that the proposed BMSCSO is stable and well-established.
Similar to this,
Table 7 provides an overview of the average and standard deviations of the specificity findings for BMSCSO, BSCSO, and all other competing methods.
Reading the specificity findings mentioned in
Table 7 reveals that BMSCSO and BLSHADE came in first and second, respectively, with each having the greatest specificity values over a total of 8 and 6 datasets out of 21, respectively. By achieving the highest specificity outcomes across four datasets, BBBO came in third. BHBA alone had the greatest specificity values in Retinopathy and Spectfheart datasets with values of 0.7210 and 0.8448, respectively. The results produced by BMFO and BPSO are respectable and superior to those of other competing algorithms like BTLBO and BAFT, even if they did not attain the best specificity results in any of the datasets. In compared to other competitors, the proposed BMSCSO has relatively low standard deviation values throughout the majority of test datasets. These results demonstrate the stability of BMSCSO’s advantage.
As demonstrated in
Table 8, the number of selected features is also taken into account when comparing the performance of the proposed BMSCSO to that of BSCSO and all other FS methods described above.
When evaluating any feature selection technique, the quantity of features selected during the classification process is just as crucial as the classification accuracy. There are many different outcomes when comparing the proposed method to the eight competing algorithms, as shown in
Table 8. The nine competing algorithms can be divided into the following groups based on the results in the average number of features: By solely accumulating the fewest amount of selected features across a total of 8 out of 21 datasets, BHBA beat its competitors. BMSCSO came in second place, receiving the minimal number of features in 6 datasets entirely. Whereas BSCSO merely decreased the amount of features in the Lymphography dataset, BMFO had the fewest features in the Retinopathy, ILPD and HeartEW datasets, and BPSO reported its superiority against these two methods by having the fewest number of selected features in the Diagnostic, ParkinsonC, PimaDiabetes and Colon datasets. In every dataset taken into consideration, the BAFT, BLSHADE, BTLBO and BBBO algorithms failed to meet any minimal number of features. A second reading of the results in
Table 8 may be used to infer that the BMSCSO outperformed its competitors since it had very small results across the majority of the datasets. This alludes that BMSCSO was able to choose around the same number of features by running the algorithm 30 separate times.
The comparisons between the proposed BMSCSO and other state-of-the-art feature selection algorithms found in the literature demonstrate the robustness of the proposed BMSCSO as noted from the results presented in
Table 4,
Table 5,
Table 6,
Table 7 and
Table 8. One can observe that algorithms like BTLBO, BBBO, and BPSO behind BMSCSO by examining these outcomes in further detail and comparing the margins between BMSCSO and its rivals. Additionally, the proposed BMSCSO’s standard divisions are small and inferior to those of other rival algorithms.
This demonstrates that the proposed algorithm’s advantage is unquestionably strong. The key to BMSCSO’s acceptable level of performance is the algorithm’s sought-after balance between its exploration and exploitation features, which is made possible by the use of both the proposed mathematical model for this algorithm and the FS process. In this, the search agents of the proposed BMSCSO were able to explore and utilize each potential location in the search space as per its mathematical model, restoring a reasonable balance between exploration and exploitation. In this regard, the search agents of BMSCSO have the option of leaving their immediate area if they find themselves trapped in local optimums.
6.4. Convergence Curves
Figure 5 exhibits the convergence curves of the proposed BMSCSO, the basic BSCSO, and the other comparative optimization algorithms for the 21 datasets based on the fitness metric measure.
In the convergence plots illustrated in
Figure 5, the y-axis displays the fitness values and the x-axis the number of iterations. These plot curves show the average convergence behavior of the best solution for the basic BSCSO, the proposed BMSCSO, and the other competing algorithms produced over a total of 30 independent runs. The algorithm that achieves the lowest fitness outcomes with the fewest iterations while also avoiding local optima is selected as the best algorithm. When exploring the search space of each feature selection problem, the behavior of the proposed BMSCSO and the other competing algorithms diverge by significant margins, as shown in
Figure 5. This is because the proposed BMSCSO algorithm successfully balances the capacities for exploitation and exploration. Re-examining the curves in
Figure 5 reveals that the convergence patterns of the proposed BMSCSO, the basic BSCSO, and the other rival algorithms in the first 40 iterations in the Diagnostic, Breast, Prognostic, Coimbra, Retinopathy, and ILPD datasets are noticeably different. Distinctly, the convergence behavior of the proposed BMSCSO is somewhat better than that of some of its competitors, namely BMFO, BPSO, BBBO, BLSHADE. After two-thirds of the iterations in the SPECT, Cleveland, Saheart, PimaDiabetes, and ProstateGE datasets, the convergence behavior of the two proposed companions (i.e., BSCSO and BMSCSO) becomes nearly similar. Anyway, the convergence behavior of the proposed BMSCSO is slightly and sometimes much better than that of the rival algorithms, namely BMFO, BPSO, BBBO, BLSHADE, BAFT, BHBA, BTLBO. The convergence curves of the HeartEW, Leukemia, and Colon datasets demonstrate how the convergence behaviors of the two proposed FS methods (that is, BSCSO and BMSCSO) and the other competing algorithms, BMFO, BBBO, BLSHADE, BAFT, BTLBO, are not similar in most iterations and becomes indistinguishable in the final few iterations. In the ParkinsonC and Spectfheart datasets, the convergence behavior of the proposed BMSCSO, however, is clearly superior to that of the standard BSCSO, as well as other competing algorithms including BTLBO and BBBO. Finally, in the datasets for Thyroid, Heart, and Hepatitis, the convergence behavior of BMSCSO is superior to that of the basic BSCSO, BTLBO, BBBO, and BAFT. It should be noted that
Table 5 shows that the proposed BMSCSO performs better than the basic BSCSO and other competing algorithms, namely BMFO, BPSO, BBBO, BLSHADE, BAFT, BHBA, BTLBO, in terms of the average fitness values in these datasets.
6.5. Exploration and Exploitation
The two most crucial characteristics of optimization algorithms are exploration and exploitation, which can help solve optimization problems more effectively [
67]. Empirical studies have demonstrated a strong correlation between optimization algorithm’s capacity for exploration and exploitation and its rate of convergence with reference to these two conceptions. In particular, exploitation techniques are known to increase convergence rate toward the global optimum but also to increase the risk of being trapped in local optimal states [
67]. On the other hand, search methods that prioritize exploration over exploitation are more likely to lead to the location of regions within the search space where the global optimum is more likely to exist. This comes at the expense of optimization algorithms’ declining convergence speed [
68]. Although it may seem minor, the question of how exploration and exploitation of solutions are accomplished in optimizations has remained an open one in recent years and has continued to be a cause of disagreement among many researchers [
69]. Although many ideas and beliefs may appear to be at odds with one another, there seems to be agreement among researchers that this sort of search techniques must have an appropriate balance between exploration and exploitation in order to function reasonably. In order to identify reasonable solutions to an optimization problem, optimizations employ a set of potential solutions to explore the search space. In general, the search process should be directed in their direction by the search agents with the best solutions. This attraction causes the gap between the search agents to widen while the effects of exploitation diminish. On the other hand, the impact of the exploration approach becomes more noticeable when the search agents’ separation widens. A diversity measurement is used to calculate the rise and decrease in distance between the search agents [
70]. According to this approach, population variety can be defined as follows [
67]:
where
n is the number of search agents,
m identifies the dimension of the problem,
stands for the dimension
j of search agent
i, and
stands for the median of dimension
j in the total population.
The distance between the dimension
j of each search agent and the dimension’s average median is referred to as the diversity in each dimension, or
. The percentage of exploration and exploitation that a specific optimization algorithm uses is known as the complete balance response. The following formulas are used to compute these values at each iteration step [
67]:
where
denotes the highest diversity value that was found throughout the whole optimization process.
The link between the diversity at each iteration and the greatest diversity obtained is represented by the percentage of exploration (XPL%). The amount of exploitation is represented by the percentage of exploitation (XPT%) [
67]. As can be seen, the factors XPL% and XPT% are complimentary and in conflict with one another. The use of the median value while evaluating the balancing response prevents inconsistencies by using a reference element. The
value discovered over the whole optimization process also has an impact on this balancing reaction. The rate of exploration and exploitation is estimated using this value as a reference.
Optimization of the BreastEW dataset is used as an example to illustrate the evaluation of the balance response of the proposed BMSCSO.
Figure 6 depicts the performance conduct produced by the proposed BMSCSO in optimizing the BreastEW dataset over 100 iterations with respect to the balance assessment presented by Equations (
35) and (
36).
Five points, denoted as (A), (B), (C), (D), and (E), in
Figure 6 have been selected to demonstrate the diversity of solutions and the balance evaluations of each of them. Point (A) denotes an early stage of the proposed BMSCSO, when XPL% and XPT% have balance evaluation values of 90 and 10, respectively. These percentages provide BMSCSO a clear direction to operate within while it explores the search space. This implies that the solutions maintain a significant amount of dispersion of the search space. Point (B) corresponds to 70 iterations, where the balance assessment at this point preserves a value of XPL% = 70 together with XPT% = 30. In this position, the proposed BMSCSO mostly engages in exploration with minimal exploitation. Points (C) and (D) correlate with 75 and 100 iterations, respectively, where the balancing assessments’ exploration and exploitation values are XPL% = 25, XPT% = 75, XPL% = 5, and XPT% = 95, respectively. When these percentages were reached, the proposed BMSCSO’s behavior changed to encourage more exploitation than exploration. These configurations cause the solutions to be dispersed among numerous bunches, which lessens the diversity overall. Finally, point (E) implements the BMSCSO’s final junction. The proposed BMSCSO maintains a subtle tendency toward the exploitation of the top solutions without taking into consideration any exploration approach in such a case.
6.6. Sensitivity Analysis
To pinpoint the ideal parameter values of the proposed BMSCSO FS with memory based, a detailed sensitivity analysis based on the Design of Experiment (DoE) approach was performed. The suggested technique employs k-NN as a classifier in DoE to investigate the sensitivity of critical control parameters ( and ). The essential control parameter ranges were initially created, and the values of these parameters were determined to determine whether the best values came within the scope or whether further tests were necessary. The FS experiments then used a parameter with one input of the generated DoE values in the desired range while retaining the remaining parameters at their beginning levels. These tests were carried out systematically to investigate the influence of input parameters on the accuracy values of the suggested BMSCSO and arrive at a reasonable answer. Here are the values of each parameter used in this experiment: = 0.5, 1.0, 1.5, 2.0 and = 0.5, 1.0, 1.5, 2.0.
The numerical findings in
Table 9 reveal that BMSCSO performed best when the parameters
and
of BMSCSO are set to 2.0 and 2.0, respectively. This highlights the need of using a reasonable range of these critical parameters to improve BMSCSO’s resilience. These suggested approaches are nearly stable to changes in the necessary control parameters for BMSCSO when data sets have varying degrees of dimensionality, as shown by the standard deviation values of the classification accuracy of BMSCSO. To summarize, the results in
Table 9 show that BMSCSO is often extremely sensitive to parameter choices of
and
and the best values are 2 for both of them.
6.7. Statistical Analysis
The accuracy of the proposed BSCSO and BMSCSO algorithms was assessed in the aforementioned subsections using a variety of performance indicators, and their performance on FS problems was compared with that of other optimization FS methods stated in the literature. Over the course of 30 separate runs, the optimal solutions (i.e., classification accuracy and fitness values) were calculated using the average and standard deviation as statistical metric measures. These measures provide a general overview of how well the proposed algorithms handle FS problems with varied complexity. The first metric shows an algorithm’s average performance, while the second one shows how stable the method is throughout all of the separate run times. These evaluation measures may show the overall effectiveness and power of the FS method under evaluation, but they could not compare each of the separate runs independently. In other words, they have demonstrated the potential exploitation and exploratory behaviors of the proposed FS methods, but they are unable to demonstrate the quality of these methods. To demonstrate the significance of the results and that they were not the result of chance, Friedman’s and Holm’s statistical test methods [
18,
71] were carried out. Using the null hypothesis that there is no difference in the accuracy of any of the compared FS methods, Friedman’s test is used to determine if there is a root difference in the results of the competing FS methods.
Always receiving the lowest rank is the method with the best performance, while always receiving the highest rank is the method with the poorest performance. For the findings of the FS problems being studied, the
p-value of the Friedman’s test must be determined. If this value is equal to or less than the degree of significance, which in this study is
, the null hypothesis is rejected. Achieving this instance indicates that there are statistically significant differences in how well the comparison strategies function. Following this statistical test, a post-hoc test method-in this case, the Holm’s test procedure-is used to examine pairwise comparison of the comparative algorithms. For post-hoc analysis, the method with the lowest rank obtained by Friedman’s test is typically employed as the control method. The average fitness and classification accuracy results of the proposed BMSCSO algorithm presented in
Table 4 and
Table 5 are, therefore, statistically deserving of attention and do not statistically deviate from the results of other promising algorithms, according to Friedman’s and Holm’s statistical methods, which were used to make this revelation.
Table 10 shows the ranking results of the Friedman’s method on the basis of the results presented in
Table 4.
Friedman’s test method yielded a
p-value of 9.442002E-11 based on the average classification accuracy results shown in
Table 10. The existence of a statistically significant difference in the accuracy of the compared algorithms was shown by the rejection of the null hypothesis of similar performance. BMSCSO is statistically significant and the most successful FS method among all other FS ones, according to the statistical results shown in
Table 10. As a result, BMSCSO gets the highest rank, 1.714285, with a significance level of 5%. In particular, BMSCSO was put in the first place, and was followed in order by BSCSO, BHBA, BLSHADE, BBBO, BAFT, BMFO, BPSO, and BTLBO.
Following Friedman’s test, Holm’s test method was used to assess if there are statistically significant differences between the competing methods indicated in
Table 10 and the control algorithm with the lowest rank (i.e., BMSCSO).
Table 4 and
Table 10 reflect the collected results;
Table 11 presents the statistical findings produced by Holm’s test procedures based on those results. In this table,
z stands for the statistical difference between the two FS methods being compared,
for the Friedman’s rank assigned to the control method,
for the Friedman’s rank assigned to method
i, and ES for the amount of the control algorithm’s influence on algorithm
i.
Using the Holm’s test, which is shown in
Table 11 and excludes hypotheses with a
p-value
, BMSCSO was compared to the other competing algorithms. BMSCSO is a potent FS approach for obtaining favorable results for the datasets under examination, as can be shown from the findings in
Table 11, and it performs noticeably better than other skilled methods utilized to handle these FS problems. The
p-values also divulges the BMSCSO’s level of performance on FS tasks and demonstrate the effectiveness of this approach. The results in
Table 10 and
Table 11 demonstrate, in brief, that BMSCSO has successfully avoided local optimal solutions and is regarded as statistically significant by achieving high classification accuracy scores in comparison to the obtained classification accuracy rates by the basic BSCSO and the other competing FS algorithms.
A secondary statistical comparison was done between the proposed BMSCSO and the other competing FS methods in regards to the mean fitness results where their obtained results are displayed in
Table 5. The ranking results of the Friedman’s test-based statistical analysis based on fitness values are listed in
Table 12.
As per the fitness scores exhibited in
Table 5, the
p-value determined by Friedman’s test is 6.380973E-11. Due to the outcomes presented in
Table 12, the proposed BMSCSO algorithm is ranked among the top three FS methods among all other competing ones. The proposed BSCSO method gained the first rank with a value of 5.14, which means the two proposed methods earned the top ranking among all other competing FS methods due to the nature of their local and global search strategies. In more detail, the proposed BMSCSO method gained the second-rank algorithm with a value of 4.095 with a significance level of a = 5%. The BTLBO is the second best FS method in this comparison with a rank of 3.833333 with a little difference from the rank obtained by BMSCSO. In view of the results displayed in
Table 12, BMSCSO acted much better than the other algorithms in the FS task, where it ranked first, followed successively by BHBA, BSCSO, BCMA-ES, BLSHADE, BAFT, BMFO, BPSO, and BTLBO.
Based on the fitness ratings shown in
Table 5, Friedman’s test yielded a
p-value of 6.380973E-11. According to the outcomes shown in
Table 12, the proposed BMSCSO is one of the most effective FS method out of all those in competition. BHBA received the top position as noted from the ranking results presented in
Table 12 with a value of 3.833333 and a degree of significance of
. With an average ranking score of 3.833333, slightly better than the rank attained by BHBA, the BTLBO technique is the second-best approach. In sum and in light of the ranking results presented in
Table 12, BMSCSO performed much better than the other algorithms in the FS problems, where it came in the third place, followed by BLSHADE, BBBO, BSCSO, BAFT, BMFO, and BPSO in the last rank.
These outcomes approximately match those that were previously shown in
Table 10. The control algorithm (i.e., BHBA) and the other compared algorithms are then compared using Holm’s test procedure to see if there are statistically discernible differences. The outcomes obtained after performing Holm’s test with
to the results shown in
Table 12 are provided in
Table 13.
For the outcomes in
Table 13, Holm’s technique eliminated the hypotheses with a
p-value
. These findings clearly manifest that BMSCSO outperformed many other binary algorithms in solving FS problems, as evidenced by its high performance score compared to those rival ones. These results demonstrate that BMSCSO has wonderfully avoided local solutions throughout the search space by undertaking sensible exploration and exploitation aspects.
Table 10,
Table 11,
Table 12 and
Table 13 show that the results of BMSCSO are statistically significant, demonstrating that this FS method is superior to many other FS methods with excellent results over other well-known established optimization algorithms. This ending demonstrates the excellent efficiency of the proposed BMSCSO, which is a binary extension of the SCSO method, in solving a wide range of well-known FS problems.