MSGL+: Fast and Reliable Model Selection-Inspired Graph Metric Learning †
Abstract
:1. Introduction
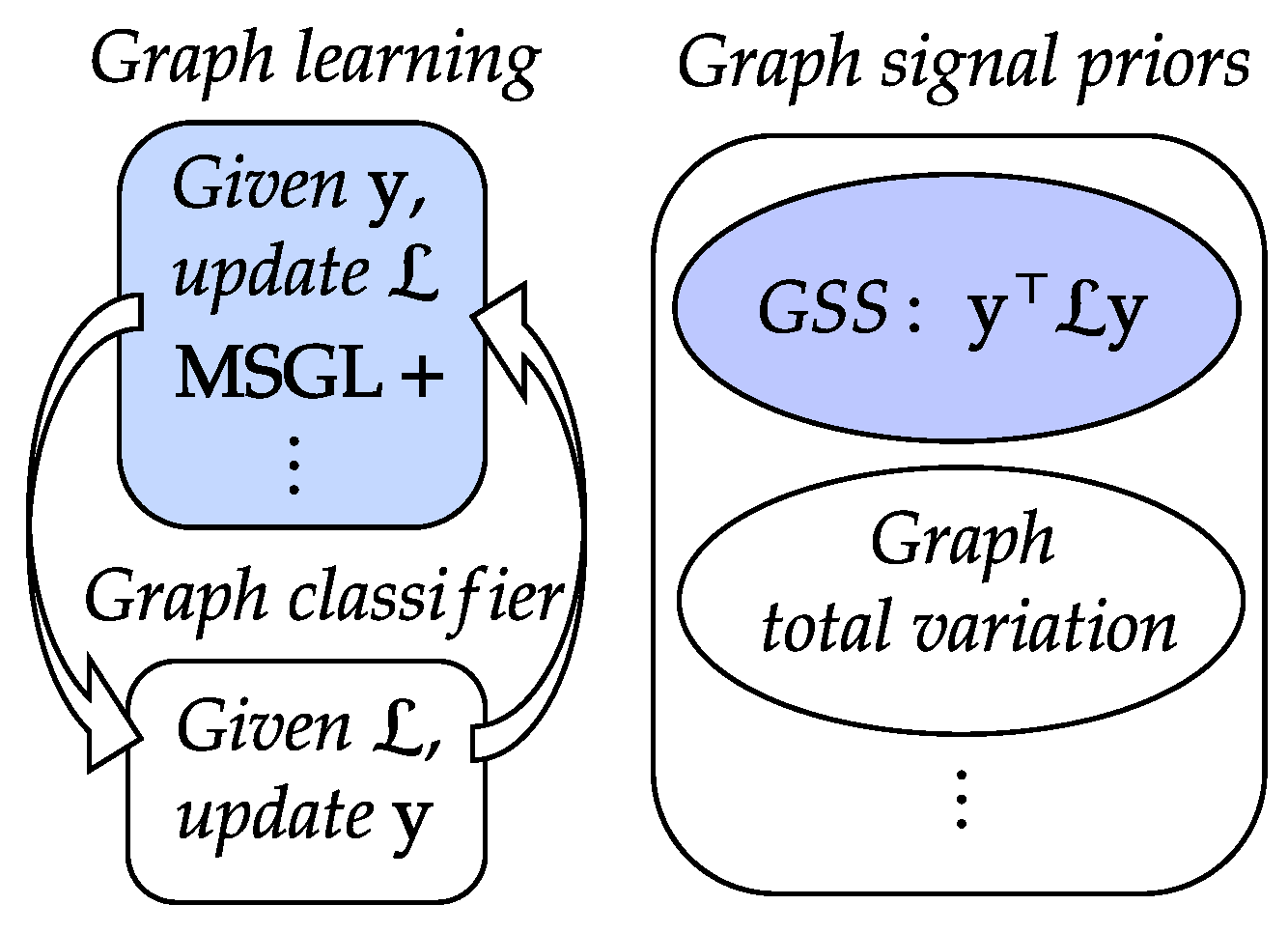
- In Equation (4), we introduce a novel graph learning framework that integrates graph kernel learning and graph signal processing. We achieve this by learning the coefficients of a polynomial kernel function that penalizes the signal’s Fourier coefficients with , where are the eigenvalues of the graph Laplacian and are the learnable parameters. This can be regarded as a generalization of the conventional smoothness prior , which uses a fixed coefficient for all eigenvalues.
- In Algorithm 1 in Section 1, we formulate the graph learning problem as a convex optimization objective that consists of a GSS prior with and a model complexity penalty with . We convert the positive definite constraint for into a group of linear constraints for . We then solve the problem efficiently using the Frank–Wolfe method, which only requires linear programs at each iteration.
Algorithm 1 The proposed MSGL+ graph metric learning method. Input: , , P. Output: . - 1:
- 2:
- Obtain eigenpairs and of via eigen decomposition.
- 3:
- Construct with , , , P, and randomly initialized via Equation (4).
- 4:
- while not converged do:
- 5:
- Solve Equation (25) for via interior-point.
- 6:
- while not converged do:
- 7:
- Solve Equation (26) for via a Newton–Raphson method.
- 8:
- end while
- 9:
- .
- 10:
- end while
- 11:
- Compute via Equation (7).
- In Equation (6) to Equation (28), we analyze the properties of the linear constraints and show that they ensure the positive definiteness of without requiring any box constraints on . We also show that we can reduce the number of optimization variables by exploring the properties of the linear constraints within the optimization objective, which leads to faster and more stable implementations than MSGL.
- In Section 3, we extend the applicability of MSGL+ to both metric matrix-defined feature-based positive graphs and graphical Lasso-learned signed graphs, which are two common types of graphs used in various applications. This extension from [31] significantly broadens the scope of MSGL+, as it can handle any symmetric, positive (semi-)definite matrix that corresponds to a (un)signed graph and learn the polynomial kernel coefficients via MSGL+.
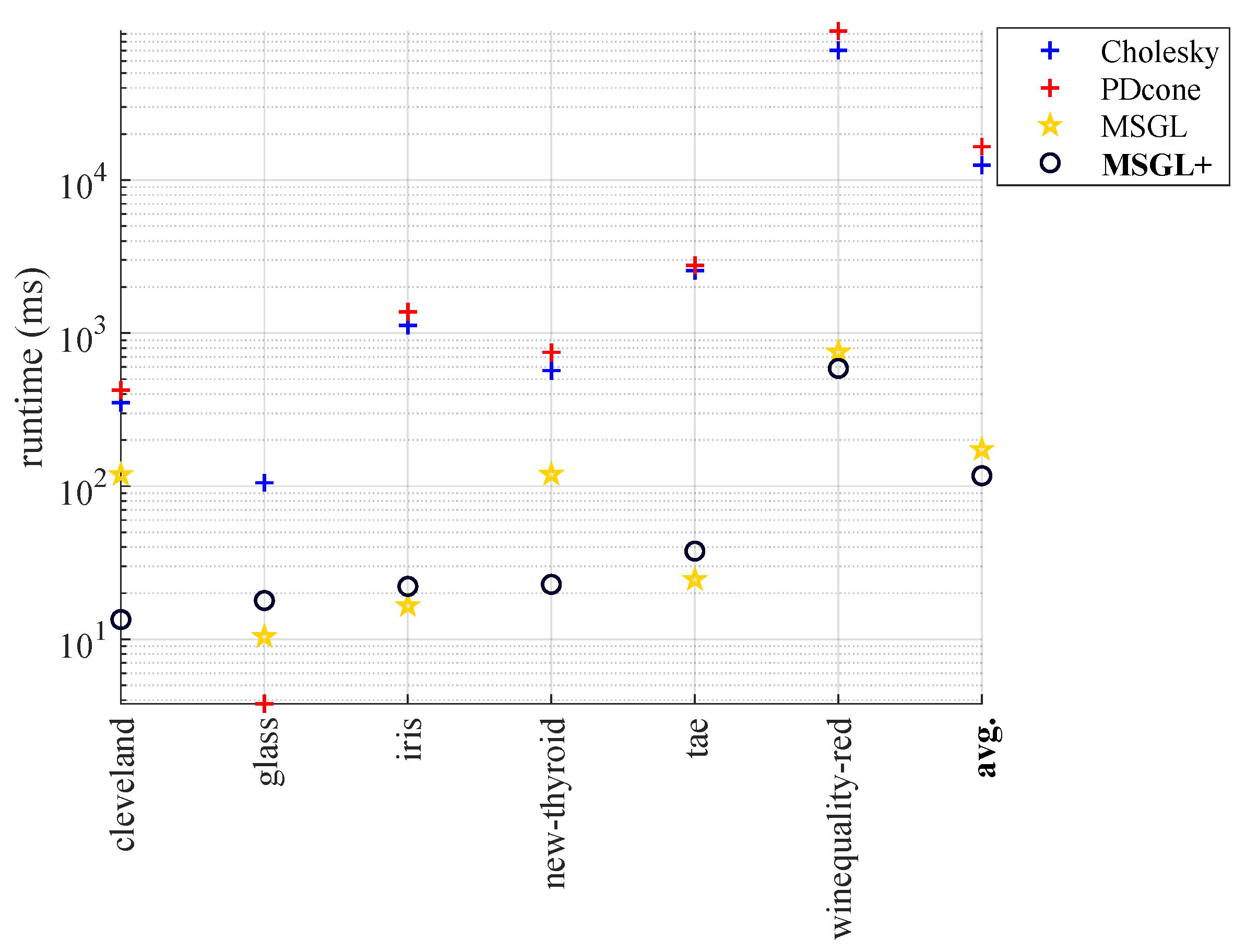
- In Section 3, we evaluate the performance of MSGL+ on various tasks, such as binary classification, multi-class classification, binary image denoising, and time-series analysis. We compare MSGL+ with several state-of-the-art graph learning methods and show that MSGL+ achieves comparable accuracy with much faster runtime.
2. Materials and Methods
2.1. Preliminaries
2.2. Problem Formulation
2.3. Algorithm Development
3. Results
3.1. Application to Binary Classification
3.2. Application to Multi-Class Classification
3.3. Application to Binary Image Denoising
3.4. Application to Time-Series Analysis
3.5. Statistical Significance of the Experiments in Terms of the Model Accuracy
4. Discussion
4.1. Accuracy
4.2. Runtime
4.3. Potential Utility in the Field of Medicine
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Nomenclature
| M | Number of features per data sample |
| N | Number of data samples |
| A data matrix | |
| Trace operator | |
| The vector of data sample labels | |
| P | 1-to-P-hop neighbor |
| Normalized graph Laplacian | |
| Polynomial function of | |
| Kernel parameter | |
| Polynomial function of | |
| The eigenvalue of | |
| A small tolerance | |
| An undirected graph | |
| Graph vertices | |
| Graph edges | |
| Adjacency matrix | |
| The -th entry of the adjacency matrix | |
| The feature vector of the i-th data sample | |
| The metric matrix | |
| Degree matrix | |
| The i-th diagonal of the degree matrix | |
| The matrix that consists of the column vectors that correspond to the eigenvectors of | |
| The i-th eigenvector of | |
| The diagonal matrix whose diagonals correspond to the eigenvalues of | |
| The matrix that consists of the column vectors that correspond to the eigenvectors of | |
| Kernel function | |
| † | pseudo-inverse |
| A dataset | |
| ∇ | Gradient |
| ⊙ | Element-wise product |
| A convex set |
Appendix A. Concise Proof of the Convexity of Equation (5)
- Convex constraints: The PD-cone constraint is convex, implying that resides within a convex set.
- The data-fit term is convex: The data-fit term exhibits both convex and concave behavior. This duality arises from the Hessian matrix of the data-fit term, which consists entirely of zero entries.
- The model complexity penalty term is convex: We express the model complexity penalty term as follows:where it comprises three components: an affine function , a logarithmic function , and another affine function . Notably, the convexity of , , and ensures that the composition of these functions preserves the convexity of the overall objective in Equation (5).
Appendix B. Relaxation of the Original Problem in Equation (5)
Appendix C. The Derivation from Equation (8) to Equation (9)
Appendix D. The Derivation from Equation (11) to Equation (12)
Appendix E. The Proof of Equation (23)
References
- Rasmussen, C.E.; Williams, C.K.I. Gaussian Processes for Machine Learning (Adaptive Computation and Machine Learning); The MIT Press: Cambridge, MA, USA, 2005. [Google Scholar]
- Ortega, A.; Frossard, P.; Kovacevic, J.; Moura, J.M.F.; Vandergheynst, P. Graph Signal Processing: Overview, Challenges, and Applications. Proc. IEEE 2018, 106, 808–828. [Google Scholar] [CrossRef]
- Ortega, A. Introduction to Graph Signal Processing; Cambridge University Press: Cambridge, UK, 2022. [Google Scholar]
- Cheung, G.; Magli, E.; Tanaka, Y.; Ng, M.K. Graph Spectral Image Processing. Proc. IEEE 2018, 106, 907–930. [Google Scholar] [CrossRef]
- Leus, G.; Marques, A.G.; Moura, J.M.; Ortega, A.; Shuman, D.I. Graph Signal Processing: History, development, impact, and outlook. IEEE Signal Process. Mag. 2023, 40, 49–60. [Google Scholar] [CrossRef]
- Zhu, X.; Ghahramani, Z.; Lafferty, J. Semi-supervised Learning Using Gaussian Fields and Harmonic Functions. In Proceedings of the International Conference on Machine Learning, Xi’an, China, 5 November 2003; pp. 912–919. [Google Scholar]
- Zhai, G.; Min, X. Perceptual image quality assessment: A survey. Sci. China Inf. Sci. 2020, 63, 1–52. [Google Scholar] [CrossRef]
- Schaeffer, S.E. Survey: Graph Clustering. Comput. Sci. Rev. 2007, 1, 27–64. [Google Scholar] [CrossRef]
- Berger, P.; Hannak, G.; Matz, G. Graph Signal Recovery via Primal-Dual Algorithms for Total Variation Minimization. IEEE J. Sel. Top. Signal Process. 2017, 11, 842–855. [Google Scholar] [CrossRef]
- Berger, P.; Buchacher, M.; Hannak, G.; Matz, G. Graph Learning Based on Total Variation Minimization. In Proceedings of the 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Calgary, AB, Canada, 15–20 April 2018; pp. 6309–6313. [Google Scholar] [CrossRef]
- Dinesh, C.; Cheung, G.; Bajić, I.V. 3D Point Cloud Super-Resolution via Graph Total Variation on Surface Normals. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019; pp. 4390–4394. [Google Scholar] [CrossRef]
- Dabush, L.; Routtenberg, T. Verifying the Smoothness of Graph Signals: A Graph Signal Processing Approach. arXiv 2023, arXiv:2305.19618. [Google Scholar]
- Belkin, M.; Matveeva, I.; Niyogi, P. Regularization and semisupervised learning on large graphs. In Proceedings of the Learning Theory, COLT, Banff, AB, Canada, 1–4 July 2004; Lecture Notes in Computer Science. Shawe-Taylor, J., Singer, Y., Eds.; Springer: Berlin/Heidelberg, Germany, 2004; Volume 3120, pp. 624–638. [Google Scholar]
- Dong, X.; Thanou, D.; Rabbat, M.; Frossard, P. Learning Graphs From Data: A Signal Representation Perspective. IEEE Signal Process. Mag. 2019, 36, 44–63. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Sparse Inverse Covariance Estimation with the Graphical Lasso. Proc. Biostat. 2008, 9, 432–441. [Google Scholar] [CrossRef]
- Yang, C.; Cheung, G.; Stankovic, V. Alternating Binary Classifier and Graph Learning from Partial Labels. In Proceedings of the Asia-Pacific Signal and Information Processing Association Annual Summit and Conference, Honolulu, HI, USA, 12–15 November 2018; pp. 1137–1140. [Google Scholar]
- Yang, C.; Cheung, G.; Hu, W. Signed Graph Metric Learning via Gershgorin Disc Perfect Alignment. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 44, 7219–7234. [Google Scholar] [CrossRef]
- Zhi, Y.C.; Ng, Y.C.; Dong, X. Gaussian Processes on Graphs Via Spectral Kernel Learning. IEEE Trans. Signal Inf. Process. Netw. 2023, 9, 304–314. [Google Scholar] [CrossRef]
- Zhou, X.; Belkin, M. Semi-supervised Learning by Higher Order Regularization. In Proceedings of the International Conference on Artificial Intelligence and Statistics, JMLR Workshop and Conference Proceedings, Lauderdale, FL, USA, 11–13 April 2011; pp. 892–900. [Google Scholar]
- Yang, C.; Hu, M.; Zhai, G.; Zhang, X.P. Graph-Based Denoising for Respiration and Heart Rate Estimation During Sleep in Thermal Video. IEEE Internet Things J. 2022, 9, 15697–15713. [Google Scholar] [CrossRef]
- Gong, X.; Li, X.; Ma, L.; Tong, W.; Shi, F.; Hu, M.; Zhang, X.P.; Yu, G.; Yang, C. Preterm infant general movements assessment via representation learning. Displays 2022, 75, 102308. [Google Scholar] [CrossRef]
- Tong, W.; Yang, C.; Li, X.; Shi, F.; Zhai, G. Cost-Effective Video-Based Poor Repertoire Detection for Preterm Infant General Movement Analysis. In Proceedings of the 2022 5th International Conference on Image and Graphics Processing (ICIGP ’22), Beijing, China, 7–9 January 2022; Association for Computing Machinery: New York, NY, USA, 2022; pp. 51–58. [Google Scholar] [CrossRef]
- Li, X.; Yang, C.; Tong, W.; Shi, F.; Zhai, G. Fast Graph-Based Binary Classifier Learning via Further Relaxation of Semi-Definite Relaxation. In Proceedings of the 2022 5th International Conference on Image and Graphics Processing (ICIGP ’22), Beijing, China, 7–9 January 2022; Association for Computing Machinery: New York, NY, USA, 2022; pp. 89–95. [Google Scholar] [CrossRef]
- Yang, C.; Cheung, G.; Zhai, G. Projection-free Graph-based Classifier Learning using Gershgorin Disc Perfect Alignment. arXiv 2021, arXiv:2106.01642. [Google Scholar]
- Yang, C.; Cheung, G.; Tan, W.t.; Zhai, G. Unfolding Projection-free SDP Relaxation of Binary Graph Classifier via GDPA Linearization. arXiv 2021, arXiv:2109.04697. [Google Scholar]
- Fang, X.; Xu, Y.; Li, X.; Lai, Z.; Wong, W.K. Learning a Nonnegative Sparse Graph for Linear Regression. IEEE Trans. Image Process. 2015, 24, 2760–2771. [Google Scholar] [CrossRef] [PubMed]
- Dornaika, F.; El Traboulsi, Y. Joint sparse graph and flexible embedding for graph-based semi-supervised learning. Neural Netw. 2019, 114, 91–95. [Google Scholar] [CrossRef]
- Han, X.; Liu, P.; Wang, L.; Li, D. Unsupervised feature selection via graph matrix learning and the low-dimensional space learning for classification. Eng. Appl. Artif. Intell. 2020, 87, 103283. [Google Scholar] [CrossRef]
- Xing, E.P.; Jordan, M.I.; Russell, S.J.; Ng, A.Y. Distance Metric Learning with Application to Clustering with Side-Information. In Proceedings of the Annual Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 9–11 December 2003; pp. 521–528. [Google Scholar]
- Wu, X.; Zhao, L.; Akoglu, L. A Quest for Structure: Jointly Learning the Graph Structure and Semi-Supervised Classification. In Proceedings of the ACM International Conference on Information and Knowledge Management, Torino, Italy, 22–26 October 2018. [Google Scholar]
- Yang, C.; Wang, F.; Ye, M.; Zhai, G.; Zhang, X.P.; Stankovic, V.; Stankovic, L. Model Selection-inspired Coefficients Optimization for Polynomial-Kernel Graph Learning. In Proceedings of the 2021 Asia-Pacific Signal and Information Processing Association Annual Summit and Conference (APSIPA ASC), Tokyo, Japan, 14–17 December 2021; pp. 344–350. [Google Scholar]
- Egilmez, H.; Pavez, E.; Ortega, A. Graph Learning From Data Under Laplacian and Structural Constraints. Proc. IEEE J. Sel. Top. Signal Process. 2017, 11, 825–841. [Google Scholar] [CrossRef]
- Jaggi, M. Revisiting Frank-Wolfe: Projection-Free Sparse Convex Optimization. In Proceedings of the International Conference on Machine Learning, Atlanta, GA, USA, 16–21 June 2013; pp. 427–435. [Google Scholar]
- Mahalanobis, P.C. On the generalized distance in statistics. Proc. Natl. Inst. Sci. India 1936, 2, 49–55. [Google Scholar]
- Hoffmann, F.; Hosseini, B.; Oberai, A.A.; Stuart, A.M. Spectral analysis of weighted Laplacians arising in data clustering. Appl. Comput. Harmon. Anal. 2022, 56, 189–249. [Google Scholar] [CrossRef]
- Anis, A.; El Gamal, A.; Avestimehr, A.S.; Ortega, A. A Sampling Theory Perspective of Graph-Based Semi-Supervised Learning. IEEE Trans. Inf. Theory 2019, 65, 2322–2342. [Google Scholar] [CrossRef]
- Sakiyama, A.; Tanaka, Y.; Tanaka, T.; Ortega, A. Eigendecomposition-Free Sampling Set Selection for Graph Signals. IEEE Trans. Signal Process. 2019, 67, 2679–2692. [Google Scholar] [CrossRef]
- Shuman, D.I.; Narang, S.K.; Frossard, P.; Ortega, A.; Vandergheynst, P. The Emerging Field of Signal Processing on Graphs: Extending High-dimensional Data Analysis to Networks and Other Irregular Domains. Proc. IEEE Signal Process. Mag. 2013, 30, 83–98. [Google Scholar] [CrossRef]
- Romero, D.; Ma, M.; Giannakis, G.B. Kernel-Based Reconstruction of Graph Signals. IEEE Trans. Signal Process. 2017, 65, 764–778. [Google Scholar] [CrossRef]
- Smola, A.J.; Kondor, R. Kernels and Regularization on Graphs. In Proceedings of the Learning Theory and Kernel Machines, Washington, DC, USA, 24–27August 2003; Schölkopf, B., Warmuth, M.K., Eds.; Springer: Berlin/Heidelberg, Germany, 2003; pp. 144–158. [Google Scholar]
- Boyd, S.; Vandenberghe, L. Convex Optimization; Cambridge University Press: Cambridge, UK, 2009. [Google Scholar]
- Papadimitriou, C.; Steiglitz, K. Combinatorial Optimization; Dover Publications, Inc.: Mineola, NY, USA, 1998. [Google Scholar]
- Raphson, J. Analysis Aequationum Universalis; University of St Andrews: St Andrews, UK, 1690. [Google Scholar]
- Hu, W.; Gao, X.; Cheung, G.; Guo, Z. Feature graph learning for 3D point cloud denoising. IEEE Trans. Signal Process. 2020, 68, 2841–2856. [Google Scholar] [CrossRef]
- Yang, C.; Cheung, G.; Hu, W. Graph Metric Learning via Gershgorin Disc Alignment. In Proceedings of the IEEE International Conference on Acoustics, Speech and Signal Processing, Barcelona, Spain, 4–8 May 2020. [Google Scholar]
- Golub, G.H.; Van Loan, C.F. Matrix Computations, 3rd ed.; The Johns Hopkins University Press: Baltimore, MD, USA, 1996. [Google Scholar]
- Weinberger, K.Q.; Saul, L.K. Distance metric learning for large margin nearest neighbor classification. J. Mach. Learn. Res. 2009, 10, 207–244. [Google Scholar]
- Russell, S.; Norvig, P. Artificial Intelligence: A Modern Approach, 3rd ed.; Prentice Hall Press: Hoboken, NJ, USA, 2009. [Google Scholar]
- Dong, M.; Wang, Y.; Yang, X.; Xue, J. Learning Local Metrics and Influential Regions for Classification. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 1522–1529. [Google Scholar] [CrossRef]
- Mcnemar, Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika 1947, 12, 153–157. [Google Scholar] [CrossRef]
- Fagerland, M.W.; Lydersen, S.; Laake, P. The McNemar test for binary matched-pairs data: Mid-p and asymptotic are better than exact conditional. BMC Med. Res. Methodol. 2013, 13, 91. [Google Scholar] [CrossRef]
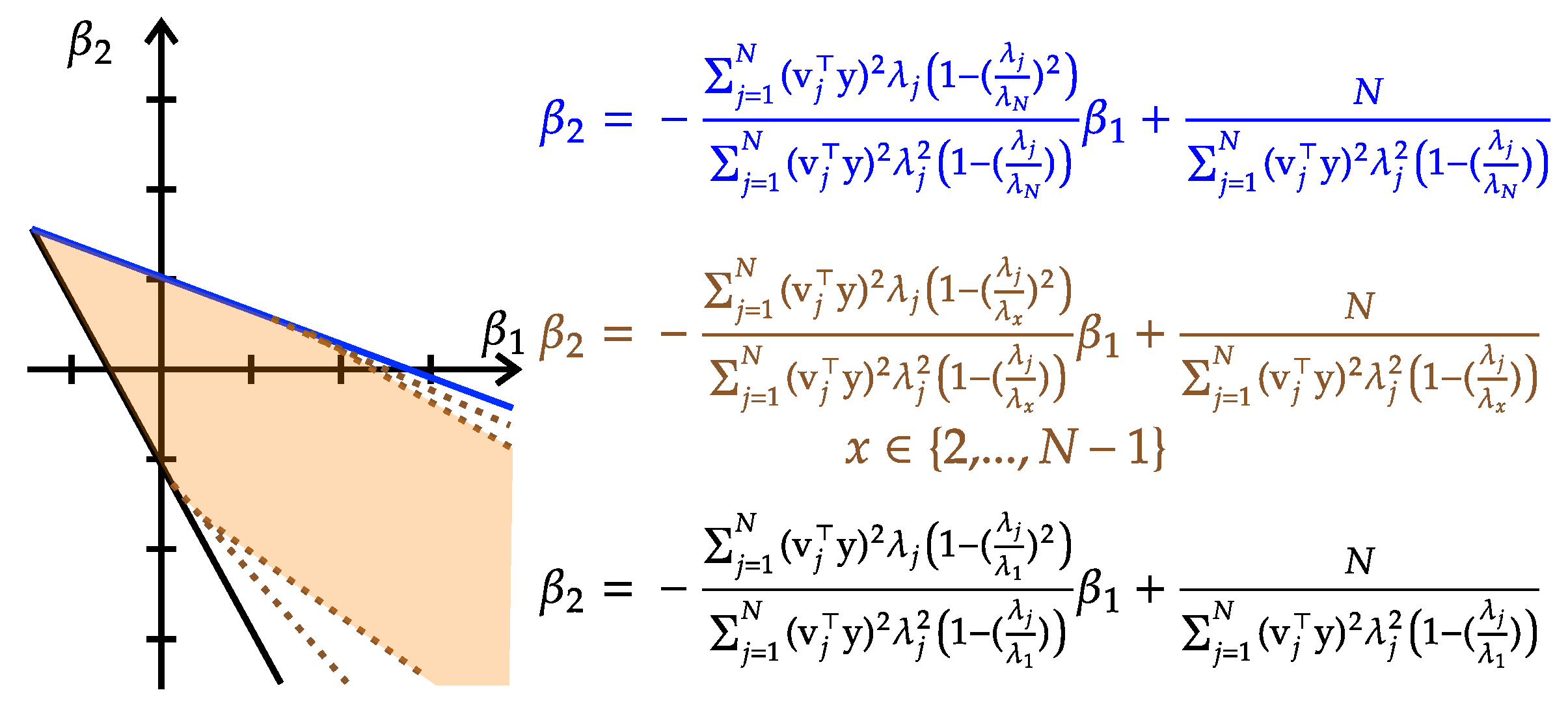



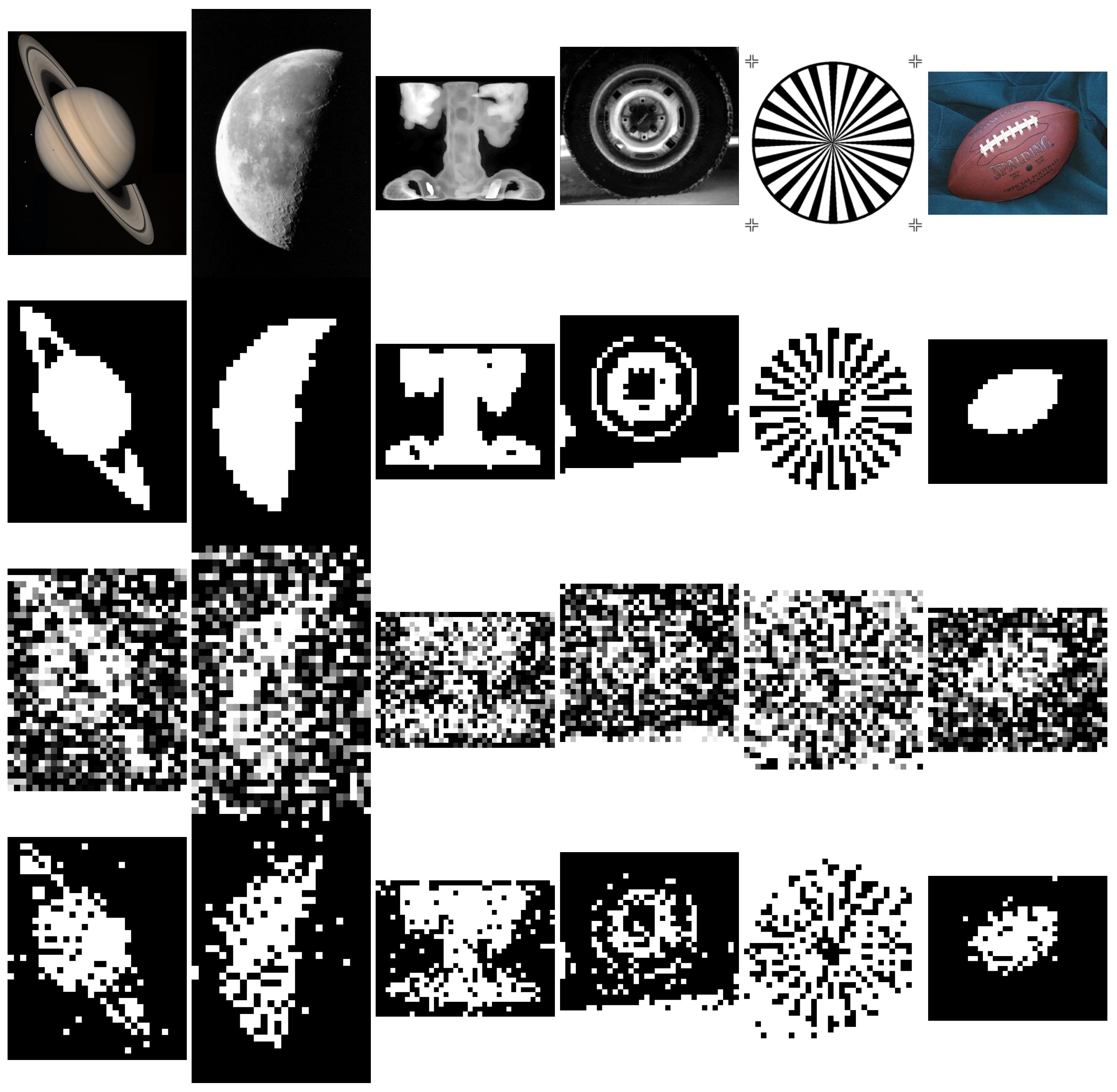

| Method | Objective | Var. | Number of Var. | Time Complexity |
|---|---|---|---|---|
| Chol. [46], 1996 | ||||
| PDcone [29], 2003 | ||||
| HBNB [44], 2020 | ||||
| PGML [45], 2020 | ||||
| SGML [17], 2022 | ||||
| MSGL [31], 2021 | Equation (29) | P | ||
| MSGL+ (prop.) | Equation (9) |
| Binary Classification Dataset | |
|---|---|
| Australian | (690, 14) |
| Breast-cancer | (683, 10) |
| Diabetes | (768, 8) |
| Fourclass | (862, 2) |
| German | (1000, 24) |
| Haberman | (306, 3) |
| Heart | (270, 13) |
| ILPD | (583, 10) |
| Liver-disorders | (145, 5) |
| Monk1 | (556, 6) |
| Pima | (768, 8) |
| Planning | (182, 12) |
| Voting | (435, 16) |
| WDBC | (569, 30) |
| Sonar | (208, 60) |
| Madelon | (2200, 500) |
| Colon-cancer | (62, 2000) |
| Binary Dataset | Baseline | Chol. | PDcone | HBNB | SGML | PGML | MSGL | MSGL+ |
|---|---|---|---|---|---|---|---|---|
| Australian | 85.16 | 85.48 | 85.88 | 85.27 | 84.58 | 84.65 | 85.81 | 85.88 |
| Breast-cancer | 97.18 | 97.73 | 97.88 | 97.77 | 97.69 | 97.51 | 97.14 | 97.36 |
| Diabetes | 71.32 | 74.61 | 74.74 | 74.32 | 73.01 | 73.70 | 75.94 | 76.04 |
| Fourclass | 77.75 | 76.83 | 77.49 | 77.58 | 77.96 | 77.76 | 77.20 | 77.23 |
| German | 70.00 | 75.90 | 74.33 | 70.00 | 71.88 | 70.00 | 75.15 | 75.45 |
| Haberman | 73.92 | 73.18 | 73.92 | 73.75 | 73.10 | 73.76 | 73.67 | 73.18 |
| Heart | 84.44 | 84.91 | 84.63 | 82.87 | 81.39 | 83.70 | 84.44 | 84.26 |
| ILPD | 71.32 | 71.41 | 71.37 | 71.32 | 71.37 | 71.32 | 71.37 | 71.45 |
| Liver-disorders | 71.72 | 70.52 | 72.07 | 71.55 | 71.90 | 72.76 | 72.07 | 72.93 |
| Monk1 | 72.13 | 88.09 | 77.84 | 71.37 | 66.78 | 73.35 | 77.66 | 80.00 |
| Pima | 70.88 | 76.38 | 75.95 | 75.24 | 74.00 | 74.13 | 76.67 | 76.80 |
| Planning | 71.34 | 71.34 | 71.34 | 71.34 | 70.79 | 71.34 | 71.34 | 71.34 |
| Voting | 90.00 | 95.69 | 95.69 | 95.92 | 95.69 | 96.20 | 94.31 | 94.31 |
| WDBC | 95.04 | 97.93 | 97.89 | 96.44 | 96.44 | 95.30 | 96.49 | 96.53 |
| Sonar | 73.30 | 81.11 | 84.49 | 75.46 | 73.06 | 73.30 | 79.55 | 79.43 |
| number of best | 2 | 6 | 4 | 1 | 1 | 2 | 1 | 6 |
| average | 78.37 | 81.41 | 81.03 | 79.35 | 78.64 | 79.25 | 80.59 | 80.81 |
| Multi-Class Classification Dataset | |
|---|---|
| cleveland | (303, 13, 5) |
| glass | (214, 9, 6) |
| iris | (150, 4, 3) |
| new-thyroid | (215, 5, 3) |
| tae | (151, 5, 3) |
| winequality-red | (1599, 11, 6) |
| Multi-Class Classification Dataset | Baseline | Chol. | PDcone | MSGL | MSGL+ |
|---|---|---|---|---|---|
| cleveland | 55.58 | 60.37 | 59.22 | 55.58 | 58.72 |
| glass | 59.94 | 63.46 | 64.05 | 59.94 | 66.85 |
| iris | 84.83 | 86.00 | 85.33 | 85.33 | 85.33 |
| new-thyroid | 87.33 | 91.98 | 91.40 | 92.21 | 92.33 |
| tae | 55.57 | 55.91 | 55.57 | 55.57 | 54.60 |
| winequality-red | 56.69 | 58.51 | 58.51 | 56.69 | 58.59 |
| number of best | 0 | 3 | 0 | 0 | 3 |
| average | 66.66 | 69.37 | 69.01 | 67.55 | 69.40 |
| Dataset | Baseline | Chol. | PDcone | HBNB | SGML | PGML | MSGL | MSGL+ |
|---|---|---|---|---|---|---|---|---|
| cameraman | 81.70 | 83.61 | 83.73 | 83.14 | 82.56 | 82.54 | 84.00 | 83.91 |
| saturn | 80.08 | 82.28 | 82.28 | 81.42 | 81.36 | 81.51 | 83.26 | 83.24 |
| moon | 80.85 | 82.78 | 82.96 | 82.11 | 82.29 | 82.17 | 83.27 | 83.29 |
| spine | 73.36 | 71.74 | 72.63 | 73.22 | 70.97 | 72.92 | 74.79 | 74.77 |
| tire | 74.71 | 77.51 | 76.92 | 75.00 | 77.22 | 75.47 | 79.75 | 79.96 |
| rice | 76.56 | 76.46 | 76.52 | 76.56 | 76.52 | 76.56 | 77.03 | 77.07 |
| testpat1 | 72.59 | 76.90 | 75.56 | 73.22 | 74.41 | 73.26 | 77.91 | 77.80 |
| canoe | 83.04 | 83.31 | 83.04 | 83.04 | 83.02 | 83.04 | 88.85 | 88.93 |
| AT3_1m4_02 | 88.34 | 89.02 | 88.86 | 89.00 | 88.49 | 88.40 | 89.15 | 89.17 |
| fabric | 76.43 | 76.37 | 76.62 | 76.56 | 75.83 | 76.64 | 76.83 | 76.87 |
| gantrycrane | 69.21 | 69.72 | 69.61 | 68.99 | 68.60 | 69.15 | 70.85 | 70.79 |
| eight | 77.25 | 77.40 | 77.25 | 77.25 | 77.23 | 77.25 | 78.34 | 78.45 |
| circuit | 75.45 | 75.00 | 75.74 | 75.59 | 74.79 | 75.51 | 76.23 | 76.21 |
| mri | 79.82 | 80.55 | 80.68 | 80.63 | 80.35 | 80.55 | 80.68 | 80.68 |
| paper1 | 73.03 | 72.87 | 73.07 | 73.28 | 70.92 | 73.65 | 74.12 | 74.10 |
| football | 82.79 | 85.11 | 84.05 | 82.89 | 82.89 | 83.10 | 90.19 | 90.21 |
| glass | 74.54 | 74.71 | 74.81 | 74.90 | 74.71 | 74.52 | 75.73 | 75.77 |
| pears | 70.97 | 71.66 | 71.48 | 70.91 | 70.99 | 70.83 | 73.04 | 72.88 |
| concordaerial | 79.94 | 80.04 | 80.16 | 80.22 | 80.14 | 79.88 | 80.24 | 80.22 |
| autumn | 87.62 | 88.46 | 87.78 | 87.68 | 85.69 | 88.46 | 87.80 | 87.80 |
| best count | 0 | 1 | 1 | 0 | 0 | 1 | 10 | 10 |
| average | 77.91 | 78.78 | 78.69 | 78.28 | 77.95 | 78.27 | 80.10 | 80.11 |
| Time-Series Dataset | |
|---|---|
| CanadaVehicle | (218, 24) |
| CanadaVote | (340, 3154) |
| USVote | (100, 1320) |
| Method | Lasso | Lasso + MSGL | Lasso + MSGL+ |
|---|---|---|---|
| CanadaVehicle | 55.74 | 60.12 | 80.43 |
| CanadaVote | 94.90 | 95.33 | 95.33 |
| USVote | 85.54 | 88.84 | 92.44 |
| number of best | 0 | 1 | 3 |
| average | 78.73 | 81.43 | 89.4 |
| Experiment Type | Binary (Section 3.1)/Multi-Class Classification (Section 3.2), and Binary Image Denoising (Section 3.3) | Time-Series Classification (Section 3.4) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| method | baseline | Chol. | PDcone | HBNB | SGML | PGML | MSGL | Lasso | Lasso +MSGL |
| 5% SL | 36.34% | 16.59% | 16.83% | 28.86% | 33.71% | 31.43% | 4.88% | 53.33% | 40.00% |
| 25% SL | 61.22% | 38.78% | 41.95% | 52.86% | 57.71% | 52.86% | 15.37% | 56.67% | 40.00% |
| 50% SL | 77.56% | 54.63% | 60.24% | 69.14% | 74.29% | 69.14% | 32.44% | 56.67% | 40.00% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.; Zheng, F.; Zou, Y.; Xue, L.; Jiang, C.; Liu, S.; Zhao, B.; Cui, H. MSGL+: Fast and Reliable Model Selection-Inspired Graph Metric Learning. Electronics 2024, 13, 44. https://doi.org/10.3390/electronics13010044
Yang C, Zheng F, Zou Y, Xue L, Jiang C, Liu S, Zhao B, Cui H. MSGL+: Fast and Reliable Model Selection-Inspired Graph Metric Learning. Electronics. 2024; 13(1):44. https://doi.org/10.3390/electronics13010044
Chicago/Turabian StyleYang, Cheng, Fei Zheng, Yujie Zou, Liang Xue, Chao Jiang, Shuangyu Liu, Bochao Zhao, and Haoyang Cui. 2024. "MSGL+: Fast and Reliable Model Selection-Inspired Graph Metric Learning" Electronics 13, no. 1: 44. https://doi.org/10.3390/electronics13010044
APA StyleYang, C., Zheng, F., Zou, Y., Xue, L., Jiang, C., Liu, S., Zhao, B., & Cui, H. (2024). MSGL+: Fast and Reliable Model Selection-Inspired Graph Metric Learning. Electronics, 13(1), 44. https://doi.org/10.3390/electronics13010044






