Bidirectional Efficient Attention Parallel Network for Segmentation of 3D Medical Imaging
Abstract
1. Introduction
- A novel Supreme Channel Attention (SCA) mechanism has been designed. By integrating channel information from two types of pooling operations, SCA maximizes the extraction of channel features. It also addresses the issue of errors introduced by spatial gaps between slices in previous attention networks for 3D medical images. This results in a significant performance enhancement for the network.
- A Parallel Spatial Attention mechanism is proposed to synergistically collaborate with Supreme Channel Attention (SCA). In particular, we concatenate together the results after running max pooling and average pooling in parallel and added the previous input block to the feature block that came out later. This aims to extract more detailed spatial information, which can be used to assist the network in making more accurate segmentation predictions.
- The proposed BEAP-Net adopts a configuration with one encoder and three slightly different decoders, constrained by consistency loss to ensure consistent outputs for a given input. This paper designs a cyclic pseudo-labeling scheme that utilizes prediction outputs from three different decoders. These predicted biases are then transformed into auxiliary supervision signals to facilitate model training. We aim for these signals to learn more information by leveraging their differences, thereby achieving performance enhancement.
- This paper conducts detailed ablation studies on various improvements of the network to better understand their impact on network performance.
- The BEAP-Net is evaluated on datasets from two different domains and compared with several state-of-the-art semi-supervised methods. The experiments demonstrate that the proposed approach achieves the most efficient performance with good generalization and robustness.
2. Related Work
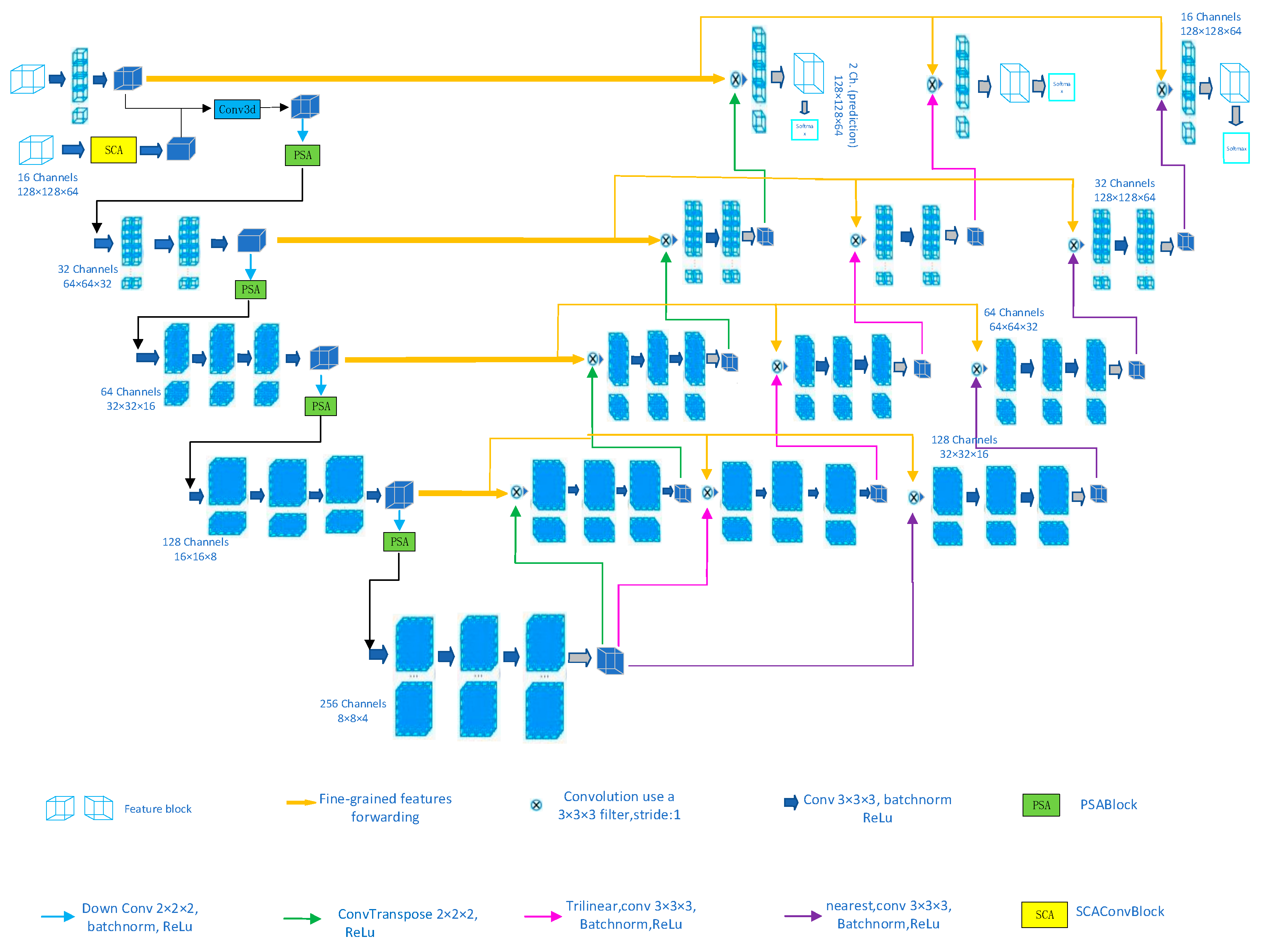
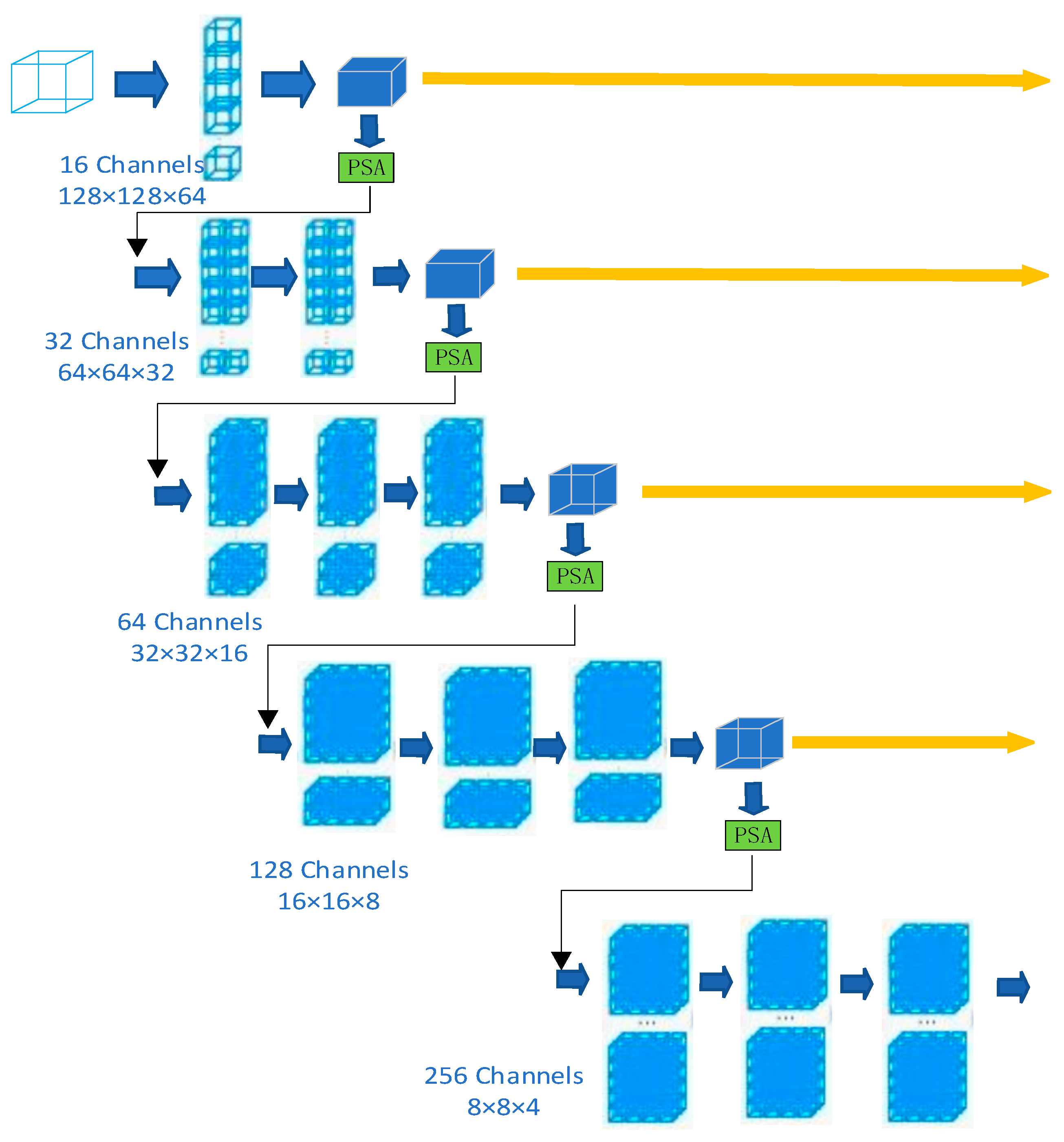
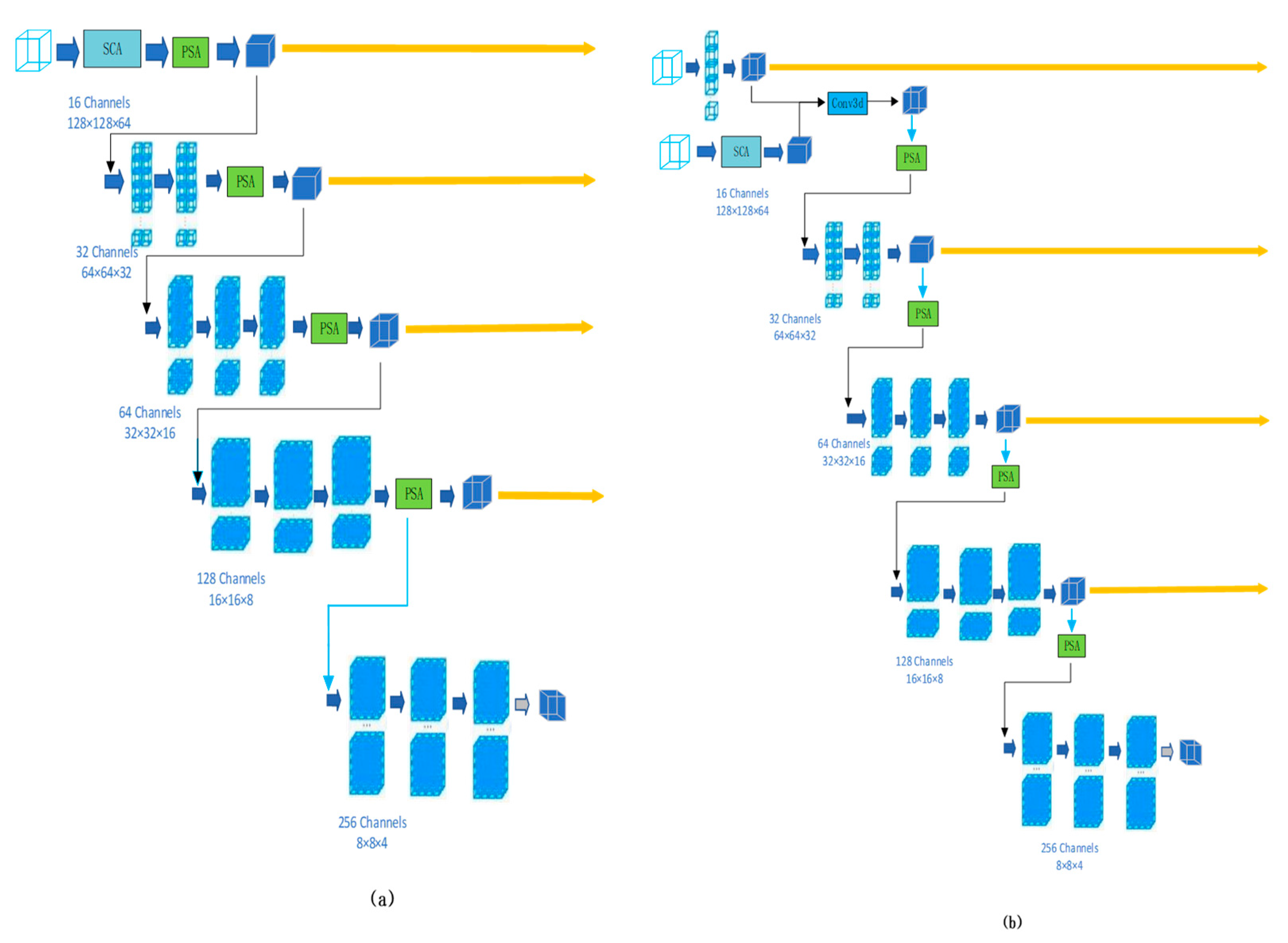
3. Methods
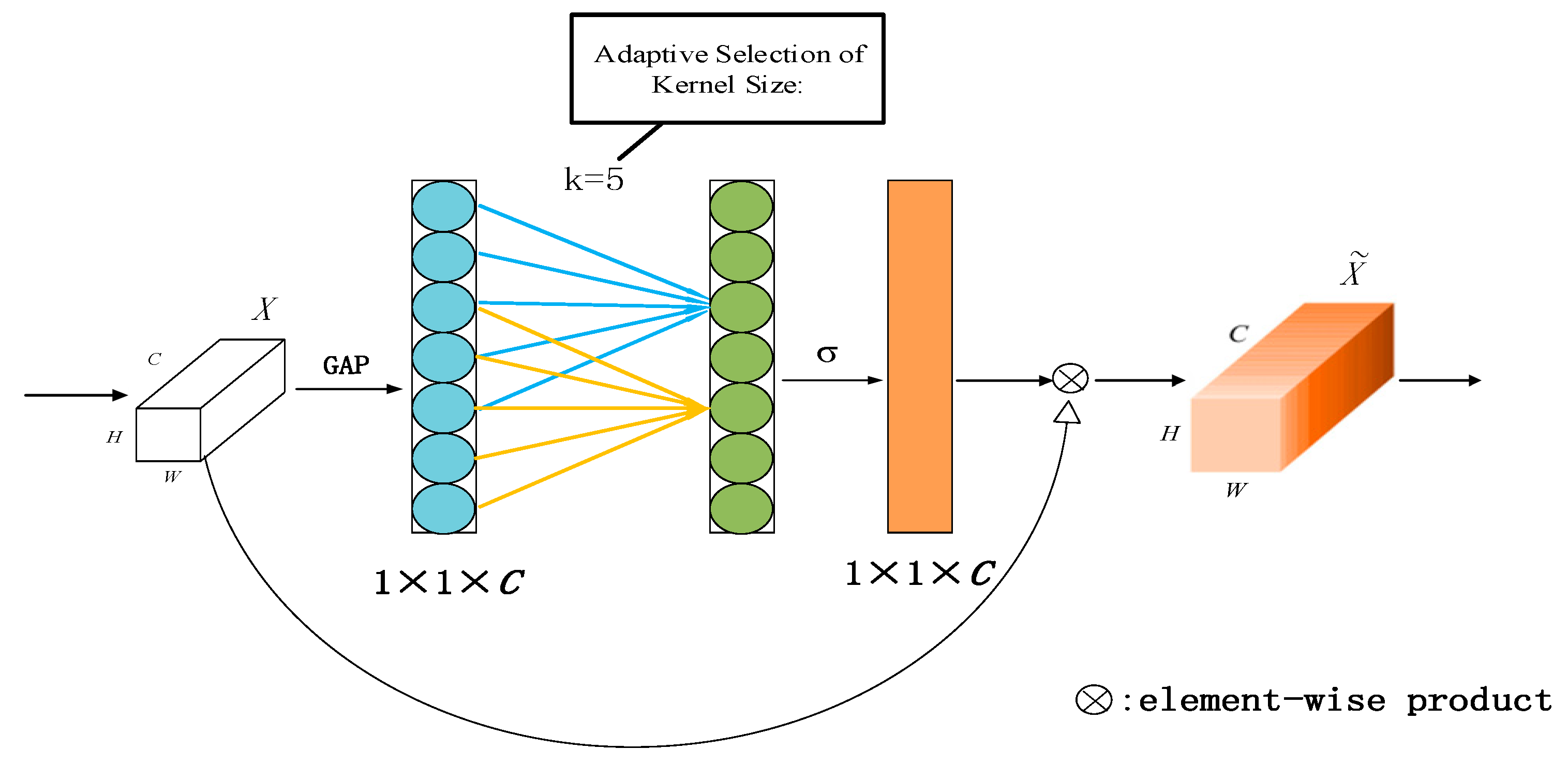
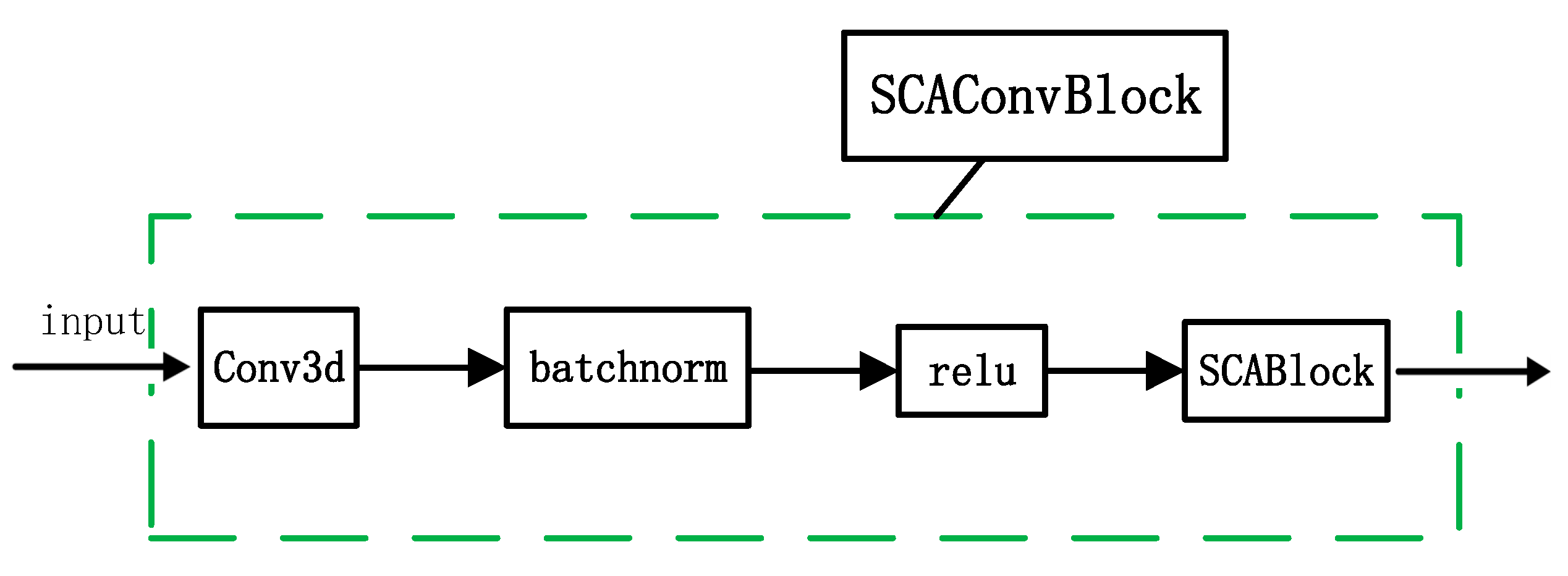
3.1. Supreme Channel Attention
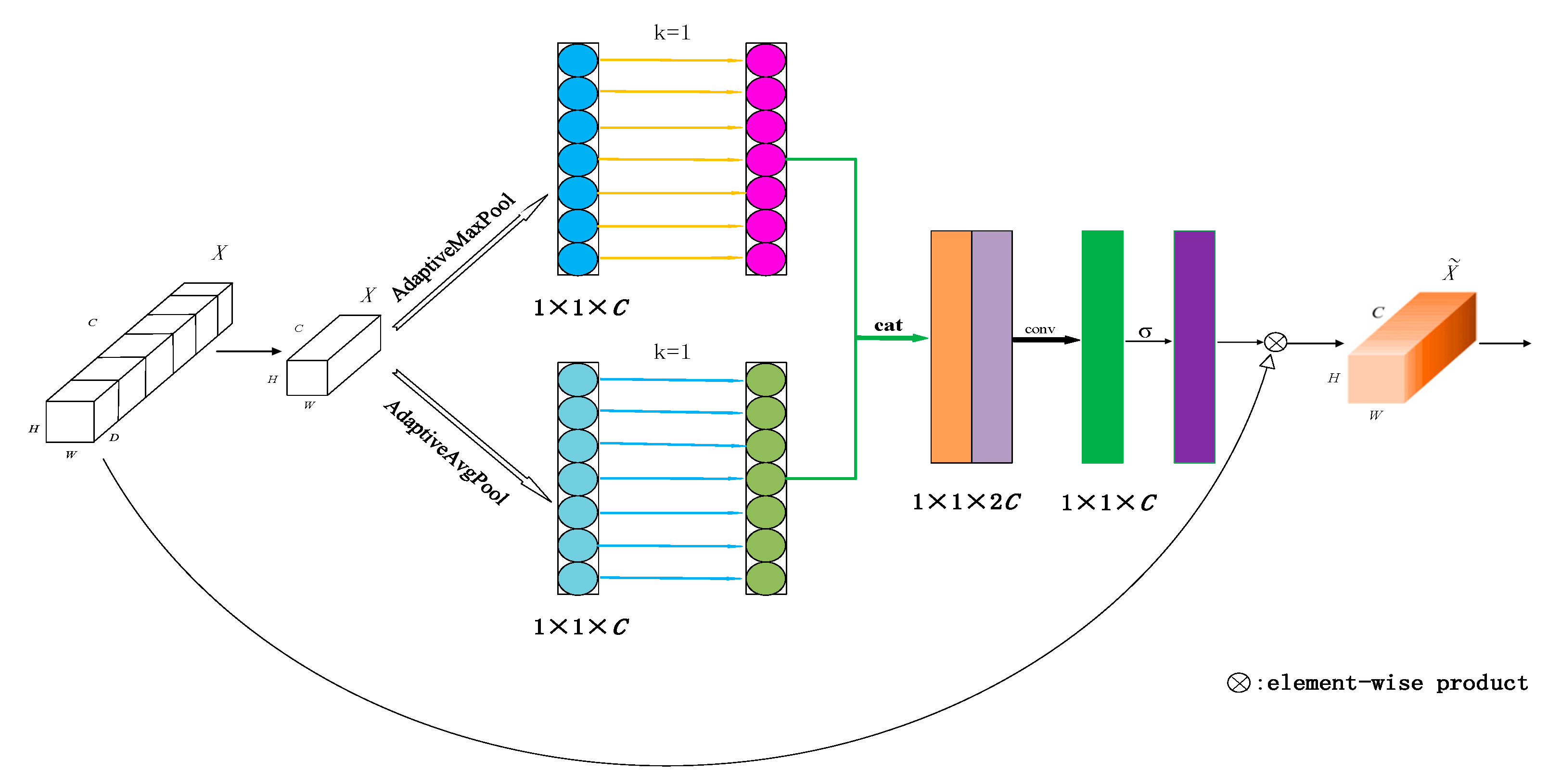
3.2. Parallel Spatial Attention
3.3. Triple Consistency Training
4. Experiment
4.1. Implementation Details
4.2. Results and Comparison with SOTA
4.3. Ablation Study
4.3.1. Effectiveness of Supreme Channel Attention
4.3.2. Effectiveness of Parallel Spatial Attention
4.3.3. Ablation Study of Triple Consistency Training
4.4. Robustness and Generalization
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Ganin, Y.; Lempitsky, V. Unsupervised domain adaptation by backpropagation. In Proceedings of the International Conference on Machine Learning, PMLR, Montreal, QC, Canada, 11–12 December 2015; pp. 1180–1189. [Google Scholar]
- Yang, J.; Dvornek, N.C.; Zhang, F.; Chapiro, J.; Lin, M.; Duncan, J.S. Unsupervised domain adaptation via disentangled representations: Application to cross-modality liver segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 255–263. [Google Scholar]
- Zhao, Z.; Xu, K.; Li, S.; Zeng, Z.; Guan, C. Mt-uda: Towards unsupervised cross-modality medical image segmentation with limited source labels. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 293–303. [Google Scholar]
- Luo, X.; Chen, J.; Song, T.; Wang, G. Semi-supervised medical image segmentation through dual-task consistency. Proc. AAAI Conf. Artif. Intell. 2021, 35, 8801–8809. [Google Scholar] [CrossRef]
- Luo, X.; Hu, M.; Song, T.; Wang, G.; Zhang, S. Semi-supervised medical image segmentation via cross teaching between cnn and transformer. In Proceedings of the International Conference on Medical Imaging with Deep Learning, Lübeck, Germany, 7–9 July 2021. [Google Scholar]
- Bai, W.; Oktay, O.; Sinclair, M.; Suzuki, H.; Rajchl, M.; Tarroni, G.; Glocker, B.; King, A.; Matthews, P.M.; Rueckert, D. Semisupervised learning for network-based cardiac mr image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Quebec City, QC, Canada, 10–14 September 2017; Springer: Berlin/Heidelberg, Germany, 2017; pp. 253–260. [Google Scholar]
- Basak, H.; Ghosal, S.; Sarkar, R. Addressing class imbalance in semi-supervised image segmentation: A study on cardiac mri. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Vancouver, BC, Canada, 8–12 October 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 224–233. [Google Scholar]
- You, C.; Zhou, Y.; Zhao, R.; Staib, L.; Duncan, J.S. SimCVD: Simple contrastive voxel-wise representation distillation for semi-supervised medical image segmentation. IEEE Trans. Med. Imaging 2022, 41, 2228–2237. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xu, M.; Ge, Z.; Cai, J.; Zhang, L. Semi-supervised left atrium segmentation with mutual consistency training. In Medical Image Computing and Computer Assisted Intervention, Proceedings of the MICCAI 2021 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part II, volume 12902 of Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2021; p. 297306. [Google Scholar]
- Lyu, F.; Ye, M.; Carlsen, J.F.; Erleben, K.; Darkner, S.; Yuen, P.C. Pseudo-label guided image synthesis for semi-supervised covid-19 pneumonia infection segmentation. IEEE Trans. Med. Imaging 2022, 42, 797–809. [Google Scholar] [CrossRef] [PubMed]
- Seibold, C.M.; Reiß, S.; Kleesiek, J.; Stiefelhagen, R. Reference-guided pseudo-label generation for medical semantic segmentation. Proc. AAAI Conf. Artif. Intell. 2022, 36, 2171–2179. [Google Scholar] [CrossRef]
- Jin, Q.; Cui, H.; Sun, C.; Zheng, J.; Wei, L.; Fang, Z.; Meng, Z.; Su, R. Semisupervised histological image segmentation via hierarchical consistency enforcement. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Vancouver, BC, Canada, 8–12 October 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 3–13. [Google Scholar]
- Hang, W.; Feng, W.; Liang, S.; Yu, L.; Wang, Q.; Choi, K.-S.; Qin, J. Local and global structure-aware entropy regularized mean teacher model for 3d left atrium segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 562–571. [Google Scholar]
- Vesal, S.; Gu, M.; Kosti, R.; Maier, A.; Ravikumar, N. Adapt everywhere: Unsupervised adaptation of point-clouds and entropy minimization for multi-modal cardiac image segmentation. IEEE Trans. Med. Imaging 2021, 40, 1838–1851. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, Y.; Tang, P.; Bai, S.; Shen, W.; Fishman, E.; Yuille, A. Semi-supervised 3d abdominal multi-organ segmentation via deep multi-planar co-training. In Proceedings of the 2019 IEEE Winter Conference on Applications of Computer Vision (WACV), Waikoloa Village, HI, USA, 7–11 January 2019; pp. 121–140. [Google Scholar]
- Wang, G.; Zhai, S.; Lasio, G.; Zhang, B.; Yi, B.; Chen, S.; Macvittie, T.J.; Metaxas, D.; Zhou, J.; Zhang, S. Semi-supervised segmentation of radiation-induced pulmonary fibrosis from lung ct scans with multi-scale guided dense attention. IEEE Trans. Med. Imaging 2022, 41, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.-P.; Zhou, T.; Ji, G.-P.; Zhou, Y.; Chen, G.; Fu, H.; Shen, J.; Shao, L. Inf-Net: Automatic covid-19 lung infection segmentation from ct images. IEEE Trans. Med. Imaging 2020, 39, 2626–2637. [Google Scholar] [CrossRef] [PubMed]
- French, G.; Laine, S.; Aila, T.; Mackiewicz, M.; Finlayson, G. Semi-supervised semantic segmentation needs strong, varied perturbations. arXiv 2019, arXiv:1906.01916. [Google Scholar]
- Ouali, Y.; Hudelot, C.; Tami, M. Semisupervised semantic segmentation with cross-consistency training. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 12674–12684. [Google Scholar]
- Sohn, K.; Berthelot, D.; Li, C.-L.; Zhang, Z.; Carlini, N.; Cubuk, E.D.; Kurakin, A.; Zhang, H.; Raffel, C. Fixmatch: Simplifying semi-supervised learning with consistency and confidence. In Proceedings of the Advances in Neural Information Processing Systems 33 (NeurIPS 2020), Online, 6–12 December 2020; Volume 33, pp. 596–608. [Google Scholar]
- Lee, D.H. Pseudo-label: The simple and efficient semi-supervised learning method for deep neural networks. In Proceedings of the ICML 2013, Atlanta, GA, USA, 16–21 June 2013; Volume 3. [Google Scholar]
- Tarvainen, A.; Valpola, H. Mean teachers are better role models: Weight-averaged consistency targets improve semi-supervised deep learning results. Adv. Neural Inf. Process. Syst. 2017, 30, 1195–1204. [Google Scholar]
- Xie, Q.; Luong, M.T.; Hovy, E.; Le, Q.V. Self-training with noisy student improves imagenet classification. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 10687–10698. [Google Scholar]
- Wang, K.; Zhan, B.; Zu, C.; Wu, X.; Zhou, J.; Zhou, L.; Wang, Y. Tripled-uncertainty guided mean teacher model for semi-supervised medical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 450–460. [Google Scholar]
- Mnih, V.; Heess, N.; Graves, A.; Kavukcuoglu, K. Recurrent Models of Visual Attention. In Advances in Neural Information Processing Systems (NIPS); Curran Associates, Inc.: Red Hook, NY, USA, 2014; Volume 27, pp. 2204–2212. [Google Scholar]
- Ba, J.; Mnih, V.; Kavukcuoglu, K. Multiple object recognition with visual attention. arXiv 2014, arXiv:1406.5679. [Google Scholar]
- Xu, K.; Ba, J.; Kiros, R.; Cho, K.; Courville, A.; Salakhudinov, R.; Zemel, R.; Bengio, Y. Show, attend and tell: Neural image caption generation with visual attention. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 2048–2057. [Google Scholar]
- Wang, Q.; Wu, B.; Wu, X.; Qiao, Y. ECA-Net: Efficient channel attention for deep convolutional neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 11534–11542. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.S. CBAM: Convolutional block attention module. Proc. Eur. Conf. Comput. Vis. (ECCV) 2018, 3, 3–19. [Google Scholar]
- Berthelot, D.; Carlini, N.; Goodfellow, I.; Papernot, N.; Oliver, A.; Raffel, C.A. Mixmatch: A holistic approach to semi-supervised learning. Adv. Neural Inf. Process. Syst. 2019, 32, 5049–5505. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Xie, S.; Girshick, R.B.; Dollár, P.; Tu, Z.; He, K. Aggregated residual transformations for deep neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 5987–5995. [Google Scholar]
- Chen, Y.; Kalantidis, Y.; Li, J.; Yan, S.; Feng, J. A2-Nets: Double attention networks. In Proceedings of the Conference on Neural Information Processing Systems (NeurIPS), Montreal, QC, Canada, 3–8 December 2018. [Google Scholar]
- Gao, Z.; Xie, J.; Wang, Q.; Li, P. Global second-order pooling convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 3242–3251. [Google Scholar]
- Fu, J.; Liu, J.; Tian, H.; Li, Y.; Bao, Y.; Fang, Z.; Lu, H. Dual attention network for scene segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 5206–5215. [Google Scholar]
- Zagoruyko, S.; Komodakis, N. Paying more attention to attention: Improving the performance of convolutional neural networks via attention transfer. In Proceedings of the International Conference on Learning Representations (ICLR), Toulon, France, 24–26 April 2017. [Google Scholar]
- Yu, L.; Wang, S.; Li, X.; Fu, C.W.; Heng, P.A. Uncertainty-Aware Self-Ensembling Model for Semi-Supervised 3D Left Atrium Segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, 13–17 October 2019. [Google Scholar]
- Kendall, A.; Gal, Y. What uncertainties do we need in bayesian deep learning for computer vision? arXiv 2017, arXiv:1703.04977. [Google Scholar]
- Xie, Q.; Dai, Z.; Hovy, E.; Luong, M.T.; Le, Q.V. Unsupervised data augmentation for consistency training. arXiv 2019, arXiv:1904.12848. [Google Scholar]
- Zhang, Y.; Xiang, T.; Hospedales, T.M.; Lu, H. Deep Mutual Learning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4320–4328. [Google Scholar]
- Xia, Y.; Liu, F.; Yang, D.; Cai, J.; Yu, L.; Zhu, Z.; Xu, D.; Yuille, A.; Roth, H. 3D semi-supervised learning with uncertainty-aware multi-view cotraining. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Snowmass Village, CO, USA, 1–5 March 2020; pp. 3646–3655. [Google Scholar]
- Xiong, Z.; Xia, Q.; Hu, Z.; Huang, N.; Bian, C.; Zheng, Y.; Vesal, S.; Ravikumar, N.; Maier, A.; Yang, X.; et al. A global benchmark of algorithms for segmenting the left atrium from late gadolinium-enhanced cardiac magnetic resonance imaging. Med. Image Anal. 2021, 67, 101832. [Google Scholar] [CrossRef] [PubMed]
- Laine, S.; Aila, T. Temporal ensembling for semi-supervised learning. arXiv 2016, arXiv:1610.02242. [Google Scholar]
- Li, S.; Zhang, C.; He, X. Shape-Aware Semi-Supervised 3D Semantic Segmentation for Medical Images. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, 4–8 October 2020; Martel, A.L., Ed.; Springer: Berlin/Heidelberg, Germany; Volume 12261, pp. 455–463. [Google Scholar]
- Zheng, H.; Lin, L.; Hu, H. Semi-supervised segmentation of liver using adversarial learning with deep atlas prior. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2019: 22nd International Conference, Shenzhen, China, 13–17 October 2019; Shen, D., Ed.; Springer: Cham, Switzerland, 2019; pp. 148–156. [Google Scholar] [CrossRef]
- Bai, Y.; Chen, D.; Li, Q.; Shen, W.; Wang, Y. Bidirectional copy-paste for semi-supervised medical image segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Vancouver, BC, Canada, 18–19 June 2023; pp. 1234–1244. [Google Scholar]
- Wang, Y.; Zhang, Y.; Tian, J.; Zhong, C.; Shi, Z.; Zhang, Y. Double-uncertainty weighted method for semi-supervised learning. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, 4–8 October 2020; Martel, A.L., Ed.; Springer: Berlin/Heidelberg, Germany; Cham, Switzerland, 2020; pp. 542–551. [Google Scholar] [CrossRef]
- Roth, H.R.; Lu, L.; Farag, A.; Shin, H.-C.; Liu, J.; Turkbey, E.B.; Summers, R.M. Deeporgan: Multi-level deep convolutional networks for automated pancreas segmentation. In Medical Image Computing and Computer-Assisted Intervention, Proceedings of the MICCAI 2015 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part I, volume 9349 of Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2015; pp. 556–564. [Google Scholar]
- Shi, Y.; Zhang, J.; Ling, T.; Lu, J.; Zheng, Y.; Yu, Q.; Qi, L.; Gao, Y. Inconsistency-aware uncertainty estimation for semi-supervised medical image segmentation. IEEE Trans. Med. Imaging 2022, 41, 608–620. [Google Scholar] [CrossRef] [PubMed]

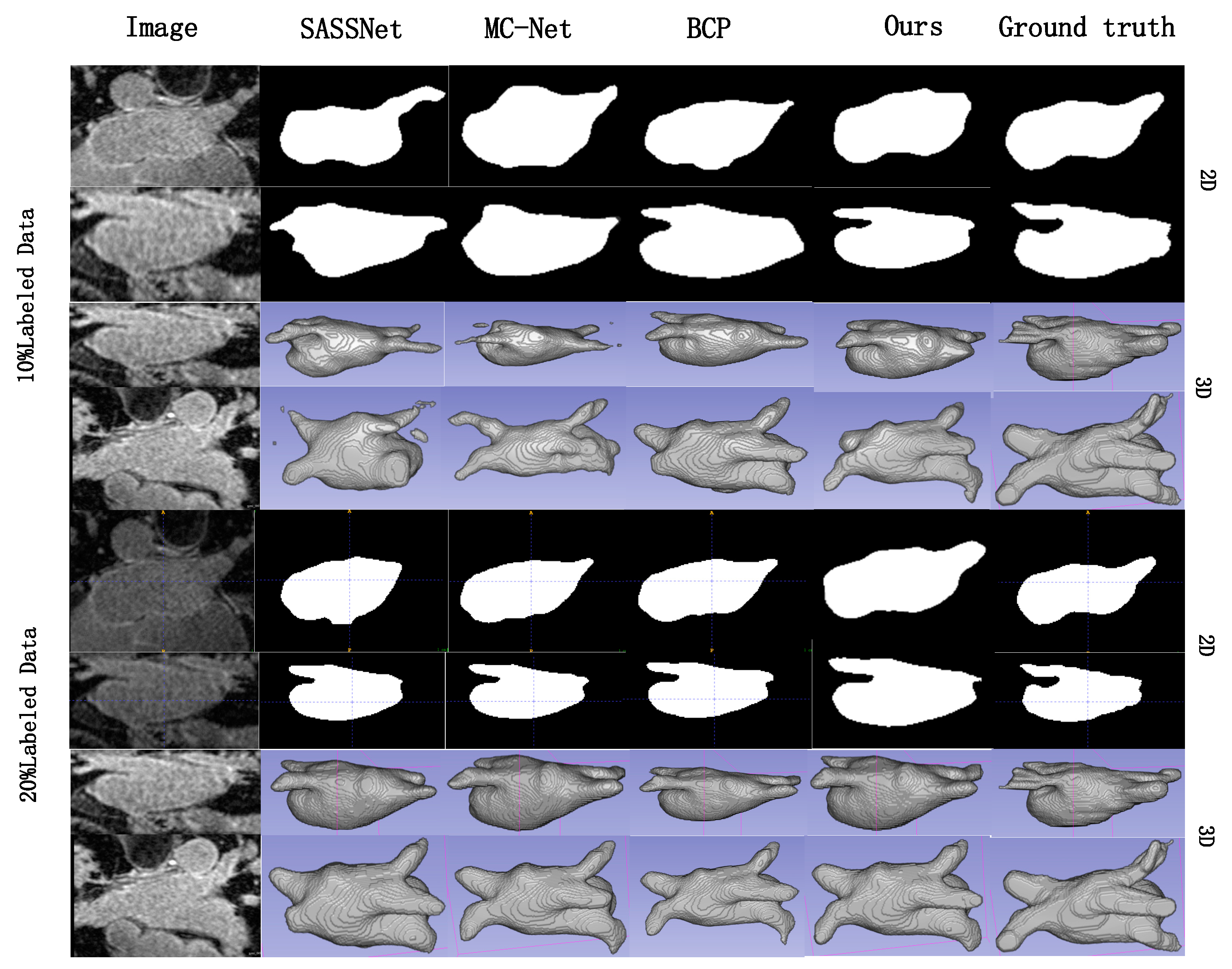


| Method | Labeled | Metrics | |||
|---|---|---|---|---|---|
| Dice ↑ | Jaccard ↑ | HD95 ↓ (Voxel) | ASD ↓ (Voxel) | ||
| V-Net | 8 (10%) | 79.99 | 68.12 | 21.11 | 5.48 |
| V-Net | 16 (20%) | 86.10 | 76.09 | 13.29 | 3.19 |
| V-Net | 80 (All) | 91.14 | 83.82 | 5.75 | 1.52 |
| DAP [48] | 81.88 | 71.23 | 15.81 | 3.79 | |
| UA-MT [43] | 84.23 | 73.48 | 13.84 | 3.35 | |
| SASSNet [47] | 87.31 | 77.72 | 9.61 | 2.55 | |
| LG-ER-MT [16] | 85.53 | 75.12 | 13.29 | 3.77 | |
| DUWM [50] | 8 (10%) | 85.90 | 75.74 | 12.67 | 3.31 |
| DTC [7] | 86.57 | 76.55 | 14.47 | 3.74 | |
| MC-Net [12] | 87.70 | 78.30 | 9.37 | 2.18 | |
| BEAP-Net (Ours) | 88.95 ↑ 1.25 | 80.18 ↑ 1.88 | 8.90 ↓ 0.47 | 1.94 ↓ 0.24 | |
| DAP [48] | 87.89 | 78.72 | 9.29 | 2.74 | |
| UA-MT [43] | 88.87 | 80.21 | 7.32 | 2.26 | |
| SASSNet [47] | 89.54 | 81.24 | 8.24 | 2.20 | |
| LG-ER-MT [16] | 89.63 | 81.30 | 7.16 | 2.06 | |
| DUWM [50] | 16 (20%) | 89.65 | 81.33 | 7.04 | 2.03 |
| DTC [7] | 89.42 | 80.97 | 7.32 | 2.10 | |
| BCP [49] | 90.56 | 82.69 | 6.03 | 1.71 | |
| MC-Net [12] | 90.34 | 82.50 | 6.01 | 1.75 | |
| BEAP-Net (Ours) | 91.52 ↑ 0.96 | 84.41 ↑ 1.72 | 5.08 ↓ 0.93 | 1.39 ↓ 0.32 | |
| Method | Labeled | Metrics | |||
|---|---|---|---|---|---|
| Dice ↑ | Jaccard ↑ | HD95 ↓ (Voxel) | ASD ↓ (Voxel) | ||
| V-Net | 8 (10%) | 82.72 | 71.73 | 13.33 | 3.27 |
| V-Net | 16 (20%) | 86.10 | 76.09 | 13.29 | 3.19 |
| V-Net | 80 (All) | 91.47 | 84.35 | 5.49 | 1.52 |
| V-Net+SE | 8 (10%) | 86.04 | 76.06 | 14.25 | 3.51 |
| V-Net+SE | 16 (20%) | 90.72 | 82.74 | 7.66 | 2.36 |
| V-Net+SCA | 8 (10%) | 90.69 | 82.56 | 7.78 | 2.42 |
| V-Net+SCA | 16 (20%) | 91.33 | 83.79 | 5.78 | 1.57 |
| Method | Labeled | Metrics | |||
|---|---|---|---|---|---|
| Dice↑ | Jaccard ↑ | HD95 ↓ (Voxel) | ASD ↓ (Voxel) | ||
| V-Net | 8 (10%) | 82.72 | 71.73 | 13.33 | 3.27 |
| V-Net | 16 (20%) | 86.10 | 76.09 | 13.29 | 3.19 |
| V-Net | 80 (All) | 91.47 | 84.35 | 5.49 | 1.52 |
| V-Net+SA | 8 (10%) | 85.06 | 75.03 | 14.35 | 3.91 |
| V-Net+SA | 16 (20%) | 90.23 | 81.76 | 7.66 | 2.54 |
| V-Net+PSA | 8 (10%) | 90.56 | 81.47 | 7.98 | 2.62 |
| V-Net+PSA | 16 (20%) | 91.16 | 83.34 | 5.88 | 1.89 |
| Method | Labeled | Metrics | |||
|---|---|---|---|---|---|
| Dice ↑ | Jaccard ↑ | HD95 ↓ (Voxel) | ASD ↓ (Voxel) | ||
| V-Net | 8 (10%) | 82.72 | 71.73 | 13.33 | 3.27 |
| V2d-Net | 8 (10%) | 85.78 | 75.40 | 14.45 | 3.84 |
| V3-Net | 8 (10%) | 85.56 | 76.69 | 13.41 | 3.93 |
| V3d-Net | 8 (10%) | 87.55 | 78.24 | 9.95 | 2.18 |
| V2d-Net | 16 (20%) | 88.96 | 80.37 | 7.63 | 2.33 |
| V3-Net | 16 (20%) | 86.64 | 79.69 | 12.07 | 2.75 |
| V3d-Net | 16 (20%) | 90.79 | 83.21 | 5.79 | 1.97 |
| Method | Labeled | Metrics | |||
|---|---|---|---|---|---|
| Dice ↑ | Jaccard ↑ | HD95 ↓ (Voxel) | ASD ↓ (Voxel) | ||
| V-Net | 6 (10%) | 55.06 | 40.48 | 32.80 | 12.67 |
| V-Net | 12 (20%) | 69.65 | 55.18 | 20.19 | 6.31 |
| V-Net | 62 (All) | 83.01 | 71.35 | 5.18 | 1.19 |
| UA-MT [43] | 6 (10%) | 68.70 | 54.65 | 13.89 | 3.23 |
| SASSNet [47] | 6 (10%) | 66.52 | 52.23 | 17.11 | 2.25 |
| DTC [7] | 6 (10%) | 66.27 | 52.07 | 15.00 | 4.43 |
| MC-Net [12] | 6 (10%) | 68.94 | 54.74 | 16.28 | 3.16 |
| BEAP-Net (Ours) | 6 (10%) | 68.99 ↑ 0.05 | 54.86 ↑ 0.12 | 14.02 ↑ 0.13 | 2.09 ↓ 0.16 |
| UA-MT [43] | 12 (20%) | 76.77 | 63.77 | 11.41 | 2.79 |
| SASSNet [47] | 12 (20%) | 77.12 | 64.24 | 8.93 | 1.41 |
| DTC [7] | 12 (20%) | 78.27 | 64.75 | 8.37 | 2.27 |
| MC-Net [12] | 12 (20%) | 79.05 | 65.82 | 10.29 | 2.71 |
| BEAP-Net (Ours) | 12 (20%) | 79.11 ↑ 0.06 | 65.94 ↑ 0.12 | 8.50 ↑ 0.13 | 1.57 ↓ 0.16 |
| Method | Labeled | Metrics | |||
|---|---|---|---|---|---|
| Dice ↑ | Jaccard ↑ | HD95 ↓ (Voxel) | ASD ↓ (Voxel) | ||
| V-Net | 80 (All) | 91.47 | 84.36 | 5.48 | 1.51 |
| BEAP-Net | 8 (10%) | 88.95 | 80.18 | 8.90 | 1.94 |
| try1 | 8 (10%) | 87.56 | 78.69 | 10.41 | 2.18 |
| try2 | 8 (10%) | 85.55 | 76.24 | 12.95 | 3.93 |
| BEAP-Net | 16 (20%) | 91.52 | 84.41 | 5.08 | 1.39 |
| try1 | 16 (20%) | 91.34 | 84.40 | 5.37 | 1.48 |
| try2 | 16 (20%) | 91.27 | 83.99 | 5.75 | 1.73 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Xv, T.; Liu, J.; Li, J.; Yang, L.; Guo, J. Bidirectional Efficient Attention Parallel Network for Segmentation of 3D Medical Imaging. Electronics 2024, 13, 3086. https://doi.org/10.3390/electronics13153086
Wang D, Xv T, Liu J, Li J, Yang L, Guo J. Bidirectional Efficient Attention Parallel Network for Segmentation of 3D Medical Imaging. Electronics. 2024; 13(15):3086. https://doi.org/10.3390/electronics13153086
Chicago/Turabian StyleWang, Dongsheng, Tiezhen Xv, Jiehui Liu, Jianshen Li, Lijie Yang, and Jinxi Guo. 2024. "Bidirectional Efficient Attention Parallel Network for Segmentation of 3D Medical Imaging" Electronics 13, no. 15: 3086. https://doi.org/10.3390/electronics13153086
APA StyleWang, D., Xv, T., Liu, J., Li, J., Yang, L., & Guo, J. (2024). Bidirectional Efficient Attention Parallel Network for Segmentation of 3D Medical Imaging. Electronics, 13(15), 3086. https://doi.org/10.3390/electronics13153086






