Artificial Intelligence-Based Algorithms in Medical Image Scan Segmentation and Intelligent Visual Content Generation—A Concise Overview
Abstract
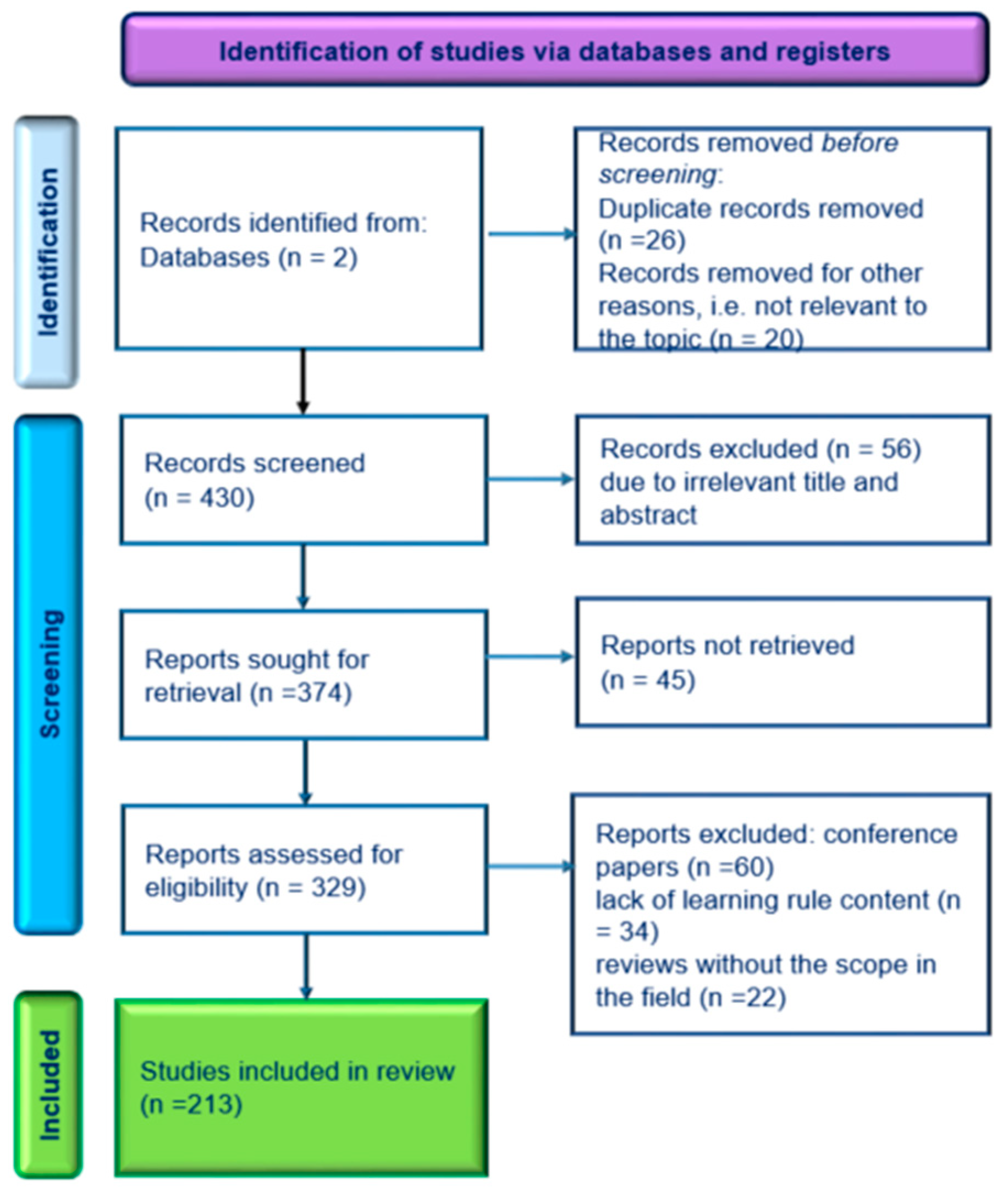
1. Introduction
2. Materials and Methods
3. Neural Communication
4. Taxonomy of Neural Network Applied in the Medical Image Segmentation Process
4.1. Convolutional Neural Network
4.2. Recurrent Neural Network
4.3. Spiking Neural Networks
5. Learning Algorithms
5.1. Backpropagation Algorithm
5.2. ANN–SNN Conversion
5.3. Supervised Hebbian Learning (SHL)
5.4. Reinforcement Learning with Supervised Models
5.5. Chronotron
5.6. Bio-Inspired Learning Algorithms
5.6.1. Spike Timing Dependent Plasticity
5.6.2. Spike-Driven Synaptic Plasticity
5.6.3. Tempotron Learning Rule
6. Neural Networks and Learning Algorithms in the Medical Image Segmentation Process
| Network Type | Neuron Model | Average Accuracy (%) | Datasets—Training/Testing/Validation Sets (%) or Training/Testing Sets (%) | Input Parameters | Learning Rule | Biological Plausibility | Ref. |
|---|---|---|---|---|---|---|---|
| ANN | Perceptron | 99.10 | Mammography images lack of information | Mammography images—33 features extracted by region of interest (ROI) | BP | low | [131] |
| CNN | Perceptron | 98.70 | Brain tumor, MRI color images 70/15/15 | MRI image scan, 12 features (mean, standard deviation (SD), entropy, energy, contract, homogeneity, correlation, variance, covariance, root mean square (RMS), skewness, kurtosis) | BP | low | [132] |
| CNN | Perceptron | 96.00 | Echocardiograms 60/40 | Disease classification, cardiac chamber segmentation, viewpoint classification in echocardiograms | lack of information | low | [133] |
| CNN | Perceptron | 94.58 | Brain tumor images 50/25/25 | Brain tumor images | lack of information | low | [134] |
| CNN | Perceptron | 91.10 | Simultaneous IVUS and OCT images | IVUS and OCT images | lack of information | low | [135] |
| CNN | Perceptron | 98.00 | 2D ultrasound 49/49/2 | Classification of the cardiac view into 7 classes | lack of information | low | [136] |
| CNN | Perceptron | 93.30 | Coronary cross-sectional images 80/20 | Detection of motion artifacts in coronary CCTA, classification of coronary cross-sectional images | lack of information | low | [137] |
| CNN | Perceptron | 99.00 | MRI image scan 60/40 | Bounding box localization of LV in short-axis MRI slices | lack of information | low | [138] |
| CNN and doc2vec | Perceptron | 96.00 | Continuous wave Doppler cardiac valve images 94/4/2 | Automatic generation of text for continuous wave Doppler cardiac valve images | lack of information | low | [139] |
| Deep CNN + complex data preparation | Perceptron | 97.00 | Vessel segmentation lack of information | Proposing a supervised segmentation technique that uses a deep neural network and structured prediction | lack of information | low | [140] |
| CNN and transformer encoders | Perceptron | 90.70 | Automated cardiac diagnosis challenge (ACDC), CT image scans from synapse 60/40 | CT image scans | BP | low | [141] |
| CNN and transformer encoders | Multilayer perceptron | 77.48 (Dice coefficient) | Multiorgan segmentation lack of information | CT image scans—synapse multiorgan segmentation dataset | BP | low | [142] |
| CNN and transformer encoders | Perception | 78.41 (Dice coefficient) | Multiorgan segmentation lack of information | CT image scans | BP | low | [99] |
| CNN and RNN | Perceptron | 95.24 (REs-Net50) 97.18(IncepnetV3) 98.03 (Dense-Net) | MRI image scan of the brain 80/20 | MRI image scan of the brain, modality, mask images | BP | low | [143] |
| CNN and RNN | Perceptron | 95.74 (REs-Net50) 97.14(DarkNet-53) | Skin image lack of information | Skin image | BP | low | [144] |
| SNN | LIF | 81.95 | Baseline T1-weighted whole-brain MRI image scan lack of information | Hippocampus section of MRI image scan | ANN–SNN conversion | low | [145] |
| SNN | LIF | 92.89 | Burn images lack of information | 256 × 256 burn image encoded into 24 × 256 × 256 feature maps | BP | low | [146] |
| SNN | LIF | 89.57 | Skin images (melanoma and non-melanoma) lack of information | Skin images converted into spikes using Poisson distribution | surrogated gradient descent | low | [147] |
| SNN | LIF | 99.60 | MRI scan of brain tumors 80/10/10 | 2D MRI scan of brain tumors | YO-LO-2-based transfer learning | low | [148] |
| SNN | LIF | 95.17 | Microscopic images of breast tumor lack of information | Microscopic images of breast tumor | SpikeProp | low | [149] |
| GAN | Perceptron | 83.70 (Dice coefficient) DRIVE dataset 82.70 (Dice coefficient) STARE dataset | Segmentation of retinal vessels lack of information | Dataset for retinal vessel segmentation: DRIVE dataset and STARE dataset | BP | low | [109] |
| GAN | Perceptron | 94.60 | Segmentation of the blood vessels of the retinal and the coronary and for the knee cartilage lack of information | Datasets for retinal vessel segmentation: DRIVE dataset and coronary dataset | BP | low | [110] |
| GAN | Perceptron | 90.71 (Dice coefficient) | Brain segmentation Brain data—MRI dataset 80/20 | Brain MRI image scan | BP | low | [109] |
7. Data Availability
8. Discussion
9. Limitations of the Study
10. Conclusions
11. Future Research Directions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| 3D | three-dimensional |
| XR | extended reality |
| VR | virtual reality |
| MR | mixed reality |
| AR | augmented reality |
| HDM | head-mounted display |
| AI | artificial intelligence |
| ML | machine learning |
| ANN | artificial neural network |
| SNN | spiking neural network |
| CNN | convolutional neural network |
| RNN | recurrent neural network |
| GAN | generative adversarial network |
| GNN | graphical neural network |
| BP | backpropagation |
| ReSuMe | reinforcement learning with supervised models |
| SHL | supervised Hebbian learning |
| STDP | spike timing-dependent plasticity |
| SDSP | spike-driven synaptic plasticity |
| SAM | segment anything model |
| YOLO | you only look once (algorithm) |
| spike-prop | supervised learning rule akin to traditional error backpropagation for a network of spiking neurons with reasonable postsynaptic potentials |
| ReLu | rectified linear unit activation function |
| MAE | mean absolute error |
| MSE | mean squared error |
| RMSE | root-mean-squared error |
| AUROC | area under receiver-operating curve |
| IoU | index of union |
| EHR | electronic health record |
| MRI | magnetic resonance imaging |
| CT | computer tomography |
| OCT | optical coherence tomography |
| IVUS | intravascular ultrasound |
| CCTA | coronary computed tomography angiography |
| LV | left ventricle |
| T1-weighted image | the basic pulse sequence in MRI, it shows the differences in the T1 relaxation times of tissue (T1 relaxation measures of how quickly the net magnetization vector recovers to its ground state) |
| ALTAI | Assessment List for Trustworthy Artificial Intelligence |
| ERPs | ethical risk points |
References
- Herculano-Houzel, S. The remarkable, yet not extraordinary, human brain as a scaled-up primate brain and its associated cost. Proc. Natl. Acad. Sci. USA 2012, 109, 10661–10668. [Google Scholar] [CrossRef]
- Shao, F.; Shen, Z. How can artificial neural networks approximate the brain? Front. Psychol. 2023, 13, 970214. [Google Scholar] [CrossRef]
- Moscato, V.; Napolano, G.; Postiglione, M.; Sperlì, G. Multi-task learning for few-shot biomedical relation extraction. Artif. Intell. Rev. 2023. online ahead of print. [Google Scholar] [CrossRef]
- Van Gerven, M. Computational Foundations of Natural Intelligence. Front. Comput. Neurosci. 2017, 11. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, J.; Gavrilova, M.; Rodolfo, F.; Kacprzyk, J. Brain-inspired systems (BIS): Cognitive foundations and applications. In Proceedings of the IEEE International Conference on Systems, Man, and Cybernetics (SMC) 2018, Miyazaki, Japan, 7–10 October 2018; pp. 991–996. [Google Scholar]
- Zhao, L.; Zhang, L.; Wu, Z.; Chen, Y.; Dai, H.; Yu, X.; Liu, Z.; Zhang, T.; Hu, X.; Jiang, X.; et al. When brain-inspired AI meets AGI. Meta-Radiology 2023, 1, 100005. [Google Scholar] [CrossRef]
- Díaz-Rodríguez, N.; Del Ser, J.; Coeckelbergh, M.; López de Prado, M.; Herrera-Viedma, E.; Herrera, F. Connecting the dots in trustworthy Artificial Intelligence: From AI principles, ethics, and key requirements to responsible AI systems and regulation. Inf. Fusion 2023, 99, 101896. [Google Scholar] [CrossRef]
- Hu, Y.-C.; Lin, Y.-H.; Lin, C.-H. Artificial Intelligence, Accelerated in Parallel Computing and Applied to Nonintrusive Appliance Load Monitoring for Residential Demand-Side Management in a Smart Grid: A Comparative Study. Appl. Sci. 2020, 10, 8114. [Google Scholar] [CrossRef]
- Hassan, N.; Miah, A.S.M.; Shin, J. A Deep Bidirectional LSTM Model Enhanced by Transfer-Learning-Based Feature Extraction for Dynamic Human Activity Recognition. Appl. Sci. 2024, 14, 603. [Google Scholar] [CrossRef]
- López-Ojeda, W.; Hurley, R.A. Digital Innovation in Neuroanatomy: Three-Dimensional (3D) Image Processing and Printing for Medical Curricula and Health Care. J. Neuropsychiatry Clin. Neurosci. 2023, 35, 206–209. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Kim, J.Y. The Metaverse for Healthcare: Trends, Applications, and Future Directions of Digital Therapeutics for Urology. Int. Neurourol. J. 2023, 27, S3–S12. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Wan, S.; Gan, W.; Chen, J.; Chao, H.-C. Metaverse in Education: Vision, Opportunities, and Challenges. In Proceedings of the 2022 IEEE International Conference on Big Data (Big Data), Osaka, Japan, 17–20 December 2022; pp. 2857–2866. [Google Scholar] [CrossRef]
- Sun, Q.; Fang, N.; Liu, Z.; Zhao, L.; Wen, Y.; Lin, H. HybridCTrm: Bridging CNN and Transformer for Multimodal Brain Image Segmentation. J. Healthc. Eng. 2021, 2021, 7467261. [Google Scholar] [CrossRef] [PubMed]
- Mazurowski, M.A.; Dong, H.; Gu, H.; Yang, J.; Konz, N.; Zhang, Y. Segment anything model form medical image analysis: An experimental study. Med. Image Anal. 2023, 89, 102918. [Google Scholar] [CrossRef] [PubMed]
- Sakshi, S.; Kukreja, V. Image Segmentation Techniques: Statistical, Comprehensive, Semi-Automated Analysis and an Application Perspective Analysis of Mathematical Expressions. Arch. Computat. Methods Eng. 2023, 30, 457–495. [Google Scholar] [CrossRef]
- Moztarzadeh, O.; Jamshidi, M.; Sargolzaei, S.; Keikhaee, F.; Jamshidi, A.; Shadroo, S.; Hauer, L. Metaverse and Medical Diagnosis: A Blockchain-Based Digital Twinning Approach Based on MobileNetV2 Algorithm for Cervical Vertebral Maturation. Diagnostics 2023, 13, 1485. [Google Scholar] [CrossRef] [PubMed]
- Huynh-The, T.; Pham, Q.-V.; Pham, M.-T.; Banh, T.-N.; Nguyen, G.-P.; Kim, D.-S. Efficient Real-Time Object Tracking in the Metaverse Using Edge Computing with Temporal and Spatial Consistency. Comput. Mater. Contin. 2023, 71, 341–356. [Google Scholar]
- Huang, H.; Zhang, C.; Zhao, L.; Ding, S.; Wang, H.; Wu, H. Self-Supervised Medical Image Denoising Based on WISTA-Net for Human Healthcare in Metaverse. IEEE J. Biomed. Health Inform. 2023, 1–9. [Google Scholar] [CrossRef]
- The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews (Published in Several Journals). 2021. Available online: http://www.prisma-statement.org/PRISMAStatement/PRISMAStatement (accessed on 8 January 2024).
- Rethlefsen, M.L.; Kirtley, S.; Waffenschmidt, S.; Ayala, A.P.; Moher, D.; Page, M.J.; Koffel, J.B. PRISMA-S: An Extension to the PRISMA Statement for Reporting Literature Searches in Systematic Reviews. Syst. Rev. 2021, 10, 39. [Google Scholar] [CrossRef]
- Adrian, E.D.; Zotterman, Y. The Impulses Produced by Sensory Nerve Endings. J. Physiol. 1926, 61, 465–483. [Google Scholar] [CrossRef]
- Adrian, E.D. The impulses produced by sensory nerve endings: Part I. J. Physiol. 1926, 61, 49. [Google Scholar] [CrossRef]
- Gerstner, W.; Kistler, W.M.; Naud, R.; Paninski, L. Neuronal Dynamics: From Single Neurons to Networks and Models of Cognition; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar]
- Rieke, F.; Warland, D.; de Ruyter van Steveninck, R.; Bialek, W. Spikes: Exploring the Neural Code; The MIT Press: Cambridge, MA, USA, 1997. [Google Scholar]
- van Hemmen, J.L.; Sejnowski, T.J. 23 Problems in Systems Neuroscience; Oxford University Press: Oxford, UK, 2006. [Google Scholar]
- Teich, M.C.; Khanna, S.M. Pulse-Number distribution for the neural spike train in the cat’s auditory nerve. J. Acoust. Soc. Am. 1985, 77, 1110–1128. [Google Scholar] [CrossRef]
- Werner, G.; Mountcastle, V.B. Neural activity in mechanoreceptive cutaneous afferents: Stimulus-response relations, Weber Functions, and Information Transmission. J. Neurophysiol. 1965, 28, 359–397. [Google Scholar] [CrossRef] [PubMed]
- Tolhurst, D.J.; Movshon, J.A.; Thompson, I.D. The dependence of Response amplitude and variance of cat visual cortical neurons on stimulus contrast. Exp. Brain Res. 1981, 41, 414–419. [Google Scholar]
- Radons, G.; Becker, J.D.; Dülfer, B.; Krüger, J. Analysis, classification, and coding of multielectrode spike trains with hidden Markov models. Biol. Cybern. 1994, 71, 359–373. [Google Scholar] [CrossRef]
- de Ruyter van Steveninck, R.R.; Lewen, G.D.; Strong, S.P.; Koberle, R.; Bialek, W. Reproducibility and variability in neural spike trains. Science 1997, 275, 1805–1808. [Google Scholar] [CrossRef] [PubMed]
- Kass, R.E.; Ventura, V. A spike-train probability model. Neural Comput. 2001, 13, 1713–1720. [Google Scholar] [CrossRef] [PubMed]
- Wójcik, D. The kinematics of the spike trains. Acta Phys. Pol. B 2018, 49, 2127–2138. [Google Scholar] [CrossRef]
- Rosenblatt, F. Principles of Neurodynamics. Perceptrons and the Theory of Bbain Mechanisms; Technical Report; Cornell Aeronautical Lab Inc.: Buffalo, NY, USA, 1961. [Google Scholar]
- Bu, T.; Fang, W.; Ding, J.; Dai, P.L.; Yu, Z.; Huang, T. Optimal ANN-SNN Conversion for High-Accuracy and Ultra-Low-Latency Spiking Neural Networks. arXiv 2023, arXiv:2303.04347. [Google Scholar] [CrossRef]
- Abbott, L.F.; Dayan, P. Theoretical Neuroscience Computational and Mathematical Modeling of Neural Systems; The MIT Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Yuan, Y.; Gao, R.; Wu, Q.; Fang, S.; Bu, X.; Cui, Y.; Han, C.; Hu, L.; Li, X.; Wang, X.; et al. Artificial Leaky Integrate-and-Fire Sensory Neuron for In-Sensor Computing Neuromorphic Perception at the Edge. ACS Sens. 2023, 8, 2646–2655. [Google Scholar] [CrossRef]
- Ghosh-Dastidar, S.; Adeli, H. Third Generation Neural Networks. In Advances in Computational Intelligence; Yu, W., Sanchez, E.N., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; Volume 116. [Google Scholar]
- Lindeberg, T. A time-causal and time-recursive scale-covariant scale-space representation of temporal signals and past time. Biol. Cybern. 2023, 117, 21–59. [Google Scholar] [CrossRef]
- Rueckauer, B.; Lungu, I.A.; Hu, Y.; Pfeiffer, M.; Liu, S.C. Conversion of Continuous-Valued Deep Networks To Efficient Event-Driven Neuromorphic Hardware. Front. Neurosci. 2017, 11, 682. [Google Scholar] [CrossRef]
- Cheng, X.; Zhang, T.; Jia, S.; Xu, B. Meta neurons improve spiking neural networks for efficient spatio-temporal learning. Neurocomputing 2023, 531, 217–225. [Google Scholar] [CrossRef]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Mehrish, A.; Majumder, N.; Bharadwaj, R.; Mihalcea, R.; Poria, S. A review of deep learning techniques for speech processing. Inf. Fusion 2023, 99, 101869. [Google Scholar] [CrossRef]
- Nielsen, M.A. Neural Networks and Deep Learning. 2015. Available online: http://neuralnetworksanddeeplearning.com/ (accessed on 8 January 2024).
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef]
- Sherstinsky, A. Fundamentals of Recurrent Neural Network (RNN) and Long Short-Term Memory (LSTM) network. Phys. D Nonlinear Phenom. 2020, 404, 132306. [Google Scholar] [CrossRef]
- Ghosh-Dastidar, S.; Adeli, H. Spiking neural networks. Int. J. Neural Syst. 2009, 19, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, K.; Vo-Ho, V.K.; Bulsara, D.; Le, N. Spiking Neural Networks and Their Applications: A Review. Brain Sci. 2022, 12b, 863. [Google Scholar] [CrossRef]
- Dampfhoffer, M.; Mesquida, T.; Valentian, A.; Anghel, L. Backpropagation-Based Learning Techniques for Deep Spiking Neural Networks: A Survey. IEEE Trans. Neural Netw. Learn. Syst. 2023, 1–16. [Google Scholar] [CrossRef]
- Ponulak, F.; Kasinski, A. Introduction to spiking neural networks: Information processing, learning and applications. Acta Neurobiol. Exp. 2011, 71, 409–433. [Google Scholar] [CrossRef]
- Wu, Y.; Deng, L.; Li, G.; Zhu, J.; Shi, L. Spatio-Temporal Backpropagation for Training High-Performance Spiking Neural Networks. Front Neurosci. 2018, 12, 331. [Google Scholar] [CrossRef]
- Pei, J.; Deng, L.; Song, S.; Zhao, M.; Zhang, Y.; Wu, S.; Wang, G.; Zou, Z.; Wu, Z.; He, W.; et al. Towards artificial general intelligence with hybrid Tianjic chip architecture. Nature 2019, 572, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Rathi, N.; Chakraborty, I.; Kosta, A.; Sengupta, A.; Ankit, A.; Panda, P.; Roy, K. Exploring Neuromorphic Computing Based on Spiking Neural Networks: Algorithms to Hardware. ACM Comput. Surv. 2023, 55, 243. [Google Scholar] [CrossRef]
- Rojas, R. The Backpropagation Algorithm. In Neural Networks; Springer: Berlin/Heidelberg, Germany, 1996; pp. 1–50. [Google Scholar]
- Singh, A.; Kushwaha, S.; Alarfaj, M.; Singh, M. Comprehensive Overview of Backpropagation Algorithm for Digital Image Denoising. Electronics 2022, 11, 1590. [Google Scholar] [CrossRef]
- Kaur, J.; Khehra, B.S.; Singh, A. Back propagation artificial neural network for diagnosis of heart disease. J. Reliab. Intell. Environ. 2023, 9, 57–85. [Google Scholar] [CrossRef]
- Hameed, A.A.; Karlik, B.; Salman, M.S. Back-propagation algorithm with variable adaptive momentum. Knowl.-Based Syst. 2016, 114, 79–87. [Google Scholar] [CrossRef]
- Cao, Y.; Chen, Y.; Khosla, D. Spiking Deep Convolutional Neural Networks for Energy-Efficient Object Recognition. Int. J. Comput. Vis. 2015, 113, 54–66. [Google Scholar] [CrossRef]
- Alemanno, F.; Aquaro, M.; Kanter, I.; Barra, A.; Agliari, E. Supervised Hebbian Learning. Europhys. Lett. 2023, 141, 11001. [Google Scholar] [CrossRef]
- Ponulak, F. ReSuMe—New Supervised Learning Method for Spiking Neural Networks; Technical Report; Poznań University of Technology: Poznań, Poland, 2005; Available online: https://www.semanticscholar.org/paper/ReSuMe-New-Supervised-Learning-Method-for-Spiking-Ponulak/b04f2391b8c9539edff41065c39fc2d27cc3d95a (accessed on 8 January 2024).
- Shrestha, A.; Ahmed, K.; Wang, Y.; Qiu, Q. Stable Spike-Timing Dependent Plasticity Rule for Multilayer Unsupervised and Supervised Learning. In Proceedings of the 2017 International Joint Conference on Neural Networks (IJCNN), Anchorage, AK, USA, 14–19 May 2017; pp. 1999–2006. [Google Scholar] [CrossRef]
- Amato, G.; Carrara, F.; Falchi, F.; Gennaro, C.; Lagani, G. Hebbian Learning Meets Deep Convolutional Neural Networks. In Proceedings of the Image Analysis and Processing—ICIAP 2019, Trento, Italy, 9–13 September 2019; Ricci, E., Rota Bulò, S., Snoek, C., Lanz, O., Messelodi, S., Sebe, N., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2019; Volume 11751, pp. 1–14. [Google Scholar] [CrossRef]
- Ponulak, F.; Kasinski, A. Supervised learning in spiking neural networks with ReSuMe: Sequence learning, classification, and spike shifting. Neural Comput. 2010, 22, 467–510. [Google Scholar] [CrossRef]
- Florian, R.V. The Chronotron: A Neuron That Learns to Fire Temporally Precise Spike Patterns. PLoS ONE 2012, 7, e40233. [Google Scholar] [CrossRef] [PubMed]
- Victor, J.D.; Purpura, K.P. Metric-space analysis of spike trains: Theory, algorithms, and applications. Network 1997, 8, 127–164. [Google Scholar] [CrossRef]
- Huang, C.; Wang, J.; Wang, S.-H.; Zhang, Y.-D. Applicable artificial intelligence for brain disease: A survey. Neurocomputing 2022, 504, 223–239. [Google Scholar] [CrossRef]
- Markram, H.; Gerstner, W.; Sjöström, P.J. A history of spike-timing-dependent plasticity. Front. Synaptic Neurosci. 2011, 3, 4. [Google Scholar] [CrossRef]
- Merolla, P.A.; Arthur, J.V.; Alvarez-Icaza, R.; Cassidy, A.S.; Sawada, J.; Akopyan, F.; Jackson, B.L.; Imam, N.; Guo, C.; Nakamura, Y.; et al. A million spiking-neuron integrated circuit with a scalable communication network and interface. Science 2014, 345, 668–673. [Google Scholar] [CrossRef]
- Chakraborty, B.; Mukhopadhyay, S. Characterization of Generalizability of Spike Timing Dependent Plasticity Trained Spiking Neural Networks. Front. Neurosci. 2021, 15, 695357. [Google Scholar] [CrossRef] [PubMed]
- Lagani, G.; Falchi, F.; Gennaro, C.; Amato, G. Spiking Neural Networks and Bio-Inspired Supervised Deep Learning: A Survey. arXiv 2023, arXiv:2307.16235. [Google Scholar] [CrossRef]
- Gütig, R.; Sompolinsky, H. The tempotron: A neuron that learns spike timing-based decisions. Nat. Neurosci. 2006, 9, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Cellina, M.; Cè, M.; Alì, M.; Irmici, G.; Ibba, S.; Caloro, E.; Fazzini, D.; Oliva, G.; Papa, S. Digital Twins: The New Frontier for Personalized Medicine? Appl. Sci. 2023, 13, 7940. [Google Scholar] [CrossRef]
- Sun, T.; He, X.; Li, Z. Digital twin in healthcare: Recent updates and challenges. Digit. Health 2023, 9, 20552076221149651. [Google Scholar] [CrossRef]
- Uhl, J.C.; Schrom-Feiertag, H.; Regal, G.; Gallhuber, K.; Tscheligi, M. Tangible Immersive Trauma Simulation: Is Mixed Reality the Next Level of Medical Skills Training? In Proceedings of the 2023 CHI Conference on Human Factors in Computing Systems (CHI ‘23), New York, NY, USA, 23–28 April 2023; Association for Computing Machinery: New York, NY, USA, 2023; p. 513. [Google Scholar] [CrossRef]
- Kshatri, S.S.; Singh, D. Convolutional Neural Network in Medical Image Analysis: A Review. Arch. Comput. Methods Eng. 2023, 30, 2793–2810. [Google Scholar] [CrossRef]
- Li, X.; Guo, Y.; Jiang, F.; Xu, L.; Shen, F.; Jin, Z.; Wang, Y. Multi-Task Refined Boundary-Supervision U-Net (MRBSU-Net) for Gastrointestinal Stromal Tumor Segmentation in Endoscopic Ultrasound (EUS) Images. IEEE Access 2020, 8, 5805–5816. [Google Scholar] [CrossRef]
- Oktay, O.; Schlemper, J.; Le Folgoc, L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning Where to Look for the Pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Munich, Germany, 5–9 October 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Crimi, A., Bakas, S., Kuijf, H., Menze, B., Reyes, M., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 3–11. [Google Scholar]
- Alom, M.Z.; Yakopcic, C.; Hasan, M.; Taha, T.M.; Asari, V.K. Recurrent residual U-Net for medical image segmentation. J. Med. Imaging 2019, 6, 014006. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Zou, D.; Xu, W.; Zhao, X.; Lu, W.; He, X. Bimodal segmentation and classification of endoscopic ultrasonography images for solid pancreatic tumor. Biomed. Signal Process. Control 2023, 83, 104591. [Google Scholar] [CrossRef]
- Urbanczik, R.; Senn, W. Reinforcement learning in populations of spiking neurons. Nat. Neurosci. 2009, 12, 250–252. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Tang, H.; Tan, K.C.; Yu, H. A brain-inspired spiking neural network model with temporal encoding and learning. Neurocomputing 2014, 138, 3–13. [Google Scholar] [CrossRef]
- Kumarasinghe, K.; Kasabov, N.; Taylor, D. Brain-inspired spiking neural networks for decoding and understanding muscle activity and kinematics from electroencephalography signals during hand movements. Sci. Rep. 2021, 11, 2486. [Google Scholar] [CrossRef]
- Niu, L.-Y.; Wei, Y.; Liu, W.-B.; Long, J.-Y.; Xue, T.-H. Research Progress of spiking neural network in image classification: A Review. Appl. Intell. 2023, 53, 19466–19490. [Google Scholar] [CrossRef]
- Yuan, F.; Zhang, Z.; Fang, Z. An Effective CNN and Transformer Complementary Network for Medical Image Segmentation. Pattern Recognit. 2023, 136, 109228. [Google Scholar] [CrossRef]
- Pregowska, A.; Osial, M.; Dolega-Dolegowski, D.; Kolecki, R.; Proniewska, K. Information and Communication Technologies Combined with Mixed Reality as Supporting Tools in Medical Education. Electronics 2022, 11, 3778. [Google Scholar] [CrossRef]
- Proniewska, K.; Dolega-Dolegowski, D.; Kolecki, R.; Osial, M.; Pregowska, A. Applications of Augmented Reality—Current State of the Art. In The 3D Operating Room with Unlimited Perspective Change and Remote Support; InTech: Rijeka, Croatia, 2023; pp. 1–23. [Google Scholar]
- Suh, I.; McKinney, T.; Siu, K.-C. Current Perspective of Metaverse Application in Medical Education, Research and Patient Care. Virtual Worlds 2023, 2, 115–128. [Google Scholar] [CrossRef]
- Liu, X.; Song, L.; Liu, S.; Zhang, Y. A Review of Deep-Learning-Based Medical Image Segmentation Methods. Sustainability 2021, 13, 1224. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Liu, J.-Y.; Wang, K.; Zhang, K.; Zhang, G.-S.; Liao, X.-F.; Yang, G. Global Transformer and Dual Local Attention Network via Deep-Shallow Hierarchical Feature Fusion for Retinal Vessel Segmentation. IEEE Trans. Cybern. 2022, 53, 5826–5839. [Google Scholar] [CrossRef]
- Kheradpisheh, S.R.; Ghodrati, M.; Ganjtabesh, M.; Masquelier, T. Bio-Inspired unsupervised learning of visual features leads to robust invariant object recognition. Neurocomputing 2016, 205, 382–392. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar] [CrossRef]
- Xiao, H.; Li, L.; Liu, Q.; Zhu, X.; Zhang, Q. Transformers in Medical Image Segmentation: A Review. Biomed. Signal Process. Control 2023, 84, 104791. [Google Scholar] [CrossRef]
- Yu, H.; Yang, L.T.; Zhang, Q.; Armstrong, D.; Deen, M.J. Convolutional Neural Networks for Medical Image Analysis: State-of-the-Art, Comparisons, Improvement, and Perspectives. Neurocomputing 2021, 444, 92–110. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2016, Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Evans, L.M.; Sozumert, E.; Keenan, B.E.; Wood, C.E.; du Plessis, A. A Review of Image-Based Simulation Applications in High-Value Manufacturing. Arch. Comput. Methods Eng. 2023, 30, 1495–1552. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.; Kaiser, L.; Polosukhin, I. Attention is all you need. Adv. Neur. Inf. Process. Syst. 2017, 30. [Google Scholar] [CrossRef]
- Tang, H.; Chen, Y.; Wang, T.; Zhou, Y.; Zhao, L.; Gao, Q.; Du, M.; Tan, T.; Zhang, X.; Tong, T. HTC-Net: A hybrid CNN-transformer framework for medical image segmentation. Biomed. Signal Process. Control 2024, 88 Pt A, 105605. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 × 16 words:Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Touvron, H.; Cord, M.; Matthijs, D.; Massa, F.; Sablayrolles, A.; Jegou, H. Training data-efficient image transformers & distillation through attention. In Proceedings of the International Conference on Machine Learning, PMLR, Virtual Event, 18–24 July 2021; pp. 10347–10357. [Google Scholar]
- Han, K.; Wang, Y.; Chen, H.; Chen, X.; Guo, J.; Liu, Z.; Tang, Y.; Xiao, A.; Xu, C.; Xu, Y. A survey on vision Transformer. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 45, 87–110. [Google Scholar] [CrossRef]
- Maurício, J.; Domingues, I.; Bernardino, J. Comparing vision Transformers and Convolutional Neural Networks for image classification: A Literature Review. Appl. Sci. 2023, 13, 5521. [Google Scholar] [CrossRef]
- Wang, H. Traffic Sign Recognition with Vision Transformers. In Proceedings of the 6th International Conference on Information System and Data Mining, Silicon Valley, CA, USA, 27–29 May 2022; Association for Computing Machinery: New York, NY, USA, 2022; pp. 55–61. [Google Scholar]
- Bakhtiarnia, A.; Zhang, Q.; Iosifidis, A. Single-layer vision Transformers for more accurate early exits with less overhead. Neural Netw. 2022, 153, 461–473. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Li, Q.; Lu, H.; Cheng, Q.; Zhang, X. GAN review: Models and medical image fusion applications. Inf. Fusion 2023, 91, 134–148. [Google Scholar] [CrossRef]
- Skandarani, Y.; Jodoin, P.-M.; Lalande, A. GANs for Medical Image Synthesis: An Empirical Study. J. Imaging 2023, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Son, J.; Park, S.J.; Jung, K.-H. Towards accurate segmentation of retinal vessels and the optic disc in Fundoscopic images with generative adversarial networks. J. Digit. Imaging 2019, 32, 499–512. [Google Scholar] [CrossRef] [PubMed]
- Güven, S.A.; Talu, M.F. Brain MRI high resolution image creation and segmentation with the new GAN method. Biomed. Signal Process. Control 2023, 80, 104246. [Google Scholar] [CrossRef]
- Scarselli, F.; Gori, M.; Tsoi, A.C.; Hagenbuchner, M.; Monfardini, G. The graph neural network model. IEEE Trans. Neural Netw. 2009, 20, 61–80. [Google Scholar] [CrossRef] [PubMed]
- Hitaj, B.; Ateniese, G.; Perez-Cruz, F. Deep Models Under the GAN: Information Leakage from Collaborative Deep Learning. In Proceedings of the 2017, CCS ‘17: Proceedings of the 2017 ACM SIGSAC Conference on Computer and Communications Security, Dallas, TX, USA, 30 October–3 November 2017; pp. 603–618. [Google Scholar] [CrossRef]
- Liang, F.; Qian, C.; Yu, W.; Griffith, D.; Golmie, N. Survey of Graph Neural Networks and Applications. Wirel. Commun. Mob. Comput. 2022, 9261537. [Google Scholar] [CrossRef]
- Jiang, X.; Hu, Z.; Wang, S.; Zhang, Y. Deep learning for medical image-based cancer diagnosis. Cancers 2023, 15, 3608. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, Y.; Che, T.; Li, S.; Wang, X. Graph neural networks for image-guided disease diagnosis: A review. iRADIOLOGY 2023, 1, 151–166. [Google Scholar] [CrossRef]
- Fabijanska, A. graph convolutional networks for semi-supervised image segmentation. IEEE Access 2022, 10, 104144–104155. [Google Scholar] [CrossRef]
- Hamilton, W.; Ying, Z.; Leskovec, J. Inductive representation learning on large graphs. Adv. Neural Inf. Process. Syst. 2017, 30, 1024–1034. [Google Scholar]
- Ahmedt-Aristizabal, D.; Armin, M.A.; Denman, S.; Fookes, C.; Petersson, L. Graph-based deep learning for medical diagnosis and analysis: Past, present and future. Sensors 2021, 21, 4758. [Google Scholar] [CrossRef]
- He, P.H.; Qu, A.P.; Xiao, S.M.; Ding, M.D. A GNN-based Network for Tissue Semantic Segmentation in Histopathology Image. In Proceedings of the 3rd International Conference on Computer, Big Data and Artificial Intelligence (ICCBDAI 2022), Zhangjiajie, China, 16–18 December 2022; Journal of Physics: Conference Series; IOP Publishing Ltd.: Bristol, UK, 2023; Volume 2504. [Google Scholar] [CrossRef]
- Jiang, W.; Luo, J. Graph Neural Network for Traffic Forecasting: A Survey. 2021. Available online: https://arxiv.org/abs/2101.11174 (accessed on 8 January 2024).
- Ayaz, H.; Khosravi, H.; McLoughlin, I.; Tormey, D.; Özsunar, Y.; Unnikrishnan, S. A random graph-based neural network approach to assess glioblastoma progression from perfusion MRI. Biomed. Signal Process. Control 2023, 86 Pt C, 105286. [Google Scholar] [CrossRef]
- Sitzmann, V.; Martel, J.N.P.; Bergman, A.W.; Lindell, D.B.; Wetzstein, G. Implicit Neural Representations with Periodic Activation Functions. arXiv 2020, arXiv:2006.09661. [Google Scholar] [CrossRef]
- Stolt-Ansó, N.; McGinnis, J.; Pan, J.; Hammernik, K.; Rueckert, D. NISF: Neural Implicit Segmentation Functions. arXiv 2023, arXiv:2309.08643. [Google Scholar] [CrossRef]
- Byra, M.; Poon, C.; Shimogori, T.; Skibbe, H. Implicit neural representations for joint decomposition and registration of gene expression images in the marmoset brain. arXiv 2023, arXiv:2308.04039. [Google Scholar] [CrossRef]
- Meta. Available online: https://segment-anything.com/ (accessed on 8 January 2024).
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.Y.; et al. Segment Anything. arXiv 2023, arXiv:2304.02643. [Google Scholar]
- He, S.; Bao, R.; Li, J.; Stout, J.; Bjornerud, A.; Grant, P.E.; Ou, Y. Computer-Vision Benchmark Segment-Anything Model (SAM) in Medical Images: Accuracy in 12 Datasets. arXiv 2023, arXiv:2304.09324. [Google Scholar]
- Zhang, Y.; Jiao, R. Towards Segment Anything Model (SAM) for Medical Image Segmentation: A Survey. arXiv 2023, arXiv:2305.03678. [Google Scholar]
- Wu, J.; Zhang, Y.; Fu, R.; Fang, H.; Liu, Y.; Wang, Z.; Xu, Y.; Jin, Y. Medical SAM Adapter: Adapting Segment Anything Model for Medical Image Segmentation. arXiv 2023, arXiv:2304.12620. [Google Scholar]
- Yi, Z.; Lian, J.; Liu, Q.; Zhu, H.; Liang, D.; Liu, J. Learning Rules in Spiking Neural Networks: A Survey. Neurocomputing 2023, 531, 163–179. [Google Scholar] [CrossRef]
- Avcı, H.; Karakaya, J. A Novel Medical Image Enhancement Algorithm for Breast Cancer Detection on Mammography Images Using Machine Learning. Diagnostics 2023, 13, 348. [Google Scholar] [CrossRef]
- Ghahramani, M.; Shiri, N. Brain tumour detection in magnetic resonance Imaging using Levenberg–Marquardt backpropagation neural network. IET Image Process. 2023, 17, 88–103. [Google Scholar] [CrossRef]
- Zhang, J.; Gajjala, S.; Agrawal, P.; Tison, G.H.; Hallock, L.H.; Beussink-Nelson, L.; Lassen, M.H.; Fan, E.; Aras, M.A.; Jordan, C.; et al. Fully automated echocardiogram interpretation in clinical practice. Circulation 2018, 138, 1623–1635. [Google Scholar] [CrossRef]
- Sajjad, M.; Khan, S.; Khan, M.; Wu, W.; Ullah, A.; Baik, S.W. Multi-grade brain tumor classification using deep CNN with extensive data augmentation. J. Comput. Sci. 2021, 30, 174–182. [Google Scholar] [CrossRef]
- Jun, T.J.; Kang, S.J.; Lee, J.G.; Kweon, J.; Na, W.; Kang, D.; Kim, D.; Kim, D.; Kim, Y.H. Automated detection of vulnerable plaque in intravascular ultrasound images. Med. Biol. Eng. Comput. 2019, 57, 863–876. [Google Scholar] [CrossRef]
- Ostvik, A.; Smistad, E.; Aase, S.A.; Haugen, B.O.; Lovstakken, L. Real-time standard view classification in transthoracic echocardiography using convolutional neural networks. Ultrasound Med. Biol. 2019, 45, 374–384. [Google Scholar] [CrossRef]
- Lossau, T.; Nickisch, H.; Wisse, T.; Bippus, R.; Schmitt, H.; Morlock, M.; Grass, M. Motion artifact recognition and quantification in coronary CT angiography using convolutional neural networks. Med. Image Anal. 2019, 52, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Emad, O.; Yassine, I.A.; Fahmy, A.S. Automatic localization of the left ventricle in cardiac MRI images using deep learning. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 683–686. [Google Scholar] [CrossRef]
- Moradi, M.; Guo, Y.; Gur, Y.; Negahdar, M.; Syeda-Mahmood, T. A Cross-Modality Neural Network Transform for Semi-automatic Medical Image Annotation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016. MICCAI 2016, Athens, Greece, 17–21 October 2016; Lecture Notes in Computer Science; Ourselin, S., Joskowicz, L., Sabuncu, M., Unal, G., Wells, W., Eds.; Springer: Cham, Switzerland, 2016; Volume 9901. [Google Scholar] [CrossRef]
- Liskowski, P.; Krawiec, K. Segmenting retinal blood vessels with deep neural networks. IEEE Trans. Med. Imaging 2016, 35, 2369–2380. [Google Scholar] [CrossRef]
- Yuan, J.; Hassan, S.S.; Wu, J.; Koger, C.R.; Packard, R.R.S.; Shi, F.; Fei, B.; Ding, Y. Extended reality for biomedicine. Nat. Rev. Methods Primers 2023, 3, 14. [Google Scholar] [CrossRef]
- Kakhandaki, N.; Kulkarni, S.B. Classification of Brain MR Images Based on Bleed and Calcification Using ROI Cropped U-Net Segmentation and Ensemble RNN Classifier. Int. J. Inf. Tecnol. 2023, 15, 3405–3420. [Google Scholar] [CrossRef]
- Manimurugan, S. Hybrid High Performance Intelligent Computing Approach of CACNN and RNN for Skin Cancer Image Grading. Soft Comput. 2023, 27, 579–589. [Google Scholar] [CrossRef]
- Yue, Y.; Baltes, M.; Abuhajar, N.; Sun, T.; Karanth, A.; Smith, C.D.; Bihl, T.; Liu, J. Spiking Neural Networks Fine-Tuning for Brain Image Segmentation. Front. Neurosci. 2023, 17, 1267639. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Li, R.; Wang, C.; Zhang, R.; Yue, K.; Li, W.; Li, Y. A Spiking Neural Network Based on Retinal Ganglion Cells for Automatic Burn Image Segmentation. Entropy 2022, 24, 1526. [Google Scholar] [CrossRef]
- Gilani, S.Q.; Syed, T.; Umair, M.; Marques, O. Skin Cancer Classification Using Deep Spiking Neural Network. J. Digit. Imaging 2023, 36, 1137–1147. [Google Scholar] [CrossRef]
- Sahoo, A.K.; Parida, P.; Muralibabu, K.; Dash, S. Efficient Simultaneous Segmentation and Classification of Brain Tumors from MRI Scans Using Deep Learning. Biocybern. Biomed. Eng. 2023, 43, 616–633. [Google Scholar] [CrossRef]
- Fu, Q.; Dong, H. Breast Cancer Recognition Using Saliency-Based Spiking Neural Network. Wirel. Commun. Mob. Comput. 2022, 2022, 8369368. [Google Scholar] [CrossRef]
- Tan, P.; Chen, X.; Zhang, H.; Wei, Q.; Luo, K. Artificial intelligence aids in development of nanomedicines for cancer management. Semin. Cancer Biol. 2023, 89, 61–75. [Google Scholar] [CrossRef]
- Malhotra, S.; Halabi, O.; Dakua, S.P.; Padhan, J.; Paul, S.; Palliyali, W. Augmented Reality in Surgical Navigation: A Review of Evaluation and Validation Metrics. Appl. Sci. 2023, 13, 1629. [Google Scholar] [CrossRef]
- Wisotzky, E.L.; Rosenthal, J.-C.; Meij, S.; Dobblesteen, J.v.D.; Arens, P.; Hilsmann, A.; Eisert, P.; Uecker, F.C.; Schneider, A. Telepresence for surgical assistance and training using eXtended reality during and after pandemic periods. J. Telemed. Telecare 2023. [Google Scholar] [CrossRef]
- Martin-Gomez, A.; Li, H.; Song, T.; Yang, S.; Wang, G.; Ding, H.; Navab, N.; Zhao, Z.; Armand, M. STTAR: Surgical Tool Tracking Using Off-the-Shelf Augmented Reality Head-Mounted Displays. IEEE Trans. Vis. Comput. Graph. 2022. [Google Scholar] [CrossRef]
- Minopoulos, G.M.; Memos, V.A.; Stergiou, K.D.; Stergiou, C.L.; Psannis, K.E. A Medical Image Visualization Technique Assisted with AI-Based Haptic Feedback for Robotic Surgery and Healthcare. Appl. Sci. 2023, 13, 3592. [Google Scholar] [CrossRef]
- Hirling, D.; Tasnadi, E.; Caicedo, J.; Caroprese, M.V.; Sjögren, R.; Aubreville, M.; Koos, K.; Horvath, P. Segmentation metric misinterpretations in bioimage analysis. Nat. Methods 2023. [Google Scholar] [CrossRef]
- Pregowska, A.; Perkins, M. Artificial Intelligence in Medical Education: Technology and Ethical Risk. 2023. Available online: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=4643763 (accessed on 8 January 2024).
- Goutte, C.; Gaussier, E. A Probabilistic Interpretation of Precision, Recall and F-Score, with Implication for Evaluation. In Advances in Information Retrieval. ECIR 2005; Losada, D.E., Fernández-Luna, J.M., Eds.; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2005; p. 3408. [Google Scholar] [CrossRef]
- Schneider, P.; Xhafa, F. (Eds.) Chapter 3—Anomaly detection: Concepts and methods. In Anomaly Detection and Complex Event Processing over IoT Data Streams; Academic Press: Cambridge, MA, USA, 2022; pp. 49–66. [Google Scholar] [CrossRef]
- Nahm, F.S. Receiver operating characteristic curve: Overview and practical use for clinicians. Korean J. Anesthesiol. 2022, 75, 25–36. [Google Scholar] [CrossRef]
- Perkins, N.J.; Schisterman, E.F. The inconsistency of “optimal” cut-points using two ROC based criteria. Am. J. Epidemiol. 2006, 163, 670–675. [Google Scholar] [CrossRef]
- Li, J.; Cairns, B.J.; Li, J.; Zhu, T. Generating synthetic mixed-type longitudinal electronic realth records for artificial intelligent applications. Digit. Med. 2023, 6, 98. [Google Scholar] [CrossRef]
- Pammi, M.; Aghaeepour, N.; Neu, J. Multiomics, artificial intelligence, and precision medicine in perinatology. Pediatr. Res. 2023, 93, 308–315. [Google Scholar] [CrossRef]
- Vardi, G. On the Implicit Bias in Deep-Learning Algorithms. Commun. ACM 2023, 66, 86–93. [Google Scholar] [CrossRef]
- Pawłowska, A.; Karwat, P.; Żołek, N. Letter to the Editor. Re: “[Dataset of breast ultrasound images by W. Al-Dhabyani, M. Gomaa, H. Khaled & A. Fahmy, Data in Brief, 2020, 28, 104863]”. Data Brief 2023, 48, 109247. [Google Scholar] [CrossRef]
- PhysioNet. Available online: https://physionet.org/ (accessed on 8 January 2024).
- National Sleep Research Resource. Available online: https://sleepdata.org/ (accessed on 8 January 2024).
- Open Access Series of Imaging Studies—OASIS Brain. Available online: https://www.oasis-brains.org/ (accessed on 8 January 2024).
- OpenNeuro. Available online: https://openneuro.org/ (accessed on 8 January 2024).
- Brain Tumor Dataset. Available online: https://figshare.com/articles/dataset/brain_tumor_dataset/1512427?file=7953679 (accessed on 8 January 2024).
- The Cancer Imaging Archive. Available online: https://www.cancerimagingarchive.net/ (accessed on 8 January 2024).
- LUNA16. Available online: https://luna16.grand-challenge.org/ (accessed on 8 January 2024).
- MICCAI 2012 Prostate Challenge. Available online: https://promise12.grand-challenge.org/ (accessed on 8 January 2024).
- IEEE Dataport. Available online: https://ieee-dataport.org/ (accessed on 8 January 2024).
- AIMI. Available online: https://aimi.stanford.edu/shared-datasets (accessed on 8 January 2024).
- fastMRI. Available online: https://fastmri.med.nyu.edu/ (accessed on 8 January 2024).
- Alzheimer’s Disease Neuroimaging Initiative. Available online: http://adni.loni.usc.edu/ (accessed on 8 January 2024).
- Pediatric Brain Imaging Dataset. Available online: http://fcon_1000.projects.nitrc.org/indi/retro/pediatric.html (accessed on 8 January 2024).
- ChestX-ray8. Available online: https://nihcc.app.box.com/v/ChestXray-NIHCC (accessed on 8 January 2024).
- Breast Cancer Digital Repository. Available online: https://bcdr.eu/ (accessed on 8 January 2024).
- Brain-CODE. Available online: https://www.braincode.ca/ (accessed on 8 January 2024).
- RadImageNet. Available online: https://www.radimagenet.com/ (accessed on 8 January 2024).
- EyePACS. Available online: https://paperswithcode.com/dataset/kaggle-eyepacs (accessed on 8 January 2024).
- Medical Segmentation Decathlon. Available online: http://medicaldecathlon.com/ (accessed on 8 January 2024).
- DDSM. Available online: http://www.eng.usf.edu/cvprg/Mammography/Database.html (accessed on 8 January 2024).
- LIDC-IDRI. Available online: https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI (accessed on 8 January 2024).
- Synapse. Available online: https://www.synapse.org/#!Synapse:syn3193805/wiki/217789 (accessed on 8 January 2024).
- Mini-MIAS. Available online: http://peipa.essex.ac.uk/info/mias.html (accessed on 8 January 2024).
- Breast Cancer His-to-Pathological Data-Base (BreakHis). Available online: https://web.inf.ufpr.br/vri/databases/breast-cancer-histopathologi-cal-database-breakhis/ (accessed on 8 January 2024).
- Messidor. Available online: https://www.adcis.net/en/third-party/messidor/ (accessed on 8 January 2024).
- Chang, X.; Ren, P.; Xu, P.; Li, Z.; Chen, X.; Hauptmann, A. A comprehensive survey of scene graphs: Generation and application. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 45, 1–26. [Google Scholar] [CrossRef]
- Ji, S.; Pan, S.; Cambria, E.; Marttinen, P.; Yu, P.S. A survey on knowledge graphs: Representation, acquisition, and applications. IEEE Trans. Neural Netw. Learn. Syst. 2021, 33, 494–514. [Google Scholar] [CrossRef]
- Li, J.; Cheng, J.; Shi, J.; Huang, F. Brief Introduction of Back Propagation (BP) Neural Network Algorithm and Its Improvement. In Advances in Computer Science and Information Engineering; Jin, D., Lin, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; Volume 169, pp. 1–10. [Google Scholar] [CrossRef]
- Johnson, X.Y.; Venayagamoorthy, G.K. Encoding Real Values into Polychronous Spiking Networks. In Proceedings of the International Joint Conference on Neural Networks (IJCNN), Barcelona, Spain, 18–23 July 2010; pp. 1–7. [Google Scholar] [CrossRef]
- Bohte, S.M.; Kok, J.N.; La Poutre, H. Error-back propagation in temporally encoded networks of spiking neurons. Neurocomputing 2002, 48, 17–37. [Google Scholar] [CrossRef]
- Rajagopal, S.; Chakraborty, S.; Gupta, M.D. Deep Convolutional Spiking Neural Network Optimized with Arithmetic Optimization Algorithm for Lung Disease Detection Using Chest X-ray Images. Biomed. Signal Process. Control 2023, 79, 104197. [Google Scholar] [CrossRef]
- Brader, J.M.; Senn, W.; Fusi, S. Learning real-world stimuli in a neural network with spike-driven synaptic dynamics. Neural Comput. 2007, 19, 2881–2912. [Google Scholar] [CrossRef]
- Masquelier, T.; Guyonneau, R.; Thorpe, S.J. Competitive STDP-based spike pattern learning. Neural Comput. 2009, 21, 1259–1276. [Google Scholar] [CrossRef]
- Lee, J.H.; Delbruck, T.; Pfeiffer, M. Training deep spiking convolutional neural Networks with STDP-based unsupervised pre-training followed by supervised fine-tuning. Front. Neurosci. 2018, 12, 435. [Google Scholar] [CrossRef]
- Lee, J.H.; Delbruck, T.; Pfeiffer, M. Enabling Spike-Based Backpropagation for Training Deep Neural Network Architectures. Front. Neurosci. 2020, 14, 119. [Google Scholar] [CrossRef]
- Wu, Y.; Deng, L.; Li, G.; Zhu, J.; Shi, L. Direct training for spiking neural networks: Faster, Larger, Better. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 1311–1318. [Google Scholar] [CrossRef]
- Neil, D.; Pfeiffer, M.; Liu, S.-C. Learning to be efficient: Algorithms for training low-latency, low-compute deep spiking neural networks. In Proceedings of the 31st Annual ACM Symposium on Applied Computing (SAC ‘2016), Pisa, Italy, 4–8 April 2016; Association for Computing Machinery: New York, NY, USA, 2016; pp. 293–298. [Google Scholar] [CrossRef]
- Lee, J.H.; Delbruck, T.; Pfeiffer, M. Training deep spiking neural networks using backpropagation. Front. Neurosci. 2016, 10, 508. [Google Scholar] [CrossRef]
- Zhan, K.; Li, Y.; Li, Q.; Pan, G. Bio-Inspired Active Learning Method in spiking neural network. Know.-Based Syst. 2023, 261, 2433. [Google Scholar] [CrossRef]
- Marcello, S.; Shunra, Y.; Ruggero, M. Neural and axonal heterogeneity improves information transmission. Phys. A Stat. Mech. Its Appl. 2023, 618, 12862. [Google Scholar] [CrossRef]
- Kanwisher, N.; Khosla, M.; Dobs, K. Using artificial neural networks to ask ‘why’ questions of minds and brains. Trends Neurosci. 2023, 46, 240–254. [Google Scholar] [CrossRef]
- Wang, J.; Chen, S.; Liu, Y.; Lau, R. Intelligent Metaverse Scene Content Construction. IEEE Access 2023, 11, 76222–76241. [Google Scholar] [CrossRef]
- UNESCO Open Data. Available online: https://unesdoc.unesco.org/ark:/48223/pf0000385841 (accessed on 8 January 2024).
- EC AI. Available online: https://digital-strategy.ec.europa.eu/en/library/assessment-list-trustworthy-artificial-intelligence-altai-self-assessment (accessed on 8 January 2024).
- Radclyffe, C.; Ribeiro, M.; Wortham, R.H. The assessment list for trustworthy artificial intelligence: A review and recommendations. Front. Artif. Intell. 2023, 6, 1020592. [Google Scholar] [CrossRef]
- EU AI Regulations. Available online: https://www.europarl.europa.eu/news/en/headlines/society/20230601STO93804/eu-ai-act-first-regulation-on-artificial-intelligence (accessed on 8 January 2024).
- Pregowska, A.; Perkins, M. Artificial Intelligence in Medical Education Part 1: Typologies and Ethical Approaches. Available online: https://ssrn.com/abstract=4576612 (accessed on 8 January 2024).
- Yao, C.; Tang, J.; Hu, M.; Wu, Y.; Guo, W.; Li, Q.; Zhang, X.-P. Claw U-Net: A UNet-Based Network with Deep Feature Concatenation for Scleral Blood Vessel Segmentation. arXiv 2020, arXiv:2010.10163. [Google Scholar] [CrossRef]
- Mo, S.; Tian, Y. AV-SAM: Segment Anything Model Meets Audio-Visual Localization and Segmentation. arXiv 2023, arXiv:2305.01836. [Google Scholar] [CrossRef]
- Himangi; Singla, M. To Enhance Object Detection Speed in Meta-Verse Using Image Processing and Deep Learning. Int. J. Intell. Syst. Appl. Eng. 2023, 11, 176–184. Available online: https://ijisae.org/index.php/IJISAE/article/view/3106 (accessed on 8 January 2024).
- Pooyandeh, M.; Han, K.-J.; Sohn, I. Cybersecurity in the AI-Based Metaverse: A Survey. Appl. Sci. 2022, 12, 12993. [Google Scholar] [CrossRef]






| Database | Data Source | Data Type | Amount of Data | Availability | |
|---|---|---|---|---|---|
| PhysioNet | [165] | EEG, X-ray images, polysomnography | Auditory evoked potential EEG biometric dataset—240 measurements from 20 subjects Brno University of Technology Smartphone PPG database (BUT PPG)—12 polysomnographic recordings CAP Sleep Database—108 polysomnographic recordings CheXmask Database: a large-scale dataset of anatomical segmentation masks for chest X-ray images—676,803 chest radiographs Electroencephalogram and eye-gaze datasets for robot-assisted surgery performance evaluation—EEG from 25 subjects Siena Scalp EEG Database—EEG from 14 subjects | Public | |
| PhysioNet | [165] | EEG, X-ray images, polysomnography | Computed tomography images for intracranial hemorrhage detection and segmentation—82 CT after traumatic brain injury (TBI) A multimodal dental dataset facilitating machine learning research and clinic service—574 CBCT images from 389 patients KURIAS-ECG: a 12-lead electrocardiogram database with standardized diagnosis ontology—EEG from 147 subjects VinDr-PCXR: an open, large-scale pediatric chest X-ray dataset for interpretation of common thoracic diseases—adult chest radiography (CXR) from 9125 subjects VinDr-SpineXR: A large annotated medical image dataset for spinal lesions detection and classification from radiographs—10,466 spine X-ray images from 5000 studies | Restricted access | |
| National Sleep Research Resource | [166] | Polysomnography | Apnea Positive Pressure Long-Term Efficacy Study—1516 subject Efficacy Assessment of NOP Agonists in Non-Human Primates—5 subjects Maternal Sleep in Pregnancy and the Fetus—106 subjects Apnea, Bariatric Surgery, and CPAP Study—49 subjects Best Apnea Interventions in Research—169 subjects Childhood Adenotonsillectomy Trial—1243 subjects Cleveland Children’s Sleep and Health Study—517 subjects Cleveland Family Study—735 subjects Cox and Fell (2020) Sleep Medicine Reviews—3 subjects Heart Biomarker Evaluation in Apnea Treatment—318 subjects Hispanic Community Health Study/Study of Latinos—16,415 subjects Home Positive Airway Pressure—373 subjects Honolulu-Asia Aging Study of Sleep Apnea—718 subjects Learn—3 subjects Mignot Nature Communications—3000 subjects MrOS Sleep Study—2237 subjects NCH Sleep DataBank—3673 subjects Nulliparous Pregnancy Outcomes Study monitoring mothers to be—3012 subjects Sleep Heart Health Study—5804 subjects Stanford Technology Analytics and Genomics in Sleep—1881 subjects Study of Osteoporotic Fractures—461 subjects Wisconsin Sleep Cohort—1123 subjects | Public on request (no commercial use) | |
| Open Access Series of Imaging Studies—OASIS Brain | [167] | MRI, Alzheimer’s disease | OASIS-1—416 subjects OASIS-2—150 subjects OASIS-3—1379 subjects OASIS-4—663 subjects | Public on request (no commercial use) | |
| OpenNeuro | [168] | MRI, PET, MEG, EEG, and iEEG data (various types of disorders, depending on the database) | 595 MRI public datasets—23 304 subjects 8 PET public datasets—19 subjects 161 EEG public dataset—6790 subjects 23 iEEG public dataset—550 subjects 32 MEG public dataset—590 subjects | Public | |
| Brain Tumor Dataset | [169] | MRI, brain tumor | MRI—233 subjects | Public | |
| Cancer Imaging Archive (TCIA) | [170] | MR, CT, positron emission tomography, computed radiography, digital radiography, nuclear medicine, other (a category used in DICOM for images that do not fit into the standard modality categories), structured reporting, pathology, various | HNSCC-mIF-mIHC-comparison—8 subjects CT-Phantom4Radiomics—1 subject Breast-MRI-NACT-Pilot—64 subjects Adrenal-ACC-Ki67-Seg—53 subjects CT Lymph Nodes—176 subjects UCSF-PDGM—495 subjects UPENN-GBM—630 subjects Hungarian-Colorectal-Screening—200 subjects Duke-Breast-Cancer-MRI—922 subjects Pancreatic-CT-CBCT-SEG—40 subjects HCC-TACE-Seg—105 subjects Vestibular-Schwannoma-SEG—242 subjects ACRIN 6698/I-SPY2 Breast DWI—385 subjects I-SPY2 Trial—719 subjects HER2 tumor ROIs—273 subjects DLBCL-Morphology—209 subjects CDD-CESM—326 subjects COVID-19-NY-SBU—1384 subjects Prostate-Diagnosis—92 subjects NSCLC-Radiogenomics—211 subjects CT Images in COVID-19—661 subjects QIBA-CT-Liver-Phantom—3 subjects Lung-PET-CT-Dx—363 subjects QIN-PROSTATE-Repeatability—15 subjects NSCLC-Radiomics—422 subjects Prostate-MRI-US-Biopsy—1151 subjects CRC_FFPE-CODEX_CellNeighs—35 subjects TCGA-BRCA—139 subjects TCGA-LIHC—97 subjects TCGA-LUAD—69 subjects TCGA-OV—143 subjects TCGA-KIRC—267 subjects Lung-Fused-CT-Pathology—6 subjects AML-Cytomorphology_LMU—200 subjects Pelvic-Reference-Data—58 subjects CC-Radiomics-Phantom-3—95 subjects MiMM_SBILab—5 subjects LCTSC—60 subjects QIN Breast DCE-MRI—10 subjects Osteosarcoma Tumor Assessment—4 subjects CBIS-DDSM—1566 subjects QIN LUNG CT—47 subjects CC-Radiomics-Phantom—17 subjects PROSTATEx—346 subjects Prostate Fused-MRI-Pathology—28 subjects SPIE-AAPM Lung CT Challenge—70 subjects ISPY1 (ACRIN 6657)—222 subjects Pancreas-CT—82 subjects 4D-Lung—20 subjects Soft-tissue-Sarcoma—51 subjects LungCT-Diagnosis—61 subjects Lung Phantom—1 subject Prostate-3T—64 subjects LIDC-IDRI—1010 subjects RIDER Phantom PET-CT—20 subjects RIDER Lung CT—32 subjects BREAST-diagnosis—88 subjects CT colonography (ACRIN 6664)—825 subjects | Public (free access, registration required) | |
| LUNA16 | [171] | CT, lung nodules | LUNA16- 888 CT scans | Public (free access to all users) | |
| MICCAI 2012 Prostate Challenge | [172] | MRI, prostate imaging | Prostate segmentation in transversal T2-weighted MR images—50 training cases | Public (free access to all users) | |
| IEEE Dataport | [173] | Ultrasound images, brain MRI, ultrawide-field fluorescein angiography images, chest X-rays, mammograms, CT, Lung Image Database Consortium, and thermal images | CNN-based image reconstruction method for ultrafast ultrasound imaging: 31,000 images OpenBHB: a multisite brain MRI Dataset for age prediction and debiasing: >5000—Brain MRI Benign Breast Tumor Dataset: 83 patients—mammograms X-ray bone shadow suppression: 4080 images STROKE: CT series of patients with M1 thrombus before thrombectomy: 88 patients Automatic lung segmentation results: NextMED project—718 of the 1012 LIDC-IDRI scans PRIME-FP20: ultrawide-field fundus photography vessel segmentation dataset—15 images Plantar Thermogram Database for the Study of Diabetic Foot Complications—122 subjects (DM group) and 45 subjects (control group) | Part public and part restricted (subscription) | |
| AIMI | [174] | Brain MRI studies, chest X-rays, echocardiograms, CT | BrainMetShare: 156 subjects CheXlocalize: 700 subjects BrainMetShare: 156 subjects COCA—coronary calcium and chest CTs: not specified CT pulmonary angiography: not specified CheXlocalize: 700 subjects CheXpert: 65,240 subjects CheXphoto: 3700 subjects CheXplanation: not specified DDI—Diverse Dermatology Images: not specified EchoNet-Dynamic: 10,030 subjects EchoNet-LVH: 12,000 subjects EchoNet-Pediatric: 7643 subjects LERA—Lower Extremity Radiographs: 182 subjects MRNet: 1370 subjects MURA: 14,863 studies Multimodal Pulmonary Embolism Dataset: 1794 subjects SKM-TEA: not specified Thyroid Ultrasound Cine-clip: 167 subjects CheXpert: 224,316 chest radiographs of 65,240 subjects | Public (free access) | |
| Fast MRI | [175] | MRI | Knee: 1500+ subjects Brain: 6970 subjects Prostate: 312 subjects | Public (free access, registration required) | |
| ADNI | [176] | MRI, PET | Scans related to Alzheimer’s disease | Public (free access, registration required) | |
| Pediatric Brain Imaging Dataset | [177] | MRI | Over 500 pediatric brain MRI scans | Public (free access to all users | |
| ChestX-ray8 | [178] | Chest X-ray images | NIH Clinical Center Chest X-Ray Dataset—over 100,000 images from more than 30,000 subjects | Public (free access to all users) | |
| Breast Cancer Digital Repository | [179] | MLO and CC images | BCDR-FM (film mammography repository): 1010 subjects BCDR-DM (full-field digital mammography repository): 724 subjects | Public (free access, registration required | |
| Brain-CODE | [180] | Neuroimaging | High-resolution magnetic resonance imaging of mouse model related to autism: 839 subjects | Restricted (application for access is required and open data releases) | |
| RadImageNet | [181] | PET, CT, ultrasound, MRI with DICOM tags | 5 million images from over 1 million studies across 500,000 subjects | Public subset available; full dataset licensable; academic access with restrictions | |
| EyePACS | [182] | Retinal fundus images for diabetic retinopathy screening | Images for training and validation set—57,146 images, test set—8790 images | Available through the Kaggle competition | |
| Medical Segmentation Decathlon | [183] | mp-MRI, MRI, CT | 10 datasets Cases (Train/Test) | Open source license, available for research use | |
| Brain | 484/266 | ||||
| Heart | 20/10 | ||||
| Hippocampus | 263/131 | ||||
| Liver | 131/70 | ||||
| Lung | 64/32 | ||||
| Pancreas | 282/139 | ||||
| Prostate | 32/16 | ||||
| Colon | 126/64 | ||||
| Hepatic Vessels | 303/140 | ||||
| Spleen | 41/20 | ||||
| DDSM | [184] | Mammography images | 2500 studies with images, subject info—2620 cases in 43 volumes categorized by case type | Public (free access) | |
| LIDC-IDRI | [185] | CT images with annotations | 1018 cases with XML and DICOM files—images (DICOM, 125GB), DICOM Metadata Digest (CSV, 314 kB), radiologist annotations/segmentations (XML format, 8.62 MB), nodule counts by patient (XLS), patient diagnoses (XLS) | Images and annotations are available for download with NBIA Data Retriever, usage under CC BY 3.0 | |
| synapse | [186] | CT scans, Zip files for raw data, registration data | CT scans—50 scans with variable volume sizes and resolutions Labeled organ data—13 abdominal organs were manually labeled Zip files for raw data—raw data: 30 training + 20 testing Registration data: 870 training–training + 600 training–testing pairs | Under IRB supervision, available for participants | |
| Mini-MIAS | [187] | Mammographic images | 322 digitized films on 2.3 GB 8 mm tape—images derived from the UK National Breast Screening Programme and digitized with Joyce-Loebl scanning microdensitometer to 50 microns, reduced to 200 microns and standardized to 1024 × 1024 pixels for the database | Free for scientific research under a license agreement | |
| Breast Cancer Histopathological Database (BreakHis) | [188] | Microscopic images of breast tumor | 9109 microscopic images of breast tumor tissue collected from 82 subjects | Free for scientific research under a license agreement | |
| Messidor | [189] | Eye fundus color numerical images | 1200 eye fundus color numerical images of the posterior pole | Free for scientific research under a license agreement | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rudnicka, Z.; Szczepanski, J.; Pregowska, A. Artificial Intelligence-Based Algorithms in Medical Image Scan Segmentation and Intelligent Visual Content Generation—A Concise Overview. Electronics 2024, 13, 746. https://doi.org/10.3390/electronics13040746
Rudnicka Z, Szczepanski J, Pregowska A. Artificial Intelligence-Based Algorithms in Medical Image Scan Segmentation and Intelligent Visual Content Generation—A Concise Overview. Electronics. 2024; 13(4):746. https://doi.org/10.3390/electronics13040746
Chicago/Turabian StyleRudnicka, Zofia, Janusz Szczepanski, and Agnieszka Pregowska. 2024. "Artificial Intelligence-Based Algorithms in Medical Image Scan Segmentation and Intelligent Visual Content Generation—A Concise Overview" Electronics 13, no. 4: 746. https://doi.org/10.3390/electronics13040746
APA StyleRudnicka, Z., Szczepanski, J., & Pregowska, A. (2024). Artificial Intelligence-Based Algorithms in Medical Image Scan Segmentation and Intelligent Visual Content Generation—A Concise Overview. Electronics, 13(4), 746. https://doi.org/10.3390/electronics13040746








