Abstract
The rapid expansion of medical information has resulted in named entity recognition (NER) and relation extraction (RE) essential for clinical decision support systems. Medical texts often contain specialized vocabulary, ambiguous abbreviations, synonyms, polysemous terms, and overlapping entities, which introduce significant challenges to the extraction process. Existing approaches, which typically rely on single models such as BiLSTM or BERT, often struggle with these complexities. Although large language models (LLMs) have shown promise in various NLP tasks, they still face limitations in handling token-level tasks critical for medical NER and RE. To address these challenges, we propose COMCARE, a collaborative ensemble framework for context-aware medical NER and RE that integrates multiple pre-trained language models through a collaborative decision strategy. For NER, we combined PubMedBERT and PubMed-T5, leveraging PubMedBERT’s contextual understanding and PubMed-T5’s generative capabilities to handle diverse forms of medical terminology, from standard domain-specific jargon to nonstandard representations, such as uncommon abbreviations and out-of-vocabulary (OOV) terms. For RE, we integrated general-domain BERT with biomedical-specific BERT and PubMed-T5, utilizing token-level information from the NER module to enhance the context-aware entity-based relation extraction. To effectively handle long-range dependencies and maintain consistent performance across diverse texts, we implemented a semantic chunking approach and combined the model outputs through a majority voting mechanism. We evaluated COMCARE on several biomedical datasets, including BioRED, ADE, RDD, and DIANN Corpus. For BioRED, COMCARE achieved F1 scores of 93.76% for NER and 68.73% for RE, outperforming BioBERT by 1.25% and 1.74%, respectively. On the RDD Corpus, COMCARE showed F1 scores of 77.86% for NER and 86.79% for RE while achieving 82.48% for NER on ADE and 99.36% for NER on DIANN. These results demonstrate the effectiveness of our approach in handling complex medical terminology and overlapping entities, highlighting its potential to improve clinical decision support systems.
1. Introduction
Recently, the rapid advancement of Internet technologies, coupled with global health crises such as the COVID-19 pandemic, has led to the emergence of a new healthcare paradigm, ‘Internet Plus Healthcare‘ [1]. This paradigm has significantly expanded online access to medical information, increasing both the quantity and complexity of healthcare-related content. Although this growing volume of medical information has facilitated knowledge sharing and collaboration, it has introduced significant challenges in efficiently extracting accurate and relevant information from unstructured medical texts [2]. In the medical domain, precise and efficient information extraction is crucial because it can directly affect patient outcomes [3].
In this context, named entity recognition (NER) and relation extraction (RE), which transform unstructured medical text into structured data to enhance information processing, have become essential tasks in medical natural language processing (NLP). NER identifies and classifies medical entities, such as diseases, symptoms, drugs, and genes, whereas RE determines the relationships between these entities. These two tasks are critical for downstream applications, including clinical decision support systems, knowledge base construction, literature-based discovery, and clinical trial matching. Automating the extraction of medical entities and their relationships enables healthcare professionals to make informed and timely decisions and uncover hidden insights in the biomedical literature, ultimately improving patient care [4,5].
However, the development of robust NER and RE techniques in the field of medicine presents several critical challenges. These challenges are summarized as follows:
- Complex medical terminology: Medical texts often contain specialized vocabulary, ambiguous abbreviations, synonyms, and polysemous terms. For example, the abbreviation ‘CA’ can indicate either ‘cancer’ or ‘cardiac arrest’, and similarly, ‘ASD’ can refer to ‘atrial septal defect’ or ‘autism spectrum disorder’. Synonyms such as ‘myocardial infarction’ and ‘heart attack’ further complicate the identification process. Moreover, polysemous terms such as ‘discharge’ can refer to either a patient’s release from a hospital or bodily fluid, leading to potential misinterpretations that affect downstream medical applications [6,7,8].
- Diverse entity types and overlapping entities: Medical texts contain a wide variety of entity types, including diseases, symptoms, drugs, procedures, and anatomical locations. These entities often overlap, making precise entity recognition more difficult [9]. For example, in the phrase ‘aspirin therapy for stroke prevention’, ‘aspirin’ is a drug entity, and ‘stroke’ is a medical condition. Additionally, overlapping entities such as ‘lung cancer screening,’ where ‘lung cancer’ is a disease entity and ‘cancer screening’ is a medical procedure, pose challenges for models to accurately identify and differentiate each component in context.
- Context-sensitive relationships: The relationships between medical terms are highly context-dependent and can extend across multiple sentences or paragraphs [10]. For instance, in a diagnostic context, ‘hypertension’ may be identified as the primary cause of ‘heart failure’, suggesting a cause-and-effect relationship. In contrast, in the treatment context, managing ‘hypertension’ might be part of a therapeutic strategy for patients already suffering from ‘heart failure’, indicating a treatment-related relationship. These complex and long-range relationships require models that can accurately capture the context across sentences or paragraphs to extract meaningful insights.
Various approaches have been proposed to address these critical challenges in NER and RE. Early methods evolved from rule-based methods to machine-learning techniques. Rule-based methods rely on manually crafted rules and dictionaries to identify medical entities and relationships; however, they are costly to maintain and struggle to handle new or out-of-vocabulary (OOV) terms effectively [11,12]. Consequently, machine learning-based methods, such as conditional random fields (CRF) and support vector machines, emerged, using annotated corpora to train models that could automatically label sequences [13,14,15]. However, these methods also faced limitations, particularly in recognizing complex entities, such as multi-word terms (e.g., ‘acute respiratory distress syndrome’) or entities embedded within other terms (e.g., ‘type 2 diabetes mellitus’) due to difficulties in capturing contextual dependencies and intricate entity structures. These challenges are more pronounced in the medical domain, where rare diseases and complex medical terminology are common.
The advent of deep learning, particularly architectures such as bidirectional long short-term memory (BiLSTM), has significantly advanced medical NER and RE by enabling models to learn contextual patterns automatically from medical texts. BiLSTM models, often paired with CRF for sequence labeling tasks, proved effective in handling complex entity structures, specifically in token-level tasks where medical entities require interpretation based on the surrounding context [16]. However, despite these improvements, the BiLSTM models struggled with long-range dependencies because of the vanishing gradient problem, which limits their ability to retain information across extended sequences.
The bidirectional encoder representation from the transformers (BERT) model addressed many of these limitations by utilizing self-attention mechanisms to capture both short- and long-range dependencies, learning intricate contextual patterns from large-scale data [17]. BERT’s parallel processing of tokens and its use of subword tokenization have proven particularly effective in understanding complex token relationships by efficiently capturing both local and long-range dependencies in texts [16]. Pretrained language models (PLMs), such as PubMedBERT and BioBERT, trained on biomedical corpora, have shown remarkable success in managing complex medical terminology, abbreviations, and overlapping entities [18,19]. However, despite these advancements, NER and RE tasks based on BERT models often rely on a single pre-trained model, which introduces several limitations. One major issue is polysemy and context dependency. A single pre-trained model, typically trained on a fixed corpus, has a limited ability to generalize across different medical subdomains. In medical texts, where terms can have multiple meanings depending on the context, single pre-trained models often struggle to disambiguate these terms accurately. This is because they are not exposed to a sufficiently broad range of contexts during training, making it difficult for them to adapt to varying terminologies or specialized usages across medical disciplines. Additionally, OOV terms remain a persistent challenge. The medical domain has evolved rapidly, with new diseases, drugs, and treatments emerging regularly. Even models trained on biomedical corpora are constrained by the vocabulary available during training, leading to performance degradation when new or previously unseen terms are encountered, particularly in clinical data containing novel terminology. Furthermore, extracting complex relationships between medical entities poses a significant challenge for BERT-based models, primarily because of input length limitations. Relationships between medical entities are often context-sensitive and can vary significantly. These relationships frequently extend across multiple sentences or paragraphs; however, BERT’s fixed input length of 512 tokens hinders its ability to capture long-range dependencies in lengthy medical documents.
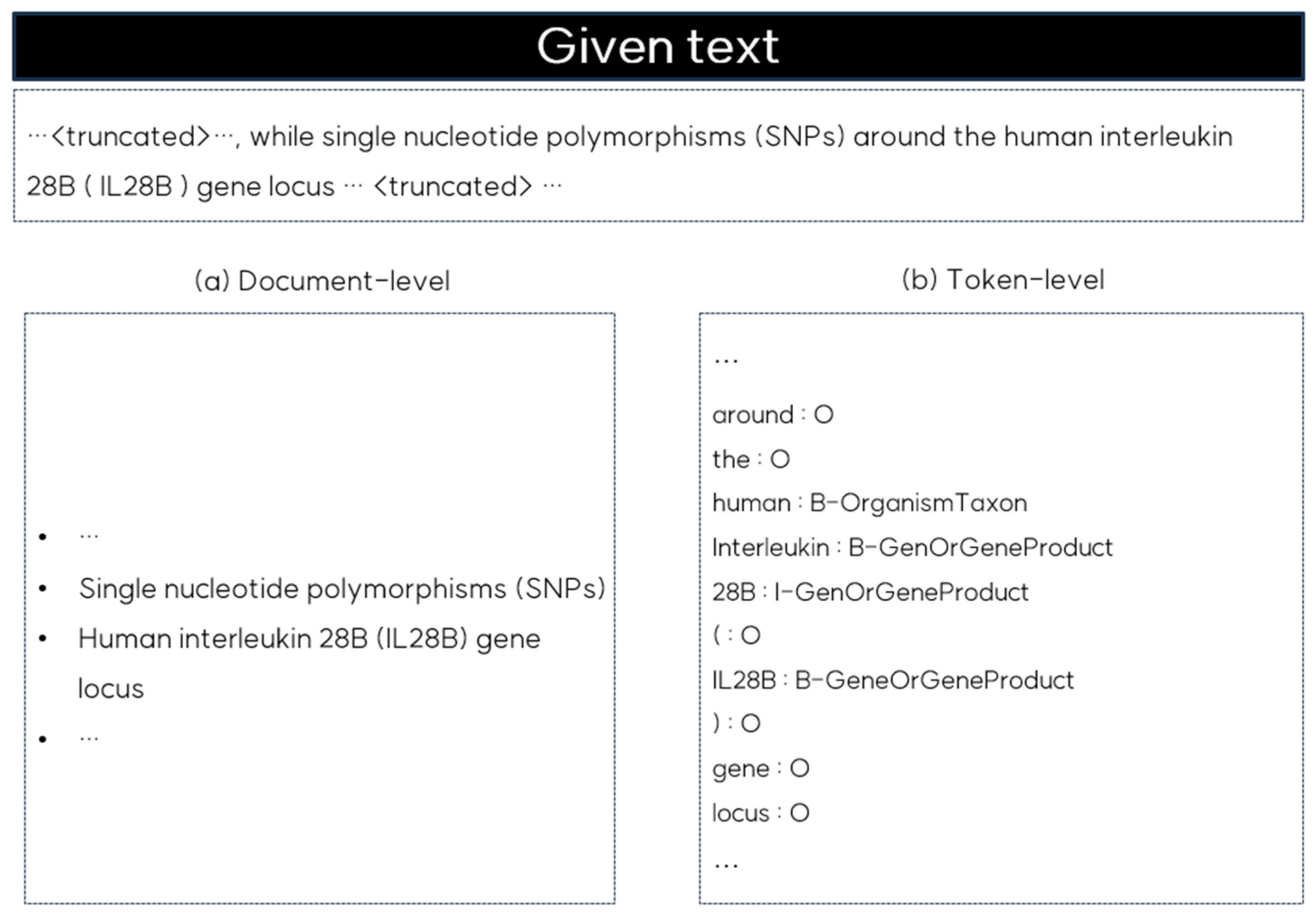
Recently, large language models (LLMs) have gained attention for their ability to perform well in zero- and few-shot learning [20,21]. These models have been successfully applied to various NLP tasks due to their capacity to generate contextually appropriate texts based on prompts without task-specific training. However, LLMs are primarily optimized for document-level understanding, and when applied to NER, they tend to identify broader entities based on the overall context, as shown in Figure 1. Document-level NER, as illustrated on the left side of Figure 1, may identify entities such as ‘single nucleotide polymorphisms’ or ‘human interleukin 28B (IL28B) gene locus’; however, it often misses the finer distinctions that token-level NER captures. Token-level NER, shown on the right side of Figure 1, annotates individual tokens such as ‘human’ as B-OrganismTaxon or ‘IL28B’ as B-GeneOrGeneProduct, providing the precision necessary in clinical and genomic contexts. This detailed level of entity recognition is crucial for understanding the specific relationships across tokens. Without this token-level granularity, the accuracy of tasks such as medical NER and RE is limited.
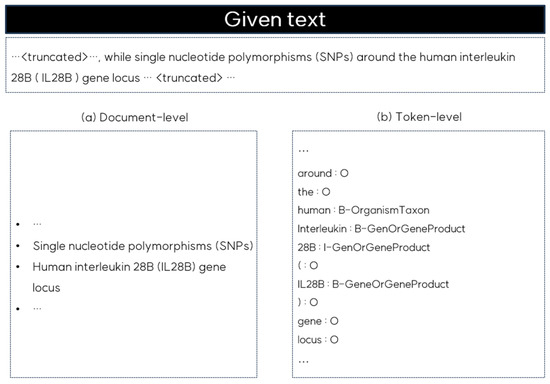
Figure 1.
Comparison of document-level and token-level NER outputs for the given text.
To address these challenges, we propose COMCARE, a collaborative ensemble framework for context-aware medical NER and RE, which integrates multiple pre-trained language models through a collaborative decision strategy. For the NER module, we employ PubMedBERT [18] and PubMed-T5 [22] to leverage their complementary strengths. PubMedBERT extracts rich contextual information from sentences, whereas PubMed-T5’s generative capabilities handle non-standard representations such as uncommon abbreviations, rarely used terms and OOV terms.
For the RE module, we integrate general-domain BERT, PubMedBERT, and PubMed-T5 to improve entity-based relation detection. This module utilizes token-level information from the NER module to enrich the relation prediction. To address long-range dependencies, we implemented a semantic chunking approach in which texts are divided into semantically coherent chunks using adaptive overlap sizes based on sentence boundaries and semantic relationships. Each chunk was independently encoded and combined into a unified context embedding. Token-level information from the NER module is directly integrated into these embeddings to create a comprehensive feature matrix, enhancing relation extraction by preserving detailed entity-specific insights. Final relationship predictions are generated using a majority voting mechanism to ensure robust extraction of context-sensitive relationships. The main contributions of this paper are as follows:
- Collaborative Decision Strategy: We present a collaborative decision strategy that fuses outputs from domain-specific models, such as PubMedBERT and PubMed-T5, leveraging their complementary strengths in handling diverse forms of medical terminology. This strategy directly addresses the challenges of complex medical terminology and overlapping entities in medical texts by combining the contextual understanding of PubMedBERT with the generative capabilities of PubMed-T5 through a token-level fusion mechanism.
- Context-Aware Relation Extraction: We integrated token-level information from the NER module with context embeddings using a semantic chunking approach, enabling the accurate capture of context-dependent relationships. This approach effectively addresses both entity-based relation detection and long-range dependencies by preserving detailed entity-specific insights while maintaining a broader contextual understanding. This integration significantly improves the ability of the model to identify and classify complex relationships across medical texts.
2. Related Work
NER and RE are essential tasks for transforming unstructured medical texts into structured data, thus enabling efficient information extraction. Consequently, numerous approaches have been proposed to address the unique challenges posed by medical texts.
2.1. Named Entity Recognition in Medical Texts
Medical NER has significantly evolved to address the challenges of processing unstructured clinical texts and complex medical terminology. Early deep-learning approaches enhanced recognition accuracy by integrating expert medical ontologies. For instance, Arbabi et al. [23] developed a neural concept recognizer (NCR) that combines CNN-based neural networks with ontologies such as human phenotype ontology (HPO) and SNOMED clinical terms (SNOMED-CT) to recognize clinical concepts in unstructured text. NCR improves upon traditional methods by predicting previously unobserved synonyms without requiring large-scale labeled training data. However, the reliance of the NCR on predefined ontologies means that it still struggles with new or rare medical terms that are not included in the ontology. To address these limitations, Zhao et al. [24] developed a neural multitask learning framework that jointly models medical NER and normalization. Their approach combined BiLSTM for sequence modeling with a CNN for character-level feature extraction, incorporating a feedback mechanism between the recognition and normalization tasks for mutual enhancement. Although their model showed notable improvements by reducing the reliance on handcrafted features, its performance remained highly dependent on the quality and coverage of the pre-trained medical word embeddings. Consequently, it struggled with OOV terms, which are frequent in medical texts, due to newly coined terminology and variations in medical language. Additionally, their model experienced difficulties in identifying polysemous medical terms and abbreviations within the context, specifically for terms that were underrepresented in pre-trained embeddings.
Recent studies have extensively leveraged pre-trained BERT models for medical NER tasks. However, these approaches continue to experience considerable challenges in handling the complexity and context dependency of medical language. Chaudhry et al. [25] developed a multitask learning framework using PubMedBERT for NER, achieving improved efficiency and accuracy on structured biomedical datasets. However, their approach struggled with contextual understanding, particularly for tissue entities in which polysemous terms required precise disambiguation. This limitation arises from the reliance on a single pre-trained model, which constrains the ability of the model to capture diverse contextual variations across different medical contexts. Li et al. [26] compared six pre-trained language models, including general-domain models such as BERT and SpanBERT and biomedical-specific models such as BioBERT, BlueBERT, PubMedBERT, and SciBERT, for NER tasks within the clinical trial eligibility criteria. Although PubMedBERT achieved the highest performance across the three corpora, significant challenges remained in handling non-flat annotations such as nested and overlapping entities. These issues arise from the challenge of capturing hierarchical and complex relationships among medical concepts within a single-model architecture. Yi et al. [27] introduced a NER method that combines part-of-speech (POS) tagging and stroke features with word vectors from a BERT-based model. Their BiLSTM-CRF architecture, using a BERT-base for pre-training, showed promising results for Chinese medical texts by incorporating language-specific features, such as stroke patterns and POS information, to help distinguish words with different semantics. However, the reliance of the model on language-specific features, such as POS tagging, combined with the stroke features of Chinese texts, limits its generalizability to other languages with differing linguistic structures. Additionally, this reliance on feature extraction introduces vulnerability to noisy or incomplete data, which is a common issue in the medical domain. Generally, clinical notes, health records, and the biomedical literature frequently contain inconsistencies, abbreviations, and unstructured formats that add significant noise and hinder accurate data extraction.
Recent efforts to overcome these limitations have led to the introduction of contrasting learning and feature attention mechanisms. Liang et al. [28] proposed textual entailment with dynamic contrastive learning (TEDC) and referred to BioNER as a textual entailment task for managing cases with limited token-level annotations. However, their binary classification framework (i.e., entity vs. non-entity) potentially oversimplifies the complex relationships between biomedical entities and encounters challenges when applied to complex and overlapping entities. Furthermore, its reliance on gazetteers (i.e., predefined entity lists) for initial training limits the entity coverage of the model. Their experiments demonstrated that these gazetteers covered only approximately 50% of the entities in the BC5CDR and NCBI-Disease datasets, limiting the ability of the model to effectively recognize novel or rare medical terms. Zhang et al. [29] integrated syntactic information through a combined feature attention mechanism, enhancing biomedical entity recognition by leveraging POS tags, syntactic constituents, and dependency relations. Although this approach improves the recognition accuracy, the model still encounters challenges with rare medical terminology and multi-level dependencies often found in complex biomedical texts. Specifically, its reliance on pre-trained BioBERT embeddings and external syntactic processing tools constrains its ability to manage uncommon terms and fully capture intricate semantic relationships, particularly in distinguishing nested or overlapping biomedical entities within multi-sentence contexts.
To address these limitations, we propose an approach that combines multiple domain-specific pre-trained models, including PubMedBERT and PubMed-T5, within an advanced ensemble framework. Our approach effectively addresses three major challenges: contextual ambiguity in medical terminology, nonstandard expressions, and nested entities. Rather than relying on single pre-trained models or simple model combinations, we incorporate BiLSTM layers to learn features from each model’s output, followed by a collaborative decision strategy that leverages the complementary strengths of each model. The CRF layer further strengthens our architecture, ensuring sequence-level consistency in entity predictions and addressing challenges with nested and overlapping entities. By integrating the generative capabilities of PubMed-T5 with the specialized contextual understanding of PubMedBERT, our framework effectively processes both standardized medical terminology and nonstandard clinical expressions. This integration specifically addresses the complexities of polysemy and OOV terms, which pose challenges to previous approaches.
2.2. Relation Extraction in Medical Texts
Medical RE has been developed using various architectures. However, the existing methods face significant limitations in addressing the unique challenges inherent in medical texts. Dewi et al. [30] developed DeepCNN, a 10-layer CNN architecture for extracting drug-drug interactions, which utilizes multi-channel word embeddings and deep layers to capture long-range dependencies. However, their approach relied on single-sentence contexts and word-level embeddings, making it difficult to handle cross-sentence relationships and complex medical terminology. Furthermore, their focus on specific drug-drug interactions limits generalizability across diverse medical entity types and relationships. Fabregat et al. [31] proposed a joint NER and RE model for biomedical texts, integrating transfer learning from negation detection and combining BiLSTM-CRF architectures with linguistic features such as POS tags, casing, and word and character embeddings. However, their BiLSTM architecture has issues with vanishing gradients in long-range dependencies. Although their character-level processing addresses some OOV terms, this approach is limited by context-dependent embeddings, thereby reducing adaptability to unfamiliar or less frequent terms. Additionally, the reliance of the model on these embeddings presents challenges for handling biomedical synonyms.
Recently, transformer architectures have been increasingly applied to medical RE tasks. Luo et al. [32] introduced BioRED, a comprehensive biomedical relation extraction dataset, showing that domain-specific models, such as PubMedBERT, outperform general BERT models because of their specialized coverage of medical vocabulary. However, their approach encounters challenges in extracting relationships involving ambiguous terminology and overlapping entities, which leads to high error rates. Additionally, the restriction of the model to abstract-level annotations limits its ability to capture long-range contextual relationships, resulting in a notably low performance in detecting novel relations. Li et al. [33] demonstrated the effectiveness of ensemble learning in biomedical NLP tasks by combining multiple domain-specific models and BioM-ELECTRA. Their approach demonstrated superior performance in both named entity recognition and document-level relation extraction on the LitCoin Challenge dataset. However, it still faces challenges with the inherent complexities of biomedical texts. The relatively lower performance of the model on ‘Association’ relationships indicates limitations in capturing complex, context-dependent relationships across document contexts. These limitations demonstrate difficulties in handling specialized terminology, overlapping entities, and context-sensitive relationships.
Despite recent advancements, many studies have not fully explored the advantages of combining general-domain BERT pre-trained models with domain-specific pre-trained models. Although domain-specific pre-trained models perform well in capturing specialized terminology, integrating general-domain BERT can enhance contextual awareness, particularly for relationships that extend beyond narrowly defined medical terms. This combination could improve the extraction of context-sensitive relationships by leveraging specialized medical knowledge and broader linguistic understanding.
To address these limitations, we integrate multiple pre-trained models, including BERT, PubMedBERT, and PubMed-T5, to capture diverse perspectives on handling complex medical terminology and relationships. Additionally, by employing a novel semantic chunking approach, we overcome the input length constraints of existing BERT models. Furthermore, integrating token-level NER information with context embeddings enhances entity-based relation detection, allowing the model to better manage overlapping entities and capture complex context-sensitive medical relationships. Our methods demonstrate significant improvements over existing RE methods.
2.3. Large Language Models in Medical Texts
The emergence of LLMs has significantly affected various fields, including healthcare, due to their advanced language processing capabilities. In medical NER and RE, LLMs have shown promising potential and notable limitations.
Lu et al. [34] highlighted that although LLMs have shown promise in document-level NER, they encounter significant challenges in token-level clinical NER. They revealed that most LLMs struggle with precise entity boundary detection and type classification at the token level, in contrast to the relatively simple task of identifying entities at the document level. A comparative analysis with BioClinicalBERT further indicates that LLMs generally underperform compared to BERT models in token-level NER tasks. This performance gap is attributed to the architectural design and pretraining objectives of LLMs, which focus on next-token prediction rather than token-level classification, limiting their effectiveness in precise entity localization and boundary detection compared with BERT models optimized for detailed text representation. Similarly, Hu et al. [35] investigated the capabilities of GPT models for clinical NER tasks by examining both the potential and limitations of prompt engineering approaches. Although their task-specific prompt framework, which incorporated annotation guidelines and few-shot examples, improved the performance of the GPT models, significant challenges remained. Their results demonstrated that even with optimized prompts, the performance of GPT-4 consistently lagged behind that of BioClinicalBERT, with the performance gap becoming more pronounced. These findings underscore the fundamental limitations of the ability of LLMs to precisely identify entity boundaries and classify complex medical terminology. Additionally, they observed that achieving competitive performance required careful prompt engineering and domain-specific knowledge integration, raising concerns about the practical applicability of LLMs in clinical settings where annotation precision is crucial.
For RE, Zhou et al. [36] introduced LEAP, an instruction-example adaptive prompting framework that dynamically integrates instructions and examples to optimize the LLM performance on biomedical RE tasks. This approach enabled models such as Llama2-MedTuned to achieve higher accuracy, demonstrating its efficacy in capturing complex relationships within biomedical texts. The adaptability of the LEAP framework to varying contexts and relationship types presents a promising solution for the limitations of traditional LLMs in extracting nuanced biomedical relationships. However, challenges persist in maintaining consistent performance across diverse datasets and in processing long-form documents with multi-sentence relationships.
To address these limitations, we propose a comprehensive approach that integrates token-level NER with RE using context embeddings, thereby enabling the precise handling of entity boundaries and classifications. By combining multiple domain-specific pre-trained models within a collaborative decision strategy, we leverage both generative and contextual capabilities to effectively address the complexities of medical terminology, overlapping entities, and cross-sentence relationships.
3. Methods
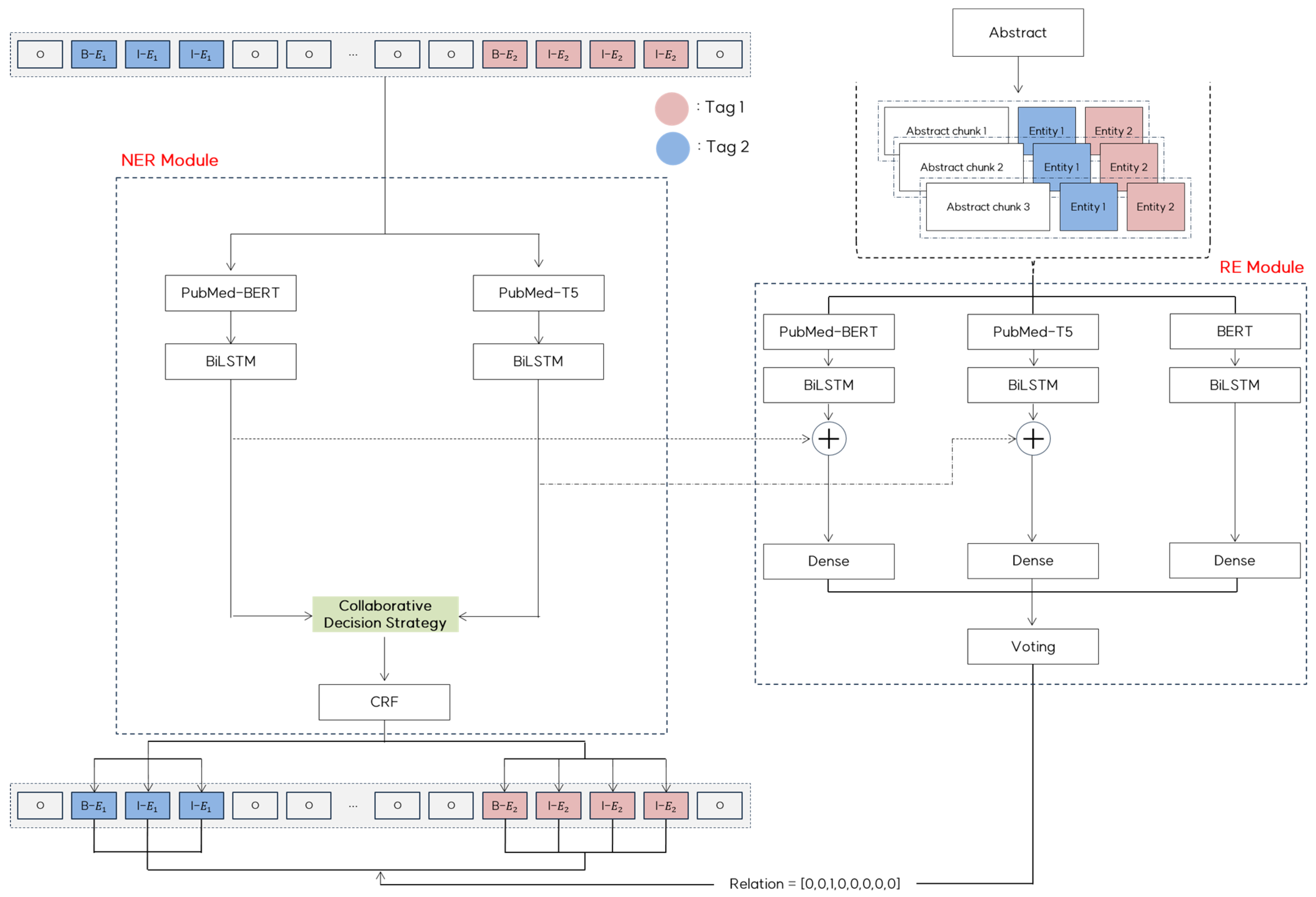
This section presents a detailed overview of COMCARE. Figure 2 illustrates our framework that leverages multiple pre-trained language models to enhance performance in handling complex and overlapping medical entities and context-sensitive relationships.
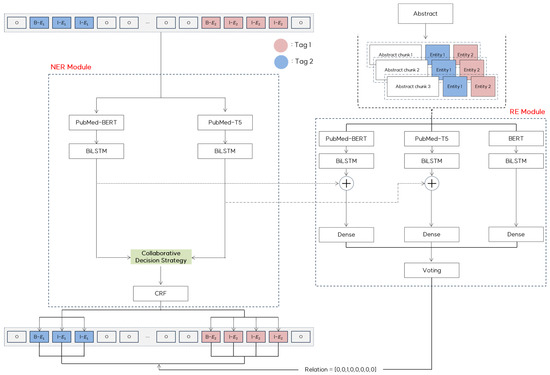
Figure 2.
COMCARE: Proposed NER and RE framework.
In the NER module, we integrate domain-specific pre-trained models to utilize their complementary strengths. PubMedBERT extracts rich contextual information from sentences, whereas PubMed-T5 addresses nonstandard representations, such as abbreviations and synonyms, through generative capabilities. The outputs from these models were processed using BiLSTM layers to capture the sequential dependencies. These outputs are then combined using a collaborative decision strategy, and the CRF layers ensure consistency across the predicted tag sequences, maintaining high precision in entity recognition.
In the RE module, we integrate predictions from BERT, PubMedBERT, and PubMed-T5 using a semantic chunking approach. To efficiently handle long documents, we utilize abstracts, which are divided into semantically coherent chunks using adaptive overlap sizes based on sentence boundaries and semantic relationships. These chunks preserve meaningful context while efficiently handling long documents. Each chunk is processed independently using three models. BiLSTM layers enhance feature learning, and token-level outputs from the NER module are incorporated into context embeddings to improve the relation extraction accuracy. The final predictions from the models are combined using a voting mechanism to ensure the robust detection of context-sensitive relationships across entities.
3.1. Medical Named Entity Recognition Module
The NER module consists of four main processing stages for effectively recognizing medical entities: (1) domain-specific token-embedding generation with pre-trained models, (2) feature learning through BiLSTM networks, (3) collaborative decision strategy, and (4) a CRF layer.
3.1.1. Domain-Specific Token Embedding Generation
The NER component of COMCARE leverages the input text using two domain-specific pre-trained models to obtain token embeddings. For entity extraction, an input sentence , where represents individual tokens and denotes the sequence length, is first tokenized using the PubMedBERT tokenizer, which employs WordPiece tokenization. The tokenized input is then processed by PubMedBERT, which is pre-trained on PubMed abstracts, to generate the embedding matrix , where = 768 represents the embedding dimension.
In parallel, we utilize a PubMed-T5 model pre-trained on PubMed summarization datasets to generate complementary token embeddings. This model was specifically selected for its generative capabilities, which make it more robust in handling nonstandard word representations than transformer encoder-only models, such as BERT. Although the PubMed-T5 model typically uses SentencePiece [37] tokenization, we applied WordPiece tokenization [38] with the PubMedBERT tokenizer for both models to maintain consistency in token-level information for the NER module. The WordPiece-tokenized input is then processed through the encoder of PubMed-T5, producing an embedding matrix with the same dimensionality as the PubMedBERT embeddings.
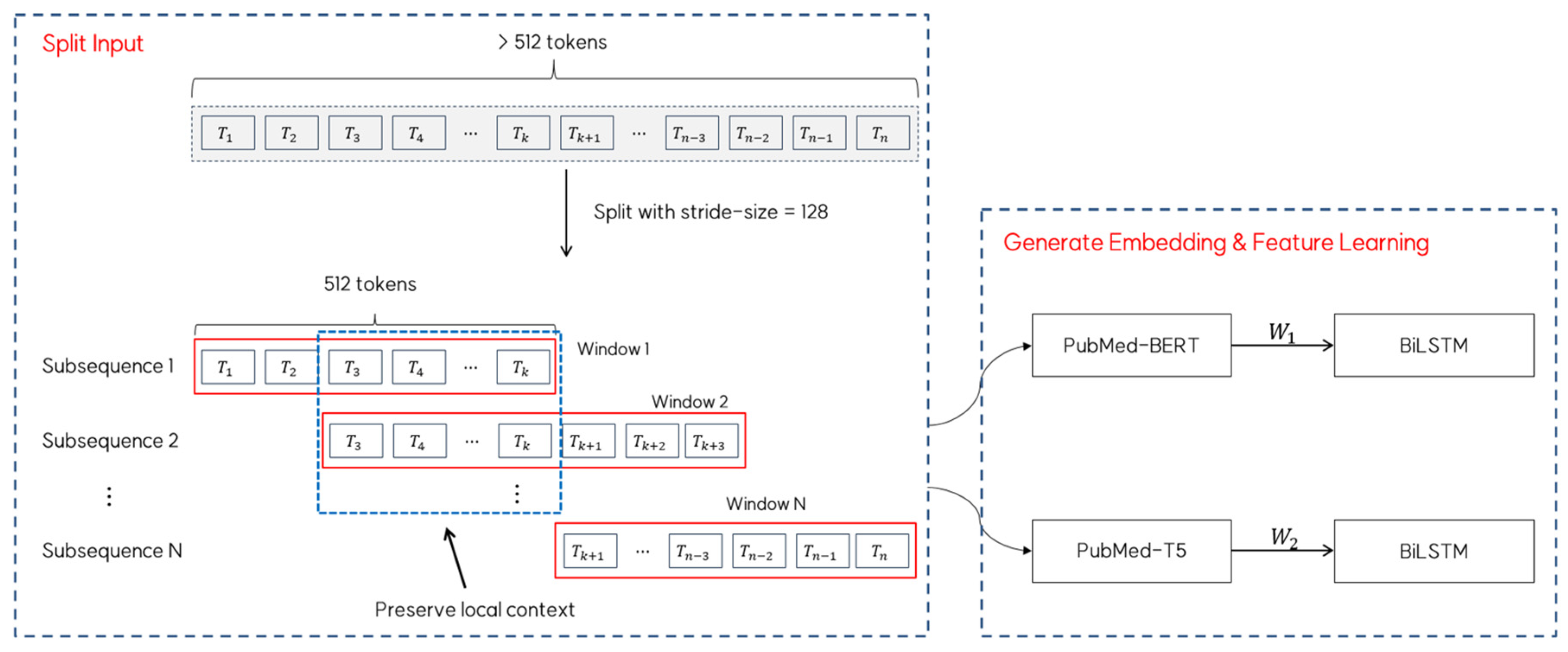
For sequences longer than the maximum length (i.e., 512), we employed a sliding-window approach with a stride of 128 tokens to ensure complete coverage while preserving the local context. In this approach, each window processes 512 tokens, and consecutive windows overlap by 384 tokens to maintain continuity in the context representation. The resulting embedding matrices and from each window are then fed into separate BiLSTM layers for feature learning. Figure 3 provides a visual representation of this sliding window mechanism, showing how the input text is segmented and processed with overlapping windows to maintain contextual coherence throughout the sequence.
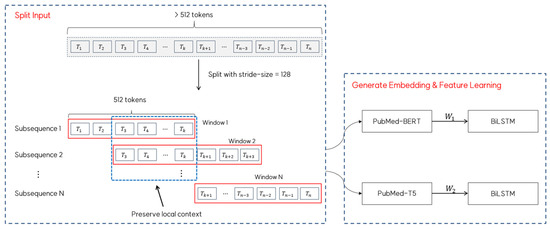
Figure 3.
Sliding window approach for long sequence processing for embedding generation.
3.1.2. Feature Learning Through BiLSTM Networks
Following embedding generation, each stream is processed through a separate BiLSTM layer to capture the contextual features of each token. The BiLSTM architecture, an extension of LSTM, processes sequences in both forward and backward directions, allowing the model to effectively capture contextual dependencies from both preceding and succeeding tokens in the sequence.
During this process, each domain-specific embedding sequence, and , is processed through separate forward and backward LSTM layers, producing forward hidden states and backward hidden states for each token. In this context, represents the m-dimensional hidden state at the time step from the forward LSTM, whereas represents the corresponding hidden state from the backward LSTM. The forward and backward hidden states are concatenated for each token to form a single unified sequence , where , capturing bidirectional information. This representation is then passed through an output layer to produce the probability matrix , where represents the probability that the -th word is assigned to the -th tag.
Subsequently, each BiLSTM layer generates emission matrices and from the PubMedBERT and PubMed-T5 streams, respectively. Each matrix has dimensions corresponding to sequence length and the number of possible entity tags, k, with each element representing the probability of a token being assigned a specific tag. These emission matrices were then combined using our collaborative decision strategy to form an ensemble matrix, .
3.1.3. Collaborative Decision Strategy
We introduce a collaborative decision strategy to effectively merge the emission matrices from two BiLSTM streams into an ensemble matrix, . Given two emission matrices, , we generate the resulting matrix , where denotes the number of tokens in the sentence and represents the number of possible output tags. This strategy was designed to leverage the complementary strengths of each model while maintaining robust performance across different types of medical entities.
Our ensemble strategy applies specific rules to minimize incorrect non-entity ‘O’ tags predictions and enhance the detection of valid medical entities. The steps of our strategy are as follows:
- Confidence-based Tag Selection: For a given token, each emission matrix and outputs probability values for all possible entity tags (e.g., O, Tag1, Tag2, Tag3). From these tag probability distributions, we identify the tag and its corresponding confidence score with the highest probability from each matrix. For instance, given an emission matrix P1 with probabilities {O:0.2, Tag1:0.3, Tag2:0.4, Tag3:0.1}, the model would select Tag2 as its prediction with a confidence score of 0.4.
- Avoidance of non-entity ‘O’ Tag Bias: When one model predicts non-entity ‘O’ tags with the highest probability and the other model predicts an entity tag with a probability above a predefined threshold, the ensemble strategy selects the entity tag. This threshold was empirically set to 0.6 by evaluating a range of potential values (from 0.1 to 1.0) on the validation set. During our experiments, we observed that lower thresholds (0.1–0.5) led to increased false positives, while higher thresholds (0.7–1.0) resulted in missed entity detections. The threshold of 0.6 provided the most effective balance between precision and recall, reducing false negatives while maintaining a high level of accuracy for detected entities. This rule helps prevent potential entities from being overlooked when only one model predicts ‘O’ tags.
- Weighted Combination: If both models assign the highest probability to the same tag, the ensemble strategy averages the probabilities as , where denotes the token index within the sequence. In cases where the models disagree and neither the confidence-based selection nor the ‘O’ tags avoidance rule applies, a weighted combination is used where . Here, denotes a weight parameter computed using a softmax function to determine the relative importance of each model’s prediction for that token.
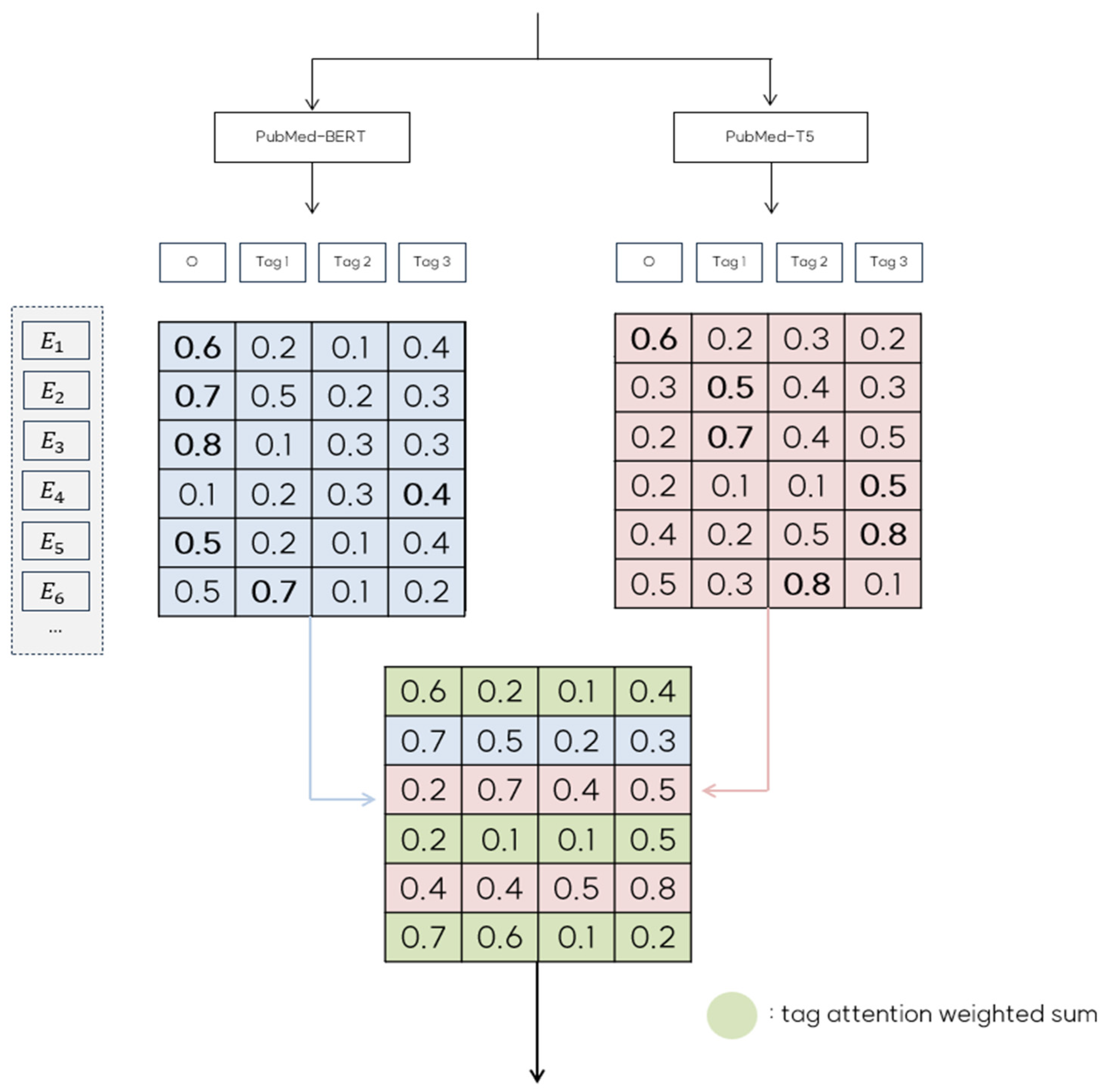
Figure 4 illustrates how emission score matrices and are combined into the final ensemble matrix EM based on specific ensemble rules.
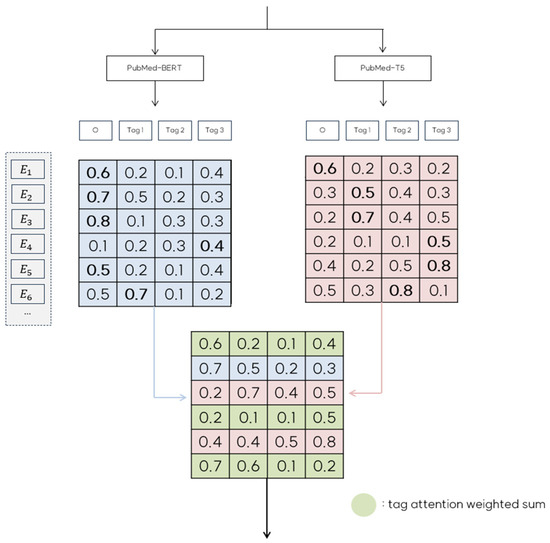
Figure 4.
An illustration of the collaborative decision strategy for the resulting emission score matrix.
For instance, in the case of row in Figure 4, predicts the ‘O’ tag with a probability of 0.8, whereas predicts ‘Tag1’ with a probability 0. 7. Because our strategy aims to minimize ‘O’ tag predictions whenever possible, we select row from and use ‘Tag1’ as the corresponding row in EM. Similarly, row predicts ‘Tag1’ with a probability of 0.5. However, the confidence score does not meet the threshold of 0.6. In this case, the prediction from is considered unreliable; therefore, we use row from as the row in EM. In cases where both models predict ‘O’ tags but differ in their predictions, as seen with , we apply a tag attention weighting mechanism to create a weighted sum of the rows from and . This combined row is then used in EM, enhancing robustness by integrating information from both models. Upon completing the collaborative decision ensemble, and are merged to form a unified emission matrix EM. This process is formalized in Algorithm 1, which details our collaborative decision ensemble strategy.
| Algorithm 1. Collaborative Decision Strategy |
| Input: Sentence S Output: Resulting ensemble matrix EM |
| 1: 2: 3: 4: 5: Initialize resulting ensemble matrix 6: for each token i in sequence do: 7: Compute attention weight 8: Get predictions from each model://Select tag and confidence score with highest probability from each model 9: 10: 11: if : 12: if : 13: 14: else if : 15: 16: else 17: //weighted combination based on attention 18: else 19: ////simple average if predictions match 20: return EM |
The resulting ensemble matrix is then passed to the CRF layer for the final sequence optimization and tag prediction. This approach leverages the strengths of both models by incorporating confidence scores and contextual information to enhance the precision and robustness of entity recognition.
3.1.4. Conditional Random Field (CRF) Layer
The final part of the NER module is the CRF layer, which captures the dependencies between adjacent labels to ensure global consistency in the predicted tag sequence. This CRF layer is crucial for medical entity recognition because it enforces specific sequential patterns that must be followed by different entity types. For instance, a ‘DiseaseOrPhenotypicFeature’ entity should begin with a ‘B-DIS’ tag, followed by ‘I-DIS’ tags, and cannot be followed by tags from different entity types, such as ‘I-DRUG’ or ‘I-GEN’. By enforcing these constraints, the CRF layer prevents invalid label sequences, thereby improving the overall accuracy and consistency of entity recognition.
For a given input sentence and its predicted tag sequence , we compute a global score that considers both the emission and transition probabilities as in the following:
where represents the emission score for token with tag , representing the transition score matrix from tag to . Note that and represent the special start and end tags, respectively, making a square matrix with dimensions where k is the number of target tags. Additional dimensions accounted for the start and end tags, ensuring that the model captured the complete sequence structure.
The probability of a specific tag sequence y given sentence S is computed using the softmax function for normalization as follows:
During training, we minimize the negative log-likelihood loss of the correct tag sequence as follows:
To determine the optimal tag sequence during inference, we apply the Viterbi algorithm to maximize the score over all possible tag sequences, where is for input sentence as in the following:
The CRF layer leverages token-level tagging information from the ensemble matrix EM and computes the loss of the NER module based on the entire sequence, thereby ensuring that the predicted tags are globally consistent. By modeling contextual dependencies and enforcing valid tag transitions, the CRF layer enables a more flexible and accurate NER, particularly in handling complex entity structures and nonstandard representations often found in medical texts.
3.2. Relation Extraction Module
Our RE module builds upon the results from the NER to identify and classify the relationships between medical entities. In this paper, we approach the RE task as a sequence classification problem, focusing on identifying the relationships between entities within the same sentence. For example, in the sentence “Phosphatidylethanolamine N-methyltransferase (PEMT) catalyzes phosphatidylcholine synthesis”, if the entities ‘PEMT’ and ‘phosphatidylcholine’ are extracted, our RE task is to determine the relationship between them. In the RE module, we leverage three models: BERT, PubMedBERT, and PubMed-T5, each providing unique perspectives for capturing medical domain relationships.
To address challenges related to document length and contextual coherence, the RE module employs three main processing stages: (1) semantic chunking and embedding generation; (2) relation prediction with BiLSTM feature learning; and (3) a majority voting mechanism.
3.2.1. Semantic Chunking and Embedding Generation
To address the challenges of long-range dependencies in medical texts, our RE module employs a semantic chunking approach that segments documents into semantically related text segments. Unlike traditional sentence-based segmentation, this method preserves the critical relationships between entities that often span multiple sentences or paragraphs. To maintain contextual continuity between consecutive chunks and minimize information loss, we implement an adaptive overlap mechanism in which the overlap size is dynamically adjusted based on semantic coherence and entity preservation.
For given abstract document D containing entity pairs , )}, we segment D into sentences and compute their embeddings using the Sentence-BERT. While clustering-based approaches could group similar content, they may disrupt the natural flow of the document and lose the important sequential context necessary for relationship extraction. Therefore, to handle the input length limitations of pre-trained language models while preserving the document’s contextual flow, we sequentially process the document to create chunks , where each chunk comprises a maximum of k tokens. During this process, the overlap size, δ, between consecutive chunks is determined through analysis of both semantic coherence and entity preservation. The overlap size δ is primarily determined by semantic coherence, which we compute using the cosine similarity between adjacent sentences at chunk boundaries using their Sentence-BERT embeddings. The overlap size is adjusted according to the following rule:
where represents the cosine similarity between adjacent sentences computed using their respective Sentence-BERT embeddings. The threshold values are defined as = 0.8 to identify strongly related sentences and = 0.3 to detect semantic boundaries. For overlap sizes, we empirically determined through validation experiments that should be set to 40% of the chunk size k for highly similar sentences to ensure sufficient context preservation, to 10% of k for dissimilar sentences where semantic relationships are minimal, and to establish a default overlap of 20% of k for moderately similar sentences, balancing context preservation. These overlap sizes were selected after testing various percentages ranging from 5% to 50% on our validation dataset.
Entity preservation is critical for maintaining accurate information about relationships between entities. During the initial chunking process, a sentence containing entity pairs is split, resulting in the entities being separated into different chunks. This separation causes the loss of contextual information required for relationship extraction. To prevent this information loss, we further adjust the overlap size when the entity pairs span chunk boundaries:
where represents the minimum distance required to include all entity pairs within a single chunk. Algorithm 2 describes the procedure for our semantic chunking process.
| Algorithm 2. Semantic Chunking |
| Input: Document D, maximum chunk size k, entity pairs EP Output: Processed chunks C |
| 1: Initialize C ← [], position ← 0 2: sentences ← split_into_sentence(D) 3: S ← compute_sentence_embedding(sentences) 4: while position < len(D) do: 5: chunk_end ← min(position + k, len(D)) |
| 6: //Semantic boundary detection 7: boundary_sim ← compute_boundary_similarity(S[position:chunk_end]) 8: //Compute overlap size based on similarity 9: if boundary_sim > high_threshold: 10: δ ← 0.4 * k//max_overlap 11: else if boundary_sim < low_threshold: 12: δ ← 0.1 * k//min_overlap 13: else: 14: δ ← 0.2 * k//α·k |
| 15: //Entity preservation adjustment 16: if contains_partial_entity_pair(D[position:chunk_end], EP): 17: entity_distance ← compute_entity_span_distance(chunk_end, EP) 18: δ ← max(δ, entity_distance) 19. chunk ← D[position:chunk_end] 20. C.append(chunk) 21. position ← chunk_end - δ 22: return C |
Each chunk is then processed independently using BERT, PubMedBERT, and PubMed-T5 models to generate embeddings. This enables a multi-perspective capture of both general and domain-specific information, in which the embeddings reflect the semantic meaning of each chunk. This approach ensures that the model captures both the broad context and specific details of the document, thereby yielding a comprehensive and rich representation of the document’s content.
3.2.2. Relation Prediction with BiLSTM Feature Learning
Following embedding generation from the three models, the chunk-level embeddings from each model are concatenated across all chunks to form a unified document-level representation , where represents the total tokens across all chunks and d is the embedding dimension. This unified representation from each model is then processed through a BiLSTM layer with 100 neurons, capturing sequential dependencies within each chunk and enhancing the token-level representations across the chunks for each model.
The resulting BiLSTM features are then concatenated with the output from the NER module, specifically from the PubMedBERT and PubMed-T5 models. We exclude the BERT outputs from this concatenation to focus on domain-specific knowledge, as PubMedBERT and PubMed-T5 are pre-trained on medical contexts. However, BERT is still essential in our framework for capturing broader and more generic language structures and contextual cues that are not specific to medical texts. This inclusion of BERT enhances the ability of the RE module to interpret medical entities within a more generalized linguistic framework, thus improving robustness when dealing with ambiguous terms or common expressions that appear in both medical and general languages.
The combined feature representation is then passed through three separate dense layers, each corresponding to one of the pre-trained models. Each dense layer independently predicts the relationship for entity pairs, and the results are aggregated using a majority voting mechanism.
3.2.3. Majority Voting Mechanism
To enhance the robustness and reliability of the relationship predictions, our RE module employs a majority voting mechanism. This mechanism consolidates the predictions across multiple chunks and models, resolves inconsistencies, and reinforces the final relationship classification. Each model outputs probability distributions over the possible relation types through a softmax layer, thereby producing confidence scores representing the predicted probabilities for each relation type. Algorithm 3 illustrates the majority voting for relation prediction.
| Algorithm 3. Majority Voting for Relation Prediction |
| Input: Model predictions P = {P_BERT, P_PubMedBERT, P_T5}, Confidence thresholds τ = {τBERT = 0.5, τPubMedBERT = 0.6, τT5 = 0.6}, Consensus threshold θ = 0.7, Validation set V Output: Final relation prediction rel* |
| 1://Weight Learning 2: Initialize performance_scores ← array of zeros[num_models] 3: for each model in models do: 4: performance_scores[model] ← compute_f1_score(model, V) 5: W ← normalize(performance_scores)//Normalize to sum to 1 |
| 6://Voting 7: Initialize vote_counts ← array of zeros[num_relation_types] 8://Aggregate votes from each model 9: for each model_pred in P do: 10: pred_type, conf ← get_max_prediction(model_pred)//conf ∈ [0, 1] 11: if conf > τ[model_type] then 12: vote_counts[pred_type] += W[model_type] * conf 13: end for |
| 14://Evaluate consensus and determine final prediction 15: if max(vote_counts) < θ: 16: //Low consensus case: prioritize domain-specific models 17: rel* ← weighted_average({P_PubMedBERT, P_T5}, W) 18: else: 19: //Strong consensus case 20: rel* ← argmax(vote_counts) 21: return rel* |
Our voting mechanism begins with weight learning, where model-specific weights W are derived based on performance metrics evaluated on validation set V. For each model, the F1 score is computed using the validation set, and these scores are normalized to sum to 1, ensuring that each model’s weight reflects its relative reliability for relation extraction. This ensures that more reliable models have a higher influence during the voting process. Thereafter, vote counts are initialized for each possible relation type. For each model’s prediction, the get_max_prediction function extracts both the predicted relation type (i.e., pred_type) and its confidence score (i.e., conf ∈ [0, 1]) from the model’s softmax output. If the confidence score exceeds the model-specific threshold (i.e., τBERT = 0.5, τPubMedBERT = 0.6, τT5 = 0.6), the corresponding vote is weighted by the model’s reliability weight (i.e., W[model_type]) and the confidence score, and added to the vote count for the predicted relation type.
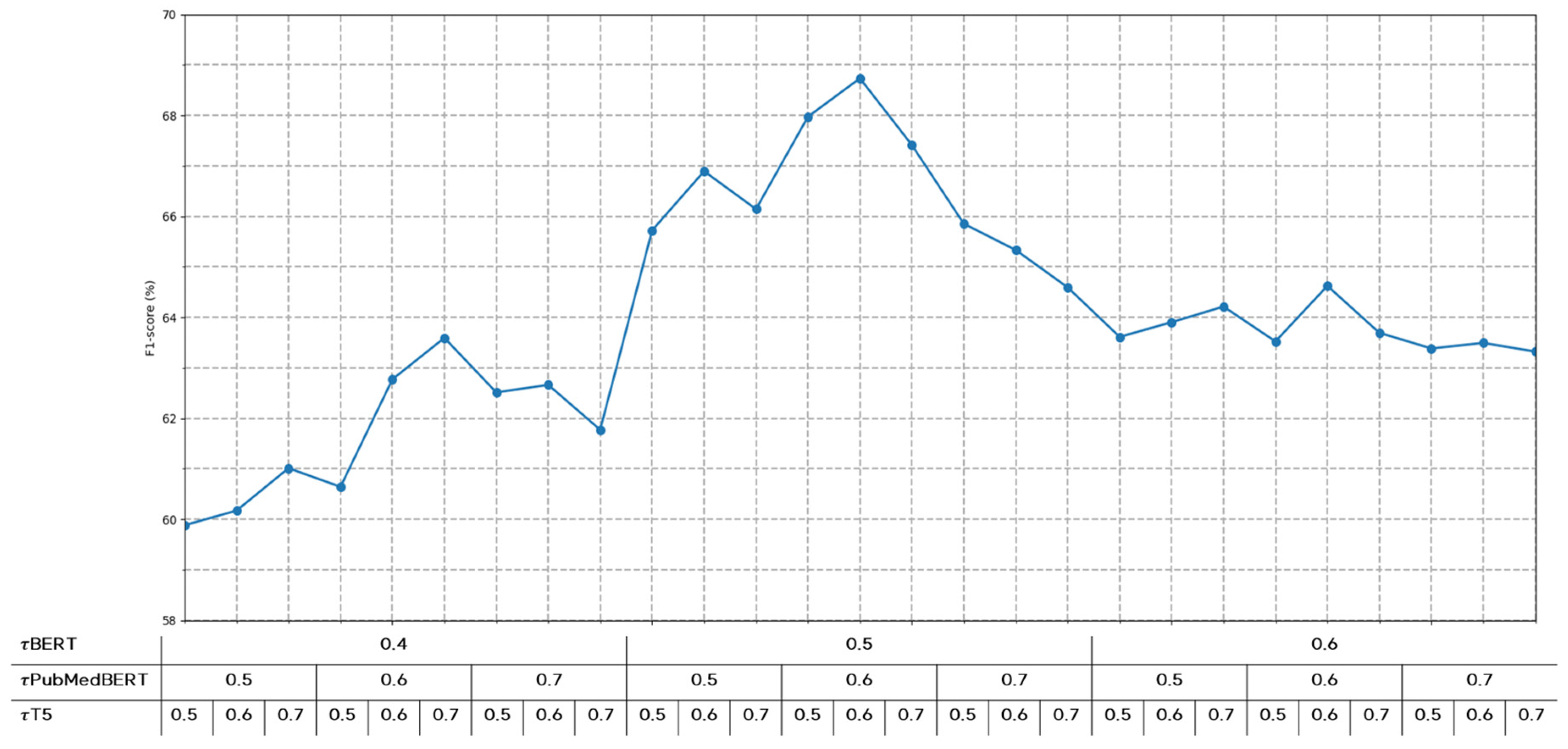
To determine optimal threshold values for each model, we conducted performance evaluations across various threshold settings, as shown in Figure 5. If the threshold is set too low, predictions with low confidence scores would frequently participate in the voting process, potentially introducing noise. Conversely, if the threshold is set too high, predictions from various models might be excluded from voting, potentially leading to insufficient information for final predictions. Therefore, through experimental validation, we identified optimal thresholds that balance noise reduction and information preservation.
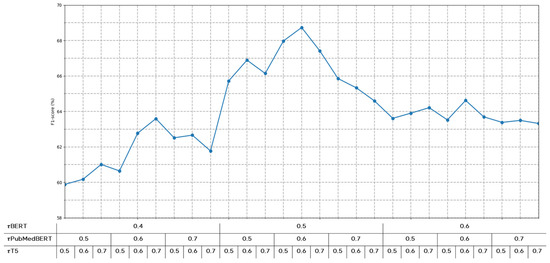
Figure 5.
Performance variations according to confidence score thresholds for each model.
In the consensus evaluation, the algorithm compares the highest vote count to a predefined consensus threshold (θ = 0.7). For strong consensus cases (i.e., max(vote_counts) ≥ θ), the relation type with the highest vote count is selected as the final prediction. However, in low consensus cases (i.e., max(vote_counts) < θ), the algorithm switches to a domain-specific strategy, computing a weighted average of predictions from PubMedBERT and PubMed-T5.
This adaptive voting mechanism enhances the ability of the system to handle complex medical relationships by combining the complementary strengths of the three models. The general BERT model provides a robust syntactic understanding of common expressions, PubMedBERT offers specialized domain knowledge, and PubMed-T5 adds versatility in processing nonstandard or rare relationships. By dynamically prioritizing domain-specific models in low-consensus scenarios, the system ensures accurate and reliable relation extraction even in challenging contexts with ambiguous or conflicting predictions.
4. Experiments
In this section, we present a comprehensive evaluation of the COMCARE framework based on extensive experiments with multiple medical datasets. First, we describe the characteristics and preparation of each dataset, followed by a detailed description of our baseline models and the implementation details. Subsequently, we present our experimental results with analyses, including ablation studies, to validate the contributions of each component.
4.1. Dataset
To ensure a robust evaluation of our framework, we conducted comprehensive experiments across four distinct medical datasets: BioRED [32], ADE [39], DIANN [40], and RDD Corpus [41]. Each dataset presents unique characteristics and challenges, allowing us to assess the performance of our framework in diverse medical contexts.
4.1.1. BioRED Dataset
The BioRED dataset comprises 600 PubMed abstracts and titles that have been professionally annotated, with six distinct entity types representing a diverse range of medical concepts. To utilize this dataset in our NER framework, we processed it through beginning, inside, and outside (BIO) tagging to create a refined dataset structure. Table 1 presents the distribution of entity types within the BioRED dataset, including CellLine, ChemicalEntity, OrganismTaxon, SequenceVariant, GeneOrGeneProduct, and DiseaseOrPhenotypicFeature. The dataset contained 13,636 annotated entities across six categories.

Table 1.
The statistics of all entity types in the BioRED Dataset.
In addition to entity annotations, BioRED includes comprehensive relationship annotations spanning eight distinct types: association, positive correlation, negative correlation, binding, cotreatment, comparison, drug interaction, and conversion. As shown in Table 2, these relationships exhibit significant variations in their distributions across different entity pair combinations. The most prevalent relationship type is ‘Association’, accounting for 2274 instances, whereas ‘Drug Interaction’ and ‘Conversion’ represent the rarest categories with only three instances each.

Table 2.
The statistics of all relation types in BioRED Dataset.
To enhance the utility of the dataset for our experimental evaluation, we implemented several modifications to the original annotations. Specifically, we identified all possible entity pairs within sentences and labeled those without explicit relationship annotations as ‘NoRelation’. This addition creates a more realistic evaluation scenario in which models must distinguish between meaningful and nonexistent relationships. We also excluded sentences containing only single entities, as they would not contribute to the relationship-extraction task.
The BioRED dataset was selected for our experiments because of its complex characteristics. The presence of multiple entity types within a single sentence makes it an ideal benchmark for evaluating the ability of our framework to accurately identify entity boundaries. Additionally, many relationships in the dataset were intricately connected based on the overall context of the text, allowing us to assess the capability of our framework to comprehend complex contexts and extract appropriate relationships.
4.1.2. ADE Corpus
The adverse drug events (ADE) corpus is a specialized dataset focused on pharmacovigilance derived from MEDLINE papers published in PubMed. Each sentence in this corpus contains information about drugs and their associated adverse effects. The corpus comprises 11,070 meticulously annotated entities, as summarized in Table 3. The annotations include 5063 drug entities, 5776 adverse effect entities, and 231 dosage entities. This balanced distribution between drug and adverse effect entities strengthens the effectiveness of the dataset for evaluating the performance of our model in capturing medical entity relationships.

Table 3.
The statistics of all entity types in ADE Corpus.
We selected the ADE corpus for the evaluation because it closely resembles real clinical data, allowing us to assess the practical applicability of our framework in clinical settings. The corpus includes complex cases where multiple drugs may be associated with single or multiple adverse effects and cases in which temporal and causal relationships must be correctly interpreted. These characteristics make it ideal for evaluating context-aware relationship extraction capabilities.
4.1.3. DIANN Corpus
The DIANN corpus was developed to evaluate the ability of the systems to detect disability mentions across the multilingual medical literature. The complete corpus comprised 1000 annotated documents evenly split between Spanish (500) and English (500) documents. For experimental purposes, we exclusively focused on the English portions of the corpus.
The English subset contained 1656 mentions, of which 583 were unique. This relatively small dataset served as an effective benchmark for assessing the performance of our framework under limited data conditions.
4.1.4. RDD Corpus
The RDD corpus was developed specifically to research NER and RE in scientific papers. The corpus contained 1000 English abstracts, with each document consisting of approximately 200 words, totaling 9657 sentences. The annotation process involved three different annotators working under expert supervision to ensure high-quality ground-truth labels.
The corpus provides comprehensive annotation coverage for both entities and relationships between rare diseases and disabilities. As shown in Table 4, the documents contained 578 annotations for rare diseases and 3678 annotations for disabilities, totaling 4256 entity annotations. Table 5 presents the statistics of the relationships within the RDD corpus, showing 1251 positive and 706 negative relationships between rare diseases and disabilities for 1957 relationships.

Table 4.
The statistics of all entity types in the RDD corpus.

Table 5.
The statistics of all relation types in the RDD corpus.
For our experimental evaluation, we carefully selected a subset containing 1957 sentences with complete annotations for both the NER and RE tasks. This subset includes 598 rare disease entities and 581 disabled entities. The RDD corpus is particularly valuable for evaluating the ability of our framework to accurately extract entity boundaries for rare diseases that are not frequently encountered.
4.2. Baseline Models
We implemented several baseline models representing different approaches for medical NER and RE tasks. The selection of the baseline models was guided by their established performance in the recent literature and their demonstrated effectiveness in processing medical texts. Each baseline model was carefully implemented and optimized to ensure a fair comparison as follows:
- CNN: The CNN implementation serves as the foundational baseline. A CNN was originally designed to process image data. However, its ability to detect meaningful patterns in specific parts of data makes it valuable for NLP tasks. CNN effectively learns specific patterns within sentences and predicts labels based on these patterns. In our experiments, we implemented the CNN model using the PyTorch framework with the following parameters: kernel_size = 3, n_filters = 5, and epochs = 100. Early stopping was consistently applied across all experiments, and Adam was used to optimize the model.
- BiLSTM: The BiLSTM network served as the sequential modeling baseline. BiLSTM is a traditional deep learning approach that is widely adopted for sequential data processing and is designed to overcome the limitations of conventional LSTM by considering sequence information from both directions. This bidirectional approach enables superior context understanding and is particularly effective for entity recognition in medical texts. Our BiLSTM implementation utilized the torch package with carefully tuned parameters: batch size of 8, learning rate of 3e-5, 100 training epochs, embedding size of 768, and hidden size of 32. The Adam optimizer was employed for model training.
- BiLSTM-CRF: The BiLSTM-CRF extends the BiLSTM architecture by incorporating a CRF layer. The addition of the CRF layer enabled the model to consider the dependencies between adjacent labels, thereby significantly improving the coherence of the predicted entity sequences. This enhancement is particularly important for capturing the structured nature of medical entity tags. The CRF layer was implemented using the pytorch-crf package, maintaining hyperparameters consistent with the base BiLSTM model while adding transition matrix optimization.
- BERT-CRF: The BERT-CRF model combines the pre-trained BERT model with a CRF layer for sequence optimization. We utilized the pre-trained BERT-base model (768 hidden dimensions and 12 attention heads) and fine-tuned it using our medical datasets. The CRF layer was added on top of the BERT final hidden states, allowing the model to optimize the tag sequences while leveraging the BERT contextual representations. We employed a learning rate of 2 ×10−5 and trained for 20 epochs with early stopping.
- BERT-BiLSTM-CRF: The BERT-BiLSTM-CRF model represents a sophisticated architecture that utilizes word embeddings obtained from BERT as inputs for the BiLSTM-CRF model. This architecture leverages BERT’s contextual understanding capabilities of BERT while maintaining the strength of BiLSTM-CRF in modeling label dependencies.
- KECI (Knowledge-Enhanced Collective Inference): The KECI is a state-of-the-art model for joint biomedical entity and relation extraction that integrates external domain knowledge [42]. The model follows three main steps: constructing an initial span graph, building a background knowledge graph, and fusing these graphs using attention mechanisms. With SciBERT as its transformer encoder, KECI processes graphs using both bidirectional and relational Graph Convolutional Networks (GCNs) while incorporating external knowledge from UMLS and MetaMap. To enhance entity representations, KECI encodes UMLS-derived definitions and relational information using SciBERT and integrates them into the entity nodes in the knowledge graph. The model’s key hyperparameters include a span length limit of 20 tokens, a learning rate of 2 × 10−5, and a batch size of 32.
- LLMs: LLMs are characterized by their extensive parameters and training data, demonstrating exceptional capabilities in language understanding, text generation, and question-answering. Following the emergence of OpenAI’s ChatGPT, interest in LLMs has surged, leading to the development of various models. In our experiments, we compared the performance of our proposed framework with GPT-3.5 and GPT-4 in zero-shot settings. For these evaluations, we carefully crafted prompts to elicit entity recognition and relation extraction responses while maintaining consistent evaluation conditions across all experiments.
4.3. Implementation Details
Our implementation of the COMCARE framework was developed using Python 3.9 with PyTorch version 2.3.1. To enable efficient computation and optimize model training, we utilized CUDA 11.4 and cuDNN 8.9.7 for GPU acceleration. All experiments were conducted on a high-performance computing system equipped with an Intel® Core™ i9-10900X CPU operating at 3.70 GHz, an NVIDIA GeForce RTX 4090 GPU with 24 GB VRAM, and 32 GB of Samsung DDR4 RAM running at 3200 MHz.
We optimized hyperparameters using Optuna [43], an automated framework that efficiently searches parameter combinations while minimizing manual tuning effort. After defining search spaces for each parameter, we evaluated multiple configurations and selected the combination that maximized the F1 score on the validation set. Table 6 shows these optimal hyperparameters.

Table 6.
Hyperparameters of the model.
For the model optimization, we employed the AdamW optimizer combined with a learning rate scheduler to dynamically adjust the learning rate during training. We implemented an early stopping mechanism with patience for three epochs, monitoring the F1 score on the validation set to prevent overfitting while ensuring optimal performance.
4.4. Experimental Results and Analysis
In this section, we present a comprehensive performance comparison between the proposed framework and baseline models across multiple medical datasets. Evaluation metrics such as precision, recall, and F1 score provide a detailed assessment of model performance on the BioRED dataset, RDD corpus, ADE corpus, and DIANN corpus. We present detailed analyses of the model performance, including comparative evaluations against baseline approaches and ablation studies examining component contributions, to validate our findings.
Before conducting the main experiments, we performed comparative experiments to identify the most suitable pre-trained model for our proposed architecture. As shown in Table 7, we evaluated various language models that were pre-trained on medical domain data. Among these, PubMedBERT achieved the highest performance on our dataset, prompting us to select it for our experiments. In contrast, BlueBERT, which was pre-trained primarily on clinical record data such as MIMIC-III, performed relatively poorly on our dataset of medical research papers. BioBERT, also pre-trained on PubMed research papers but using the original BERT vocabulary, showed comparable performance due to its extensive exposure to biomedical text. SciBERT, despite learning a domain-specific vocabulary during pre-training, demonstrated weaker performance in the medical domain, as it focused on scientific papers more broadly rather than specifically on medical texts. While BlueBERT, BioBERT, and SciBERT were all fine-tuned using domain-specific data on top of an existing BERT model, PubMedBERT was trained entirely from scratch on domain-specific data, resulting in stronger representational capabilities for the medical domain. However, the KECI model employed SciBERT as its transformer encoder, incorporating UMLS-derived definitions and relational information into its entity nodes. Based on these findings, we used PubMedBERT for all subsequent experiments and SciBERT specifically for the KECI baseline model.

Table 7.
Comparison of NER Results Across Pre-Trained Models.
Our experimental results on the BioRED and RDD datasets, presented in Table 8 and Table 9, demonstrate the superior performance of the proposed ensemble model. Detailed results for each of the 10 experimental repetitions, along with statistical significance tests, are provided in Appendix A, Table A1 and Table A2. On the BioRED dataset, our model achieved F1 scores of 93.76% for NER (precision: 95.11%, recall: 92.45%) and 68.73% for RE (precision: 72.58%, recall: 65.27%), outperforming all baseline models. The experimental results revealed several significant findings regarding model architecture and performance.

Table 8.
Performance Comparison Results on the BioRED Dataset.

Table 9.
Performance Comparison Results on the RDD Dataset.
First, models incorporating BERT consistently demonstrated superior performance compared to traditional CNN and BiLSTM approaches, as evidenced by rows 1–3 in the results tables. This performance advantage can be attributed to BERT’s masked word prediction training approach, which enables a better capture of semantic information and is particularly beneficial for token-level NER tasks.
The addition of BiLSTM layers to the BERT models, as shown in rows 4 and 6, further enhances the recognition performance. This improvement stems from the ability of the BiLSTM to utilize bidirectional context information, allowing the model to better leverage both the preceding and following token information. Furthermore, the incorporation of CRF layers (rows 3, 5, and 7) consistently improved the performance across different model architectures, demonstrating the value of modeling dependencies between entity tags for a more accurate NER.
In relation to extraction tasks, although we observed similar patterns regarding the superiority of transformer-based models, notable differences existed. Although BERT-based models still outperform traditional CNN and BiLSTM approaches, the addition of BiLSTM layers to BERT slightly decreases performance. This can be attributed to the inherent limitations of LSTM architectures in handling longer contextual dependencies required for relationship identification in RE tasks.
While KECI leverages specialized medical concept information from UMLS and MetaMap, its performance is somewhat limited by its foundation model, SciBERT, which was trained on a broad range of scientific papers rather than being specifically focused on the biomedical literature. This results in performance metrics of NER F1 scoring 90.71% and RE F1 scoring 66.17% on the BioRED dataset, and NER F1 scoring 75.51% and RE F1 scoring 83.13% on the RDD dataset. When compared to domain-specific models like PubMedBERT, which was pre-trained directly on PubMed papers, KECI shows relatively lower performance due to its less specialized understanding of biomedical domain knowledge.
The effectiveness of our ensemble learning approach is particularly evident in the consistent performance improvements across all datasets. By combining the BERT-BiLSTM-CRF architecture with a T5-BiLSTM-CRF model, our ensemble method successfully leverages the complementary strengths of different model architectures. The T5 component provides an enhanced contextual understanding of medical terminology, while the BERT-based component offers robust general language representation. This synergistic combination enables more accurate entity recognition and relationship extraction, as demonstrated by the performance gains across diverse biomedical datasets. For the RE tasks, we addressed the long-term dependency limitations of the LSTM-based models by implementing a chunk-based approach for processing abstracts, which resulted in improved F1 scores.
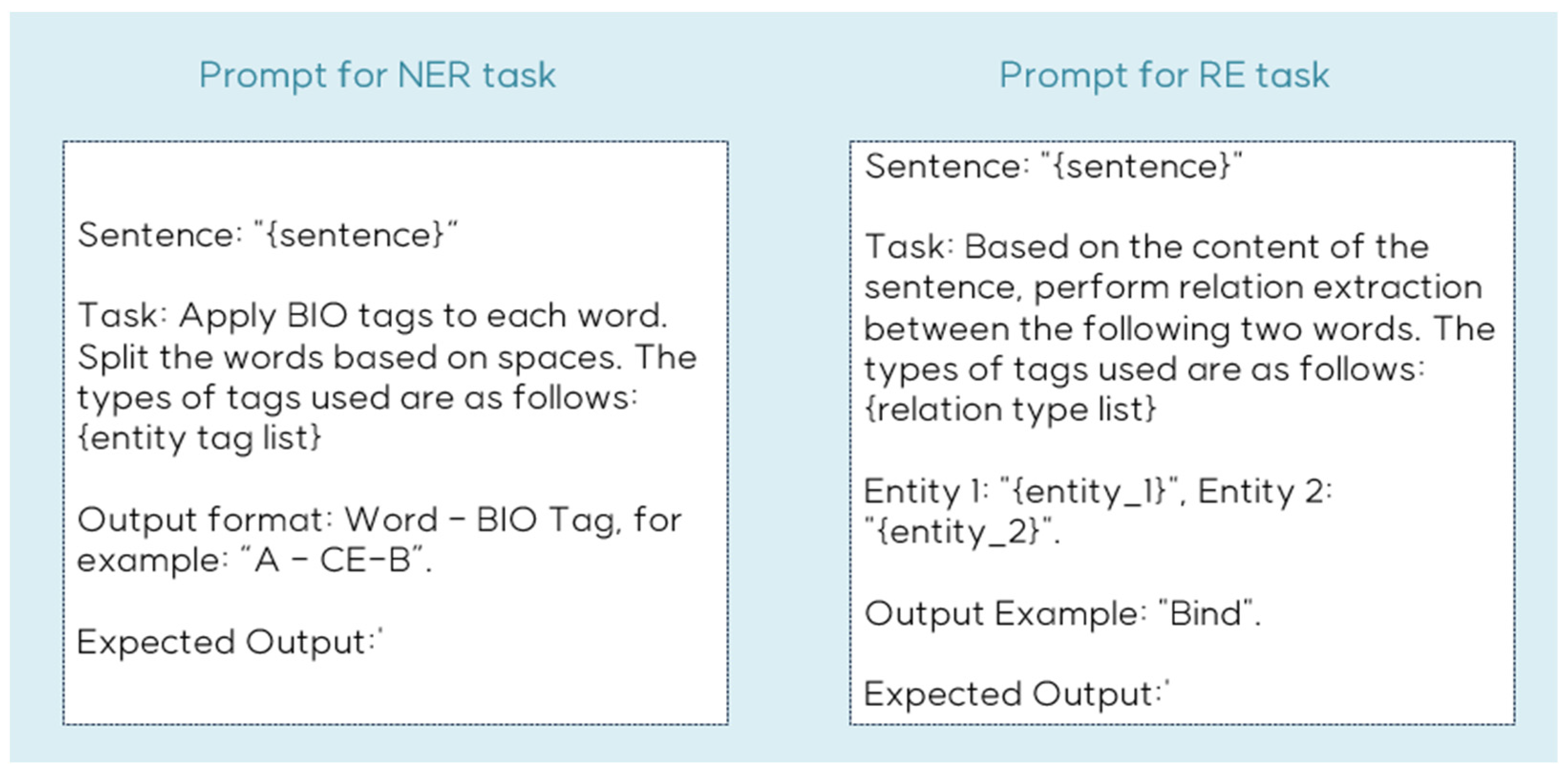
We also conducted comparative evaluations against LLMs, specifically OpenAI’s GPT-3.5-turbo and GPT-4 models, in zero-shot settings. Our proposed model demonstrated superior performance across all datasets for both NER and RE tasks compared to zero-shot LLM implementations. The prompt structure used for the LLM evaluation is illustrated in Figure 6.
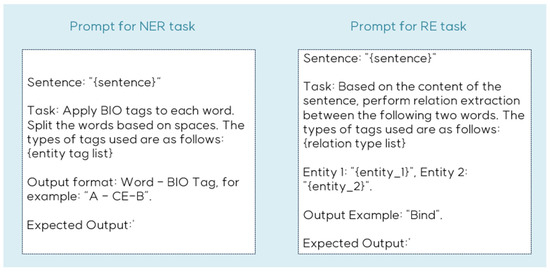
Figure 6.
Example of prompt for NER and RE task.
Table 10 and Table 11 present the comparative results for the ADE and DIANN corpus, respectively. Consistent with the results observed in the BioRED dataset and RDD corpus, our proposed model achieved improved F1 scores compared with all baseline models. Detailed results for each of the 10 experimental repetitions, along with statistical significance tests, are provided in Appendix A, Table A3 and Table A4.

Table 10.
Performance Comparison Results on the ADE Dataset.

Table 11.
Performance Comparison Results on the DIANN Dataset.
The experimental results on the ADE dataset showed similar performance patterns to those observed in the BioRED and RDD datasets. Our model achieved an F1 score of 82.48% (precision: 94.69%, recall: 73.36%), outperforming the BERT-BiLSTM-CRF baseline (F1: 81.65%). The relatively lower recall scores across all models on the ADE dataset can be attributed to the complex nature of adverse drug event relationships and the high variability in their textual descriptions.
On the DIANN dataset, our model demonstrated exceptional performance with an F1 score of 99.36% (precision: 99.94%, recall: 98.78%), showing significant improvement over both traditional and transformer-based baselines. The notably high performance across all BERT-based models on this dataset (F1 scores > 97%) suggests that the disease annotation patterns in DIANN are more consistent and well-structured compared to other biomedical datasets. This is particularly evident in the contrast between the performance on DIANN and the more challenging ADE dataset.
Interestingly, the LLM-based approaches (GPT-3.5 and GPT-4o-mini) showed particularly poor performance on both ADE and DIANN datasets, with GPT-3.5 failing to identify any entities in the DIANN dataset (0% across all metrics).
The consistent superior performance across all four datasets (BioRED, RDD, ADE, and DIANN) validates the robustness and generalizability of our ensemble approach. The model demonstrates strong adaptability to different biomedical text-mining tasks, successfully handling variations in writing styles, terminology, and relationship complexity.
Table 12 presents the results of additional experiments conducted to evaluate the performance of the various techniques implemented in our framework. Specifically, we examined the performance variations resulting from different ensemble strategies for combining information from two pre-trained models in the NER task, as well as the impact of incorporating general information alongside domain-specific information in the RE task. Additionally, we compared the performance of the individual modules and joint training of the NER and RE modules.

Table 12.
The results of the Ablation experiment.
Rows 1–5 demonstrate the performance outcomes of the different ensemble strategies when combining information from the two models in the NER module. Comparing rows 1 and 2, we observe that incorporating the T5 model alongside BERT improves the recognition performance compared to using BERT alone. This improvement can be attributed to the generative capabilities of the T5 model, which enable better recognition of polysemous terms and nonstandard word representations that are characteristic of medical terminology. However, the results in rows 3–5 reveal that increasing the influence of the T5 model beyond a certain point degrades performance. This decline occurs because while medical documents contain polysemous terms and non-standard word representations, their proportion is relatively lower compared to standard representations. In response to these findings, we developed a model that employs collaborative decision-making through our Collaborative Decision Strategy (CDS) rather than simple weighted combinations to leverage the strengths of both models.
To evaluate the effectiveness of each component in the CDS algorithm, we conducted comparative experiments, as shown in rows 6 and 7. The results demonstrate that even a simple combination based on the highest confidence scores outperforms conventional weighted ensembles. However, this approach has limitations in cases where one model assigns high confidence scores to incorrect tags. Therefore, our proposed method carefully combines information from both models using both confidence scores and attention weights, enabling more robust and accurate entity recognition. This comprehensive CDS approach demonstrated superior F1 scores compared to conventional weighted ensemble methods.
Row 9 shows the impact of incorporating general domain information alongside domain-specific model information in RE tasks on performance. The results validate the effectiveness of combining both types of information for improved relationship extraction. Additionally, the joint training of the NER and RE modules demonstrated an enhanced overall performance, highlighting the benefits of sharing information between the two tasks.
These comprehensive results validate the effectiveness of our framework in medical entity recognition and relationship extraction tasks. The framework demonstrates robust performance across diverse medical datasets, with particular strengths in handling specialized terminology and complex relationships. The superior performance compared with both traditional baselines and current LLMs supports the value of our multi-model ensemble approach with semantic chunking for medical text processing tasks.
5. Conclusions
In this paper, we present a novel framework that combines NER and RE modules to extract medical entities and their relationships from medical documents. Our NER module integrates the pre-trained BERT model, which has demonstrated superior performance in existing NER tasks, with the T5 model, which was specifically pre-trained to handle medical terminology characteristics, such as polysemy and non-standard representations. Through our collaborative decision strategy, these models are combined in a way that effectively leverages their complementary strengths, resulting in performance improvements that are particularly significant for challenging medical entities. The experimental results demonstrate that this ensemble approach not only achieves higher F1 scores than baseline models but also provides substantially improved accuracy in critical cases such as ambiguous medical terminology and rare disease names, where single models often struggle.
Our RE module addresses key challenges in relation extraction by obtaining context vectors from pre-trained models using abstract chunks as inputs. This approach effectively resolves the limitations of BERT’s input token constraints and long-term dependency issues inherent in LSTM-based models. The module processes these vectors through both domain-specific and general domain pre-trained language models to derive comprehensive contextual information for the final relation-type prediction. The experimental results confirm that our proposed RE module achieves higher F1 scores compared to approaches using pre-trained models alone, with particularly notable improvements in handling long-range dependencies and complex relationship classifications that are crucial in medical text analysis.
The effectiveness of our ensemble approach is particularly evident in its ability to maintain consistent performance across diverse medical contexts. The enhanced robustness and reliability demonstrated by our framework make it particularly valuable in clinical settings where accurate interpretation of medical terminology and relationships is crucial for patient care.
However, our research has several limitations that need to be addressed in future work. The primary limitation is the high computational complexity resulting from the use of multiple pre-trained language models. Additionally, the necessity of chunking abstracts to address input token limitations increases memory consumption during processing. Although our study prioritized accuracy in medical NER and RE tasks, and the results justify the computational overhead through improved accuracy in critical cases, future research should focus on optimizing these aspects. We plan to investigate methods for reducing the complexity and developing lightweight versions of the entire module architecture to create a framework that maintains high accuracy while achieving greater computational efficiency in NER and RE tasks.
Author Contributions
Conceptualization, M.J.; Methodology, M.J. and G.-W.K.; Software, M.J. and G.-W.K.; Formal analysis, S.-M.C. and G.-W.K.; Investigation, M.J. and G.-W.K.; Data curation, M.J.; Writing—original draft, M.J. and G.-W.K.; Supervision, S.-M.C. and G.-W.K.; Project administration, S.-M.C. and G.-W.K.; Funding acquisition, S.-M.C. and G.-W.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a research grant from the Gyeongsang National University in 2024. This work was supported by the Glocal University 30 Project Fund of Gyeongsang National University in 2024. This research was supported by the “Regional Innovation Strategy (RIS)” through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (MOE) (2021RIS-003).
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A. Detailed Comparative Results: Base Model and Proposed Model

Table A1.
Statistical significance analysis of the differences between two pairs of experiments using the BioRED dataset: The final results of the base model and the proposed model were compared. The last row indicates whether the differences are statistically significant across all considered metrics according to the Wilcoxon Test.
Table A1.
Statistical significance analysis of the differences between two pairs of experiments using the BioRED dataset: The final results of the base model and the proposed model were compared. The last row indicates whether the differences are statistically significant across all considered metrics according to the Wilcoxon Test.
| BioRED | NER | RE | |||||
|---|---|---|---|---|---|---|---|
| Precision (%) | Recall (%) | F1-Score (%) | Precision (%) | Recall (%) | F1-Score (%) | ||
| Base model (NER: BERT-BiLSTM-CRF, RE: BERT) | Seed 0 | 93.08 | 91.63 | 92.35 | 70.88 | 64.15 | 67.35 |
| Seed 1 | 93.86 | 93.01 | 93.43 | 72.31 | 66.37 | 69.21 | |
| Seed 2 | 95.19 | 91.08 | 93.09 | 70.21 | 65.66 | 67.86 | |
| Seed 3 | 95.3 | 91.54 | 93.38 | 70.4 | 63.97 | 67.03 | |
| Seed 4 | 93.91 | 91.34 | 92.61 | 72.02 | 65.03 | 68.34 | |
| Seed 5 | 94.04 | 91.42 | 92.71 | 69.66 | 65.05 | 67.28 | |
| Seed 6 | 94.17 | 90.36 | 92.23 | 70.17 | 64.96 | 67.46 | |
| Seed 7 | 94.92 | 89.96 | 92.38 | 71.04 | 65.58 | 68.2 | |
| Seed 8 | 93.18 | 91.69 | 92.43 | 71.23 | 65.32 | 68.15 | |
| Seed 9 | 93.47 | 91.12 | 92.28 | 71.47 | 66.17 | 68.72 | |
| Proposed model | Seed 0 | 95.34 | 92.56 | 93.93 | 73.7 | 63.51 | 68.23 |
| Seed 1 | 94.54 | 92.47 | 93.49 | 73.39 | 64.11 | 68.43 | |
| Seed 2 | 97.24 | 94.96 | 96.09 | 71.19 | 65.88 | 68.43 | |
| Seed 3 | 95.04 | 89.78 | 92.33 | 73.56 | 65.9 | 69.52 | |
| Seed 4 | 94.2 | 92.3 | 93.24 | 74.38 | 66.01 | 69.95 | |
| Seed 5 | 95.21 | 91.21 | 93.17 | 72.98 | 64.91 | 68.71 | |
| Seed 6 | 94.95 | 91.6 | 93.73 | 72.4 | 64.7 | 68.33 | |
| Seed 7 | 94.21 | 92.38 | 93.29 | 74.49 | 63.2 | 68.38 | |
| Seed 8 | 95.94 | 91.6 | 93.72 | 72.66 | 65.07 | 68.65 | |
| Seed 9 | 94.11 | 91.81 | 92.95 | 72.48 | 65.6 | 68.87 | |
| p-value < 0.05 | No | Yes | Yes | Yes | No | Yes | |

Table A2.
Statistical significance analysis of the differences between two pairs of experiments using the RDD corpus: The final results of the base model and the proposed model were compared. The last row indicates whether the differences are statistically significant across all considered metrics according to the Wilcoxon Test.
Table A2.
Statistical significance analysis of the differences between two pairs of experiments using the RDD corpus: The final results of the base model and the proposed model were compared. The last row indicates whether the differences are statistically significant across all considered metrics according to the Wilcoxon Test.
| RDD | NER | RE | |||||
|---|---|---|---|---|---|---|---|
| Precision (%) | Recall (%) | F1-Score (%) | Precision (%) | Recall (%) | F1-Score (%) | ||
| Base model (NER: BERT-BiLSTM-CRF, RE: BERT) | Seed 0 | 94.19 | 64.18 | 76.34 | 85.0 | 87.33 | 86.15 |
| Seed 1 | 93.98 | 64.75 | 76.67 | 83.59 | 85.41 | 84.49 | |
| Seed 2 | 95.73 | 67.79 | 79.37 | 85.2 | 85.44 | 85.32 | |
| Seed 3 | 94.96 | 64.29 | 76.67 | 85.65 | 84.56 | 85.1 | |
| Seed 4 | 95.18 | 64.01 | 76.55 | 84.32 | 84.83 | 84.58 | |
| Seed 5 | 96.27 | 64.52 | 77.26 | 84.44 | 82.56 | 83.49 | |
| Seed 6 | 93.94 | 65.36 | 77.09 | 84.92 | 85.71 | 85.31 | |
| Seed 7 | 95.04 | 65.23 | 77.36 | 86.18 | 84.68 | 85.42 | |
| Seed 8 | 94.61 | 64.53 | 76.73 | 84.82 | 85.69 | 85.25 | |
| Seed 9 | 94.43 | 67.23 | 78.54 | 85.81 | 83.89 | 84.84 | |
| Proposed model | Seed 0 | 93.96 | 67.04 | 78.25 | 87.91 | 84.6 | 86.22 |
| Seed 1 | 93.88 | 67.71 | 78.68 | 86.98 | 86.82 | 86.9 | |
| Seed 2 | 95.42 | 66.23 | 78.19 | 88.2 | 86.13 | 87.15 | |
| Seed 3 | 94.26 | 67.03 | 78.35 | 87.02 | 86.11 | 86.56 | |
| Seed 4 | 94.4 | 66.63 | 78.12 | 87.19 | 85.84 | 86.51 | |
| Seed 5 | 95.76 | 66.54 | 78.52 | 86.75 | 85.13 | 85.93 | |
| Seed 6 | 93.6 | 65.52 | 77.08 | 89.26 | 86.62 | 87.92 | |
| Seed 7 | 93.67 | 65.68 | 77.22 | 89.26 | 85.8 | 85.96 | |
| Seed 8 | 95.28 | 67.18 | 78.8 | 86.79 | 84.8 | 85.78 | |
| Seed 9 | 94.59 | 64.32 | 76.57 | 88.25 | 84.71 | 86.45 | |
| p-value < 0.05 | No | Yes | No | Yes | No | Yes | |

Table A3.
Statistical significance analysis of the differences between two pairs of experiments using the ADE corpus: The final results of the base model and the proposed model were compared. The last row indicates whether the differences are statistically significant across all considered metrics according to the Wilcoxon Test.
Table A3.
Statistical significance analysis of the differences between two pairs of experiments using the ADE corpus: The final results of the base model and the proposed model were compared. The last row indicates whether the differences are statistically significant across all considered metrics according to the Wilcoxon Test.
| ADE | NER | |||
|---|---|---|---|---|
| Precision (%) | Recall (%) | F1-Score (%) | ||
| Base model (BERT-BiLSTM-CRF) | Seed 0 | 93.08 | 91.63 | 92.35 |
| Seed 1 | 93.86 | 93.01 | 93.43 | |
| Seed 2 | 95.19 | 91.08 | 93.09 | |
| Seed 3 | 95.3 | 91.54 | 93.38 | |
| Seed 4 | 93.91 | 91.34 | 92.61 | |
| Seed 5 | 94.04 | 91.42 | 92.71 | |
| Seed 6 | 94.17 | 90.36 | 92.23 | |
| Seed 7 | 94.92 | 89.96 | 92.38 | |
| Seed 8 | 93.18 | 91.69 | 92.43 | |
| Seed 9 | 93.47 | 91.12 | 92.28 | |
| Proposed model | Seed 0 | 92.9 | 72.54 | 81.47 |
| Seed 1 | 95.26 | 74.07 | 83.34 | |
| Seed 2 | 96.67 | 74.26 | 84.0 | |
| Seed 3 | 95.27 | 71.91 | 81.96 | |
| Seed 4 | 95.71 | 74.49 | 83.78 | |
| Seed 5 | 95.9 | 73.37 | 83.13 | |
| Seed 6 | 94.28 | 73.18 | 82.4 | |
| Seed 7 | 93.84 | 73.51 | 82.44 | |
| Seed 8 | 93.01 | 75.54 | 82.37 | |
| Seed 9 | 94.81 | 73.61 | 82.87 | |
| p-value < 0.05 | Yes | Yes | Yes | |

Table A4.
Statistical significance analysis of the differences between two pairs of experiments using the DIANN corpus: The final results of the base model and the proposed model were compared. The last row indicates whether the differences are statistically significant across all considered metrics according to the Wilcoxon Test.
Table A4.
Statistical significance analysis of the differences between two pairs of experiments using the DIANN corpus: The final results of the base model and the proposed model were compared. The last row indicates whether the differences are statistically significant across all considered metrics according to the Wilcoxon Test.
| DIANN | NER | |||
|---|---|---|---|---|
| Precision (%) | Recall (%) | F1-Score (%) | ||
| Base model (BERT-BiLSTM-CRF) | Seed 0 | 97.23 | 97.39 | 97.31 |
| Seed 1 | 99.45 | 97.48 | 98.45 | |
| Seed 2 | 98.62 | 97.38 | 97.99 | |
| Seed 3 | 98.68 | 96.73 | 97.69 | |
| Seed 4 | 99.5 | 96.07 | 97.75 | |
| Seed 5 | 99.04 | 97.61 | 98.32 | |
| Seed 6 | 98.88 | 95.88 | 97.35 | |
| Seed 7 | 99.2 | 99.32 | 99.26 | |
| Seed 8 | 98.54 | 95.44 | 96.97 | |
| Seed 9 | 98.16 | 97.18 | 97.67 | |
| Proposed model | Seed 0 | 99.36 | 97.79 | 98.57 |
| Seed 1 | 99.86 | 98.3 | 99.07 | |
| Seed 2 | 99.29 | 98.95 | 99.12 | |
| Seed 3 | 99.61 | 99.78 | 99.69 | |
| Seed 4 | 98.31 | 99.83 | 99.06 | |
| Seed 5 | 99.76 | 98.87 | 99.31 | |
| Seed 6 | 99.42 | 98.61 | 99.01 | |
| Seed 7 | 98.9 | 97.87 | 98.38 | |
| Seed 8 | 98.95 | 98.96 | 98.85 | |
| Seed 9 | 99.07 | 98.66 | 98.86 | |
| p-value < 0.05 | Yes | Yes | Yes | |
References
- Yang, F.; Shu, H.; Zhang, X. Understanding “Internet Plus Healthcare” in China: Policy Text Analysis. J. Med. Internet Res. 2020, 23, e23779. [Google Scholar] [CrossRef] [PubMed]
- Wiest, I.C.; Ferber, D.; Zhu, J.; van Treeck, M.; Meyer, S.K.; Juglan, R.; Carrero, Z.I.; Paech, D.; Kleesiek, J.; Ebert, M.P.; et al. Privacy-preserving large language models for structured medical information retrieval. NPJ Digit. Med. 2024, 7, 257. [Google Scholar] [CrossRef] [PubMed]
- Elgaar, M.; Cheng, J.; Vakil, N.; Amiri, H.; Celi, L.A. MedDec: A Dataset for Extracting Medical Decisions from Discharge Summaries. In Findings of the Association for Computational Linguistics: ACL 2024; Association for Computational Linguistics: Bangkok, Thailand, 2024; pp. 16442–16455. [Google Scholar]
- da Silva, D.P.; da Rosa Fröhlich, W.; de Mello, B.H.; Vieira, R.; Rigo, S.J. Exploring named entity recognition and relation extraction for ontology and medical records integration. Inform. Med. Unlocked 2023, 43, 101381. [Google Scholar] [CrossRef]
- Navarro, D.F.; Ijaz, K.; Rezazadegan, D.; Rahimi-Ardabili, H.; Dras, M.; Coiera, E.W.; Berkovsky, S. Clinical named entity recognition and relation extraction using natural language processing of medical free text: A systematic review. Int. J. Med. Inform. 2023, 177, 105122. [Google Scholar] [CrossRef]
- Moscato, V.; Postiglione, M.; Sansone, C.; Sperlí, G. TaughtNet: Learning Multi-Task Biomedical Named Entity Recognition From Single-Task Teachers. IEEE J. Biomed. Health Inform. 2023, 27, 2512–2523. [Google Scholar] [CrossRef]
- Grossman Liu, L.; Grossman, R.H.; Mitchell, E.G.; Weng, C.; Natarajan, K.; Hripcsak, G.; Vawdrey, D.K. A deep database of medical abbreviations and acronyms for natural language processing. Sci. Data 2021, 8, 149. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Ma, X. A novel neural network model fusion approach for improving medical named entity recognition in online health expert question-answering services. Expert Syst. Appl. 2023, 223, 119880. [Google Scholar] [CrossRef]
- Jonker, R.A.; Almeida, T.; Antunes, R.; Almeida, J.R.; Matos, S. Multi-head CRF classifier for biomedical multi-class named entity recognition on Spanish clinical notes. Database 2024, 2024, baae068. [Google Scholar] [CrossRef]
- Noriega-Atala, E.; Hein, P.D.; Thumsi, S.S.; Wong, Z.; Wang, X.; Hendryx, S.M.; Morrison, C.T. Extracting Inter-Sentence Relations for Associating Biological Context with Events in Biomedical Texts. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 17, 1895–1906. [Google Scholar] [CrossRef] [PubMed]
- Popovski, G.; Kochev, S.; Korousic-Seljak, B.; Eftimov, T. FoodIE: A Rule-based Named-entity Recognition Method for Food Information Extraction. In Proceedings of the International Conference on Pattern Recognition Applications and Methods, Prague, Czech Republic, 19–21 February 2019. [Google Scholar]
- Gorinski, P.J.; Wu, H.; Grover, C.; Tobin, R.; Talbot, C.; Whalley, H.C.; Sudlow, C.L.; Whiteley, W.; Alex, B. Named Entity Recognition for Electronic Health Records: A Comparison of Rule-based and Machine Learning Approaches. arXiv 2019, arXiv:1903.03985. [Google Scholar]
- Khan, W.; Daud, A.; Shahzad, K.; Amjad, T.; Banjar, A.T.; Fasihuddin, H.A. Named Entity Recognition Using Conditional Random Fields. Appl. Sci. 2022, 12, 6391. [Google Scholar] [CrossRef]
- Suthaharan, S. Support Vector Machine. In Machine Learning Models and Algorithms for Big Data Classification; Integrated Series in Information Systems; Springer: Boston, MA, USA, 2016; Volume 36. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, T.; Tsai, C.; Lu, Y.; Yao, L. Evolution and emerging trends of named entity recognition: Bibliometric analysis from 2000 to 2023. Heliyon 2024, 10, e30053. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Xu, W.; Yu, K. Bidirectional LSTM-CRF Models for Sequence Tagging. arXiv 2015, arXiv:1508.01991. [Google Scholar]
- Devlin, J.; Chang, M.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. In Proceedings of the North American Chapter of the Association for Computational Linguistics, Minneapolis, MN, USA, 2–7 June 2019. [Google Scholar]
- Gu, Y.; Tinn, R.; Cheng, H.; Lucas, M.R.; Usuyama, N.; Liu, X.; Naumann, T.; Gao, J.; Poon, H. Domain-Specific Language Model Pretraining for Biomedical Natural Language Processing. ACM Trans. Comput. Healthc. (HEALTH) 2020, 3, 1–23. [Google Scholar] [CrossRef]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2019, 36, 1234–1240. [Google Scholar] [CrossRef]
- OpenAI. ChatGPT (3.5-turbo, 4o-mini) [Large Language Model]. 2024. Available online: https://chat.openai.com/ (accessed on 7 December 2024).
- Touvron, H.; Lavril, T.; Izacard, G.; Martinet, X.; Lachaux, M.; Lacroix, T.; Rozière, B.; Goyal, N.; Hambro, E.; Azhar, F.; et al. LLaMA: Open and Efficient Foundation Language Models. arXiv 2023, arXiv:2302.13971. [Google Scholar]
- Raffel, C.; Shazeer, N.M.; Roberts, A.; Lee, K.; Narang, S.; Matena, M.; Zhou, Y.; Li, W.; Liu, P.J. Exploring the Limits of Transfer Learning with a Unified Text-to-Text Transformer. J. Mach. Learn. Res. 2019, 21, 1–67. [Google Scholar]
- Arbabi, A.; Adams, D.R.; Fidler, S.; Brudno, M. Identifying Clinical Terms in Medical Text Using Ontology-Guided Machine Learning. JMIR Med. Inform. 2019, 7, e12596. [Google Scholar] [CrossRef]
- Zhao, S.; Liu, T.; Zhao, S.; Wang, F. A Neural Multi-Task Learning Framework to Jointly Model Medical Named Entity Recognition and Normalization. arXiv 2018, arXiv:1812.06081. [Google Scholar] [CrossRef]
- Chaudhry, M.; Kazmi, A.; Jatav, S.; Verma, A.; Samal, V.; Paul, K.; Modi, A. Reducing Inference Time of Biomedical NER Tasks using Multi-Task Learning. In Proceedings of the 19th International Conference on Natural Language Processing, New Delhi, India; 2022; pp. 116–122. [Google Scholar]
- Li, J.; Wei, Q.; Ghiasvand, O.A.; Chen, M.; Lobanov, V.S.; Weng, C.; Xu, H. A comparative study of pre-trained language models for named entity recognition in clinical trial eligibility criteria from multiple corpora. BMC Med. Inform. Decis. Mak. 2022, 22, 235. [Google Scholar] [CrossRef]
- Yi, F.; Liu, H.; Wang, Y.; Wu, S.; Sun, C.; Feng, P.; Zhang, J. Medical Named Entity Recognition Fusing Part-of-Speech and Stroke Features. Appl. Sci. 2023, 13, 8913. [Google Scholar] [CrossRef]
- Liang, T.; Xia, C.; Zhao, Z.; Jiang, Y.; Yin, Y.; Yu, P. Transferring From Textual Entailment to Biomedical Named Entity Recognition. IEEE/ACM Trans. Comput. Biol. Bioinform. 2023, 20, 2577–2586. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, A.L. Biomedical named entity recognition with the combined feature attention and fully-shared multi-task learning. BMC Bioinform. 2022, 23, 458. [Google Scholar] [CrossRef]
- Dewi, I.N.; Dong, S.; Hu, J. Drug-drug interaction relation extraction with deep convolutional neural networks. In Proceedings of the 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Kansas City, MO, USA, 13–16 November 2017; pp. 1795–1802. [Google Scholar]
- Fabregat, H.; Duque, A.; Martínez-Romo, J.; Araujo, L. Negation-based transfer learning for improving biomedical Named Entity Recognition and Relation Extraction. J. Biomed. Inform. 2023, 138, 104279. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Lai, P.; Wei, C.; Arighi, C.N.; Lu, Z. BioRED: A rich biomedical relation extraction dataset. Brief. Bioinform. 2022, 23, bbac282. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wei, Q.; Huang, L.; Li, J.; Hu, Y.; Chuang, Y.; He, J.; Das, A.; Keloth, V.K.; Yang, Y.; et al. Ensemble pretrained language models to extract biomedical knowledge from literature. J. Am. Med. Inform. Assoc. 2024, 31, 1904–1911. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Li, R.; Wen, A.; Wang, J.; Wang, L.; Liu, H. Large Language Models Struggle in Token-Level Clinical Named Entity Recognition. arXiv 2024, arXiv:2407.00731. [Google Scholar]
- Hu, Y.; Ameer, I.; Zuo, X.; Peng, X.; Zhou, Y.; Li, Z.; Li, Y.; Li, J.; Jiang, X.; Xu, H. Improving large language models for clinical named entity recognition via prompt engineering. J. Am. Med. Inform. Assoc. 2023, 31, 1812–1820. [Google Scholar] [CrossRef]
- Zhou, H.; Li, M.; Xiao, Y.; Yang, H.; Zhang, R. LEAP: LLM instruction-example adaptive prompting framework for biomedical relation extraction. J. Am. Med. Inform. Assoc. 2024, 31, 2010–2018. [Google Scholar] [CrossRef]
- Kudo, T.; Richardson, J. SentencePiece: A simple and language independent subword tokenizer and detokenizer for Neural Text Processing. In Proceedings of the Conference on Empirical Methods in Natural Language Processing, Brussels, Belgium, 31 October–4 November 2018. [Google Scholar]
- Wu, Y.; Schuster, M.; Chen, Z.; Le, Q.V.; Norouzi, M.; Macherey, W.; Krikun, M.; Cao, Y.; Gao, Q.; Macherey, K.; et al. Google’s Neural Machine Translation System: Bridging the Gap between Human and Machine Translation. arXiv 2016, arXiv:1609.08144. [Google Scholar]
- Gurulingappa, H.; Rajput, A.M.; Roberts, A.; Fluck, J.; Hofmann-Apitius, M.; Toldo, L. Development of a benchmark corpus to support the automatic extraction of drug-related adverse effects from medical case reports. J. Biomed. Inform. 2012, 45, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, H.; Araujo, L.; Martínez-Romo, J. Deep neural models for extracting entities and relationships in the new RDD corpus relating disabilities and rare diseases. Comput. Methods Programs Biomed. 2018, 164, 121–129. [Google Scholar] [CrossRef]
- Fabregat, H.; Martínez-Romo, J.; Araujo, L. Overview of the DIANN Task: Disability Annotation Task. In Proceedings of the IberEval@SEPLN, Seville, Spain, 18 September 2018. [Google Scholar]
- Lai, T.; Ji, H.; Zhai, C.; Tran, Q.H. Joint Biomedical Entity and Relation Extraction with Knowledge-Enhanced Collective Inference. In Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing; Volume 1: Long Papers; Association for Computational Linguistics: Seattle, WA, USA, 2021; pp. 6248–6260. [Google Scholar]
- Akiba, T.; Sano, S.; Yanase, T.; Ohta, T.; Koyama, M. Optuna: A next-generation hyperparameter optimization framework. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Anchorage, AK, USA, 4–8 August 2019; pp. 2623–2631. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).