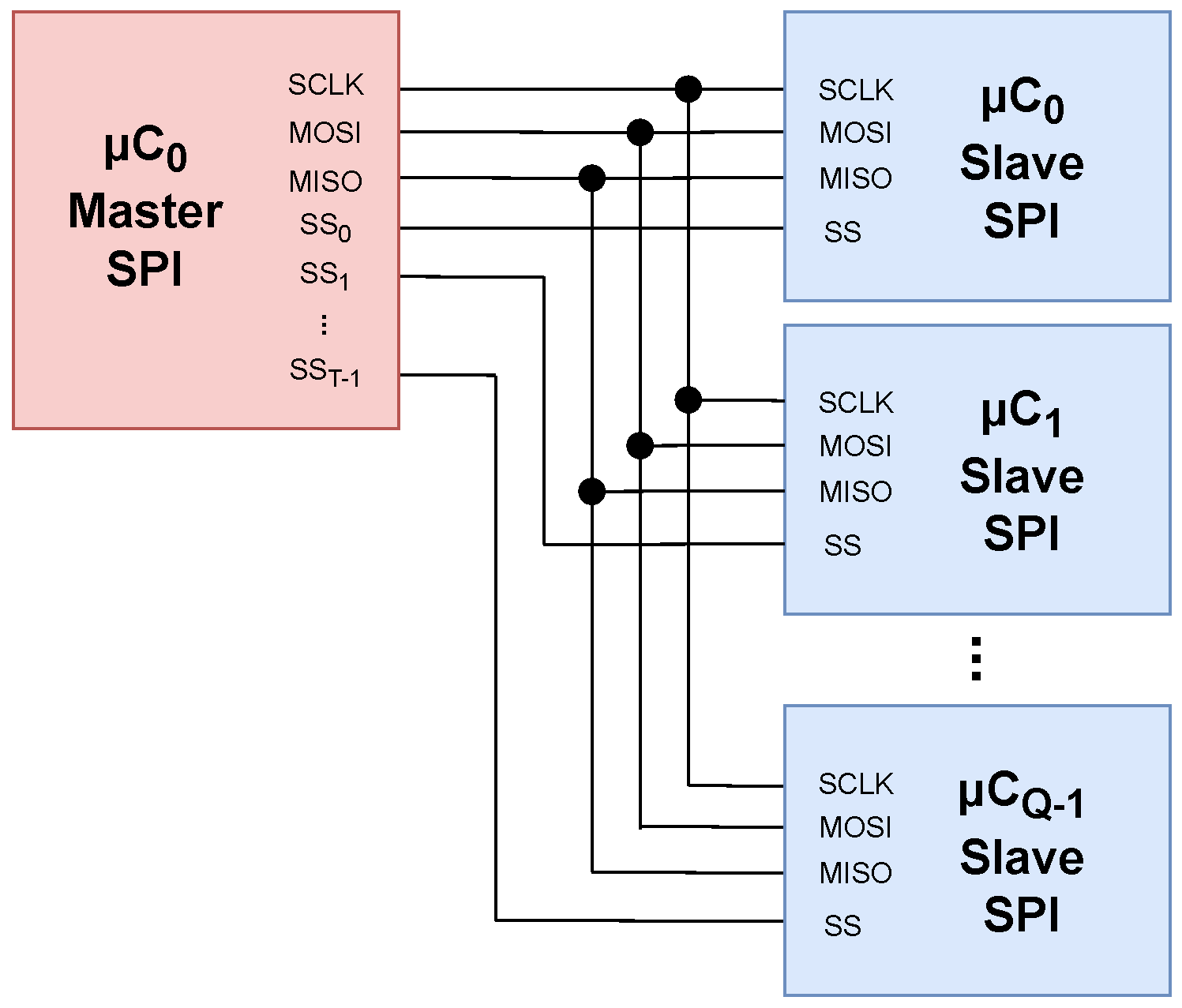
5.1. Memory Consumption
The first results collected were the program and data memory consumption. The program memory is non-volatile and is used to store the instructions to be executed by the processor, that is, the compiled program. The data memory is volatile and used to store variables during the run of the program. Also, the data memory can be divided into two segments:
Static memory: the memory consumed by global and static variables and is kept allocated during the whole program execution. That means this section of the memory cannot be freed and used by other variables.
Stack memory: the memory used by local variables and that can be allocated and freed according to their lifetime (for example, a local variable defined inside of a function will be freed when the function is finished).
The measurement of static memory is straightforward because the compiler can calculate it. The stack memory, in turn, needs to be calculated empirically. Therefore, both results are shown separately as for the master as for the slave node. To simplify the measurements, all experiments were performed with a fixed number of generations , since this affects only the processing time. Also, the evaluation function used was , with dimension , to avoid the use of external libraries. Finally, the crossover was configured as one-point and the number of mutated individuals was .
After running some experiments with the parameters above, the program memory consumption for the master and slave implementations is shown in
Table 2 and
Table 3, respectively. The compiled program consumes only a small portion of the 32 KB available and practically does not scale, using only about 11% of program memory in the master node and 7% in the slave for almost all scenarios. This result is important because it allows this distributed genetic algorithm implementation to be deployed as part of other projects.
The results regarding data memory are divided into static and stack memory. For all the scenarios tested above, the static memory was always 8 bytes. This was expected because this project does not use global or static variables so that almost all data memory can be used dynamically as stack memory. The results of stack memory, in turn, are shown in
Table 4 and
Table 5, sequentially. The numbers obtained in this work are similar to those obtained in [
13], because after dividing the global population both microcontrollers got the same population size used in the experiments in that work.
The numbers were also plotted in a chart in
Figure 10 and
Figure 11, and the best approximation of a linear function was done for all the cases. The stack memory consumption seems to increase linearly with the population size
N and at a slower rate with the increase of the individual size
M. While not presented, the same linear increase is expected for the number of dimensions
D in the individuals since another dimension is equivalent to add another individual. For a typical situation using 2 microcontrollers with a population size of 128, with individuals mapped into 16 bits, the total memory consumption is around 31% for the master and 28% for the slave. This low usage is important because it leaves about 70% of the memory available and allows this DGA implementation to reside together with other projects in the microcontrollers.
Therefore, it is important to consider the peculiarities of each application of this implementation. For this scenario with 2 microcontrollers, a global population of 512 individuals mapped into 32-bit would not be viable because the data memory would not be enough (by following the trend, it would be necessary more than 3.2 KB at least in each µC). As possible solutions, the population
N or the precision
M could be reduced or more microcontrollers could be added to provide more resources. The only problem with this last approach is that it would double the costs with hardware since the number of nodes
Q must be a power of 2 as explained in Equation (
7).
5.2. Processing Time
The second results collected from this distributed genetic algorithm implementation was the processing time. The methodology used in [
13], which was mostly based on measuring the number of clock cycles using the Atmel Studio 7 debugger, is not interesting for this work because the communication between multiple microcontrollers may not keep the algorithm fully deterministic. As shown in
Figure 6, some part of this implementation is not synchronized and some nodes may finish the run before others. Another issue that can happen is when the master sends a byte with a command and because of some error the slave didn’t receive it properly, then the slave will not send the acknowledge message and will wait for a resending of the command again. Thus, the following results present the real run time, experimentally measured with an external timer.
To evaluate the processing time, the following evaluation functions were used:
For all these functions, the following GA parameters were fixed: population size
, individuals mapped into
bits, number of generations
, and number of mutated individuals
. The results are presented in
Table 6. The processing time seems to not change so much with the type of evaluation function and this can be noticed when comparing functions
,
and
, which have different mathematical operations but same number of dimensions and similar run times. The same happened for
and
, which have one dimension as common characteristic. Thus, this suggests the time spent with the communication is being predominantly the part that is consuming more time.
To understand how the SPI communication is affecting the global time, several experiments were done to evaluate how the processing time changes according to the size of population N, number of generations K, and the number of dimensions D-all related with the number of bits per individual M. For the experiments to analyze N, K,and M, the function was used. For evaluate D, it was used the evaluation function , by adding or removing more terms when the dimension was greater than 2. For example, to evaluate the version with 4 dimensions, the terms and are added to the function and so on.
The results for processing time for
N,
K, and
D are presented in
Table 7,
Table 8 and
Table 9, respectively. For the results of
N and
K, by observing the lines from the top to the bottom, the value of
M does not affect so much the processing time and the difference in time when using 16-bit and 32-bit individuals is small. However, by analyzing the columns from left to right, it was noticed a sort of linear increase of processing time proportional as to
N as to
K in both
Table 7 and
Table 8. This impression can be proved in
Figure 12 and
Figure 13, wherein both cases the points seem to represent a first-degree polynomial function.
Finally, the results for the number of dimensions
D are presented in
Table 9. The value of
M seems to affect more the time than the other 2 previous cases (
N and
K). On the other hand, even though the increase in the number of dimensions
D affects the consumption of data memory, it produces only a slight increase in the processing time. A first-degree polynomial function is plotted in
Figure 14 and shows how the increase is expected for different values of
D.
Therefore, the results of processing time are importing to show how it increases based on important parameters of the distributed genetic algorithm. All the main four variables analyzed above (N, K, D, and M) influence directly on the time spent with communication between the nodes, which is the main overhead in this case, because an 8-bit microcontroller can transfer only one byte (8 bits) at once via SPI. The variables D and M define how large is each individual in terms of bytes and N and K how many transfers need to be done during a run of the distributed GA. For that reason, it is crucial to select the proper GA parameters to have control over the processing time.
5.4. Comparison with Standalone Version
A final experiment was to investigate how the distributed genetic algorithm proposed in this work is compared to the standalone version, that is, the genetic algorithm that runs in one single 8-bit microcontroller, which is presented in [
13]. There are two motivations for this result:
Verify if it is possible to accelerate the genetic algorithm for a certain application by adding more microcontrollers;
Evaluate if, by using multiple microcontrollers configured with lower voltage and lower clock frequency, it is possible to save energy and have a similar performance to the standalone version.
By analyzing the results presented in
Section 5.2, there is a large overhead due to the SPI communication between the microcontrollers, which is consuming a lot of processing time even using those simple evaluation functions. Thus, in order to have some advantage with multiple cores, the evaluation function needs to be complex enough so that the processing time spent with it is much higher than the time spent with the data transfer between the nodes. To not change the original evaluation functions, they were changed in such a way to consume more clock cycles but generating the same result. This idea is expressed in the Algorithm 4.
| Algorithm 4 Redefinition of Evaluation Function to Become Slower |
▹ Define how many times the evaluation function will repeat (2000 times, for example).
- 1:
-
▹ The original evaluation function will run REPEAT times. - 2:
function EFM () - 3:
- 4:
for to do - 5:
- 6:
end for - 7:
return - 8:
end function
|
For the following experiments, the GA was set up with the following parameters: population size
, number of generations
, individual size
, number of mutated individual
, and evaluation function
, which was set up to repeat 1000, 2000, 4000 and 8000 times using the strategy proposed in Algorithm 4. By measuring the number of clock cycles that this function needs to run for each case, the processing time of the modified evaluation function,
, can be calculated as
where
is the number of clock cycles to run the modified evaluation function and CLK the clock frequency of the microcontroller. This processing time is used below for the different scenarios. The values of
were collecting via experiments in Atmel Studio 7 and are shown in
Table 10.
The first measures were done using both standalone and distributed version of the GA running at the same clock speed and voltage. As shown in
Table 11 and
Figure 21, when the evaluation function is not complex enough, the overhead due SPI communication makes the distributed GA slower than the standalone GA. However, as the evaluation function becomes more complex, the distributed GA becomes faster. In fact, this can also be noticed by analyzing both polynomial functions that fit those points shown in
Figure 21, which is in the format
, and is defined as
for the standalone version, and
for the distributed version, where
t represents the processing time in seconds and
represents the evaluation function clock cycles. When the number of clock cycles
is large enough, the distributed version will run approximately 2 times faster than standalone as demonstrated as follows
Another important analysis from Equation (
23) is the high overhead. By applying the Genetic Algorithm parameters and the SPI clock frequency, defined in 125 kHz, to Equations (
14) and (
20), it is possible to see how the theoretical model compares to the experiments. The
is calculated as follows
where the value of
would be the maximum overhead in seconds for the worst-case scenario, that is, if all individuals were selected from the slave. However, since this is unlikely to happen, then the
seconds in Equation (
23) is reasonable and under the theoretical limit.
Finally, to validate the theoretical model presented in Equation (
18), by applying the results from the experiment shown in Equation (
22) and from Equation (
25), the expected equation for the distributed versions would be
where
is the estimated processing time for the distributed GA with the same configuration. The result of Equation (
26) is similar to the one obtained experimentally in Equation (
23). It is important to emphasize again that the
is calculated for the worst-case scenario (all individuals selected from the slave) and that is why the second term
is greater than
.
Figure 22 illustrates how the theoretical model is reasonable when compared to the experiments, by showing that the theoretical model (blue line) has approximately the same inclination of the experimental result (cyan line). What makes the theoretical model to be higher is because it represents the time for the DGA when the overhead is maximum (the worst-case scenario). For most practical applications, the overhead will be lower than this and the line will be shifted vertically to a lower position.
The second experiment was with the distributed version set up with reduced voltage and lower clock frequency for the same GA configuration used above. The motivation for this configuration is to take advantage of how dynamic power is defined for CMOS systems, which is present in regular microcontrollers [
29,
30]. By reducing the frequency and voltage, it is possible to reduce the power and consequently energy consumption in a higher rate. This idea can be verified in the equation that defines the power,
P, as the sum of the dynamic power,
, and static power,
, in a CMOS integrated circuit and is defined as
where
C is the capacitance of the transistor gates,
f the operating frequency,
V the power supply voltage, and
the static power which depends mostly on the number of transistors and how they are organized spatially. Thus, by reducing the voltage
V in the system, the reduction in the dynamic power will be in a quadratic level.
The behavior of Equation (
27) can also be found out in the datasheet of the microcontroller ATmega328P [
28]. The
Figure 23 shows what is the current
consumed by the µC for different combinations of frequency (from 0 to 20 MHz) and voltage (from 2.7 V to 5.5 V). Since power can be also defined as
then the power will be reduced for low values of voltage and frequency as well (power reduces from right to left and from top to bottom in
Figure 23).
To run this last experiment, both microcontrollers in the DGA were arranged to run at 8 MHz at a voltage of 2.7 V. This is the minimum operational voltage for this frequency, as shown in
Figure 24. The processing time for the same configuration in the previous experiment in shown in
Table 12. As expected, by running at a slower clock frequency made the processing time increase, and even in situations where the evaluation function is complex, the processing time for the distributed GA is always slower than the standalone GA running at 16 MHz, as expressed in
Table 11.
The comparison between these new results with the standalone version is presented in
Figure 25. Both lines seem to be parallel, which suggests the distributed version with 2 nodes and half of the clock speed will never be faster than the standalone version. In fact, the first-degree polynomial functions that fit these points are calculated as follows
where
is the processing time for the DGA running at reduced clock speed. This equation has almost the same inclination of Equation (
22) and the small difference may be a consequence of error/lack of precision of the measurements. Thus, this result suggests that for this GA configuration the DGA will always be about 16.43 s slower than the standalone GA, no matter how complex is the evaluation function. However, for long runs, the time difference will decrease relatively. For example, if the standalone GA takes 5 min, the DGA will take 5 min plus 16.43 s, which is only about 5% slower.
Even though the distributed genetic algorithm with 2 microcontrollers running at a half frequency of the standalone version is always slower, the main advantage of this structure is the save of power and consequently energy. This is one of the most common goals in embedded systems because they normally run on batteries and need to be power-efficient. The equation of energy consumption,
E, is the product of the power equation by the elapsed time, defined as follows
where
is the elapsed time. Since the elapsed time for the standalone and distributed versions on lower frequency, represented by
and
respectively, were calculated in Equations (
22) and (
29), after applying them to Equation (
30), the energy consumption equations for both cases are determined as
and
where
,
, and
are respectively the energy, power, and time consumption in the standalone system;
,
, and
are respectively the energy, power, and time consumption in the distributed system with reduced clock speed; and
Q is the number of nodes in the distributed system (
in these results).
From
Figure 23 and
Figure 24 and considering both GA versions running at the lowest operating voltage for the specified frequency, the standalone version at 16 MHz needs approximately 4.5 V and consumes a current of 7.5 mA, and the distributed version needs approximately 2.7 V and consumes a current of 2.3 mA. By using these values in Equations (
31) and (
32), respectively, the final expressions for energy are defined as
and
where the unity mAh means milliampere hour.
By plotting the energy consumption equations in
Figure 26, the equation of the distributed version grows slower than the standalone version. The energy consumption in the distributed genetic algorithm for this configuration will be lower than the standalone GA when the evaluation function has at least 73,244 clock cycles, as demonstrated in
where the value of
that solves this equation is 73244.
For example, when the evaluation function requires around 1,000,000 clock cycles, the standalone genetic algorithm needs approximately 130 s and 4400 mAh and the distributed GA approximately 147 s to run but only 1832 mAh, which is less than half of the energy spent by the standalone one. When the number of clock cycles is big enough, the distributed version will consume merely 37.1% regarding the standalone as demonstrated in
Therefore, the results presented in this section show some possible scenarios where the distributed genetic algorithm can have some advantages over a regular GA running on a single microcontroller. For situations where the evaluation function is not too complex, the standalone version is still the best option because it runs faster and consumes less energy. However, if it is complex enough, this proposed DGA, even having a large overhead due to the SPI communication, can be used either to accelerate the execution by running the microcontrollers in high frequency or to save power by reducing voltage and frequency. Finally, similar results are expected in case of employing more microcontrollers (4, 8, etc.) and with more cores, the global clock could be even more reduced to 4 MHz, 2 MHz, and so on.