In Silico Identification of Dysregulated miRNAs Targeting KRAS Gene in Pancreatic Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Differential Expression of miRNAs in PCs
2.2. Identification of KRAS-Targeted miRNAs
2.3. Differential Expression of KRAS-Targeted miRNAs in PCs
2.4. miRNA- KRAS mRNA Co-Expression Analysis
3. Results
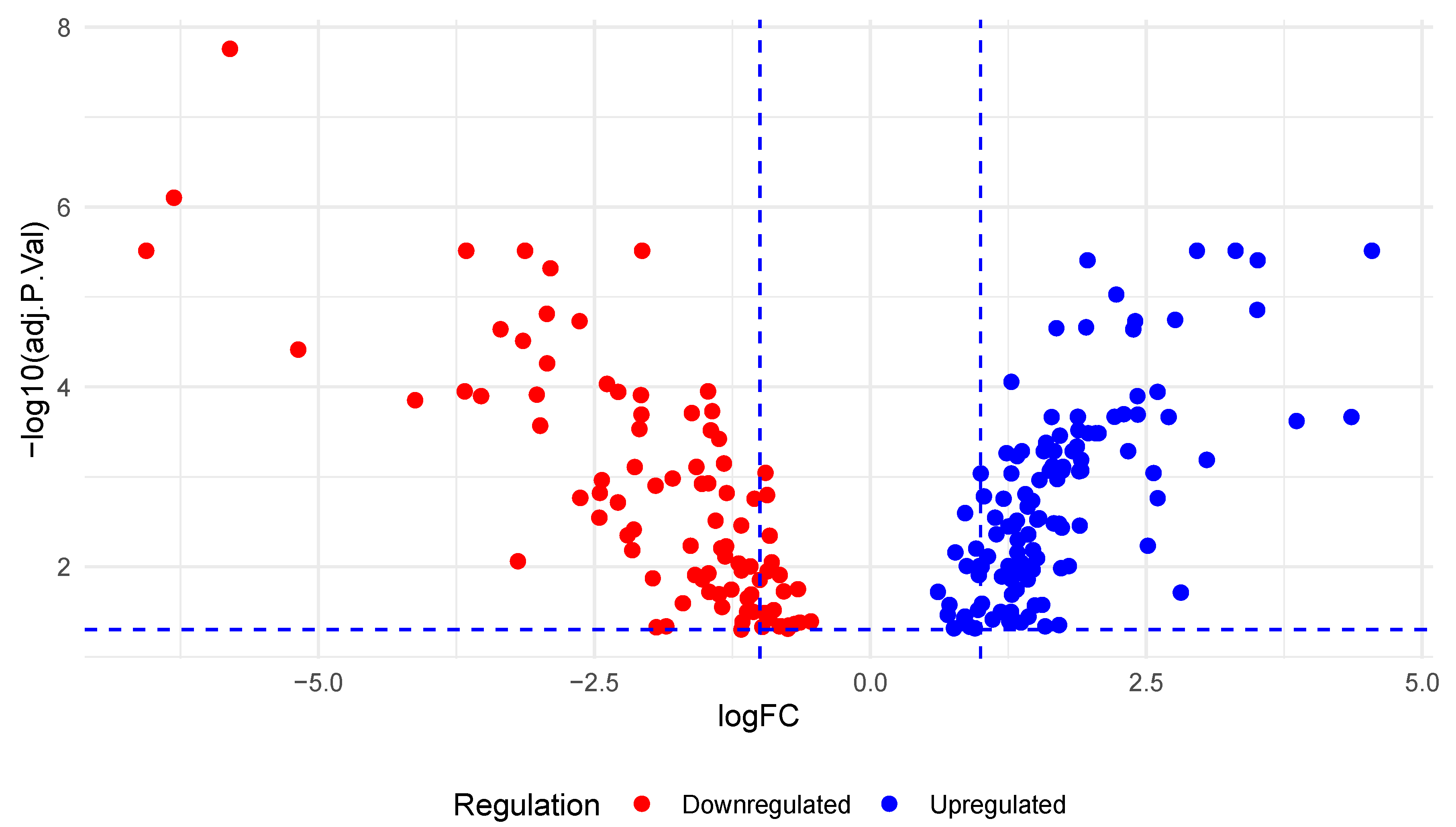
3.1. Differential Expression of miRNAs in PC
3.2. Identification of KRAS-Targeted miRNAs
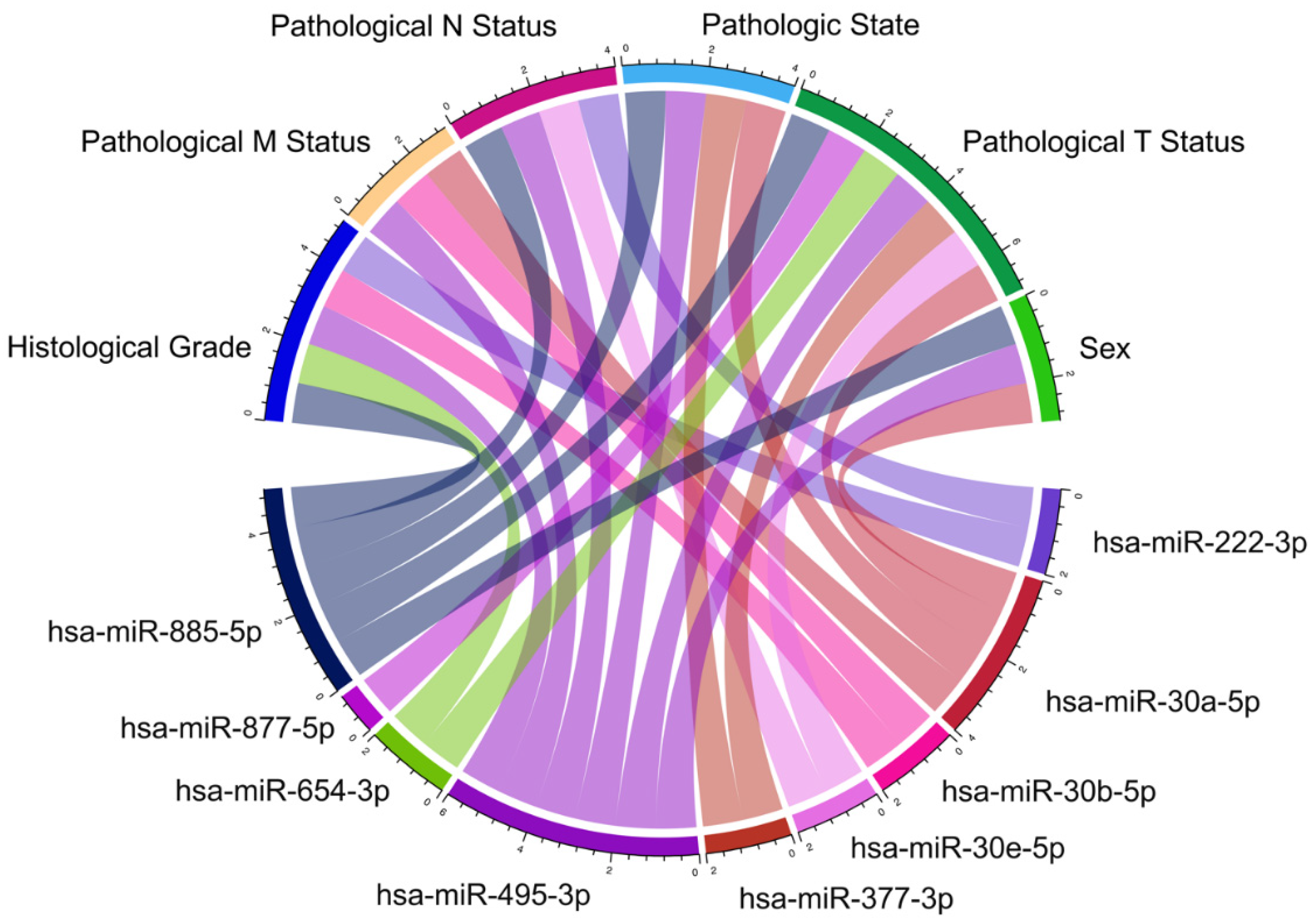
3.3. Differential Expression of KRAS-Targeting miRNAs in PC
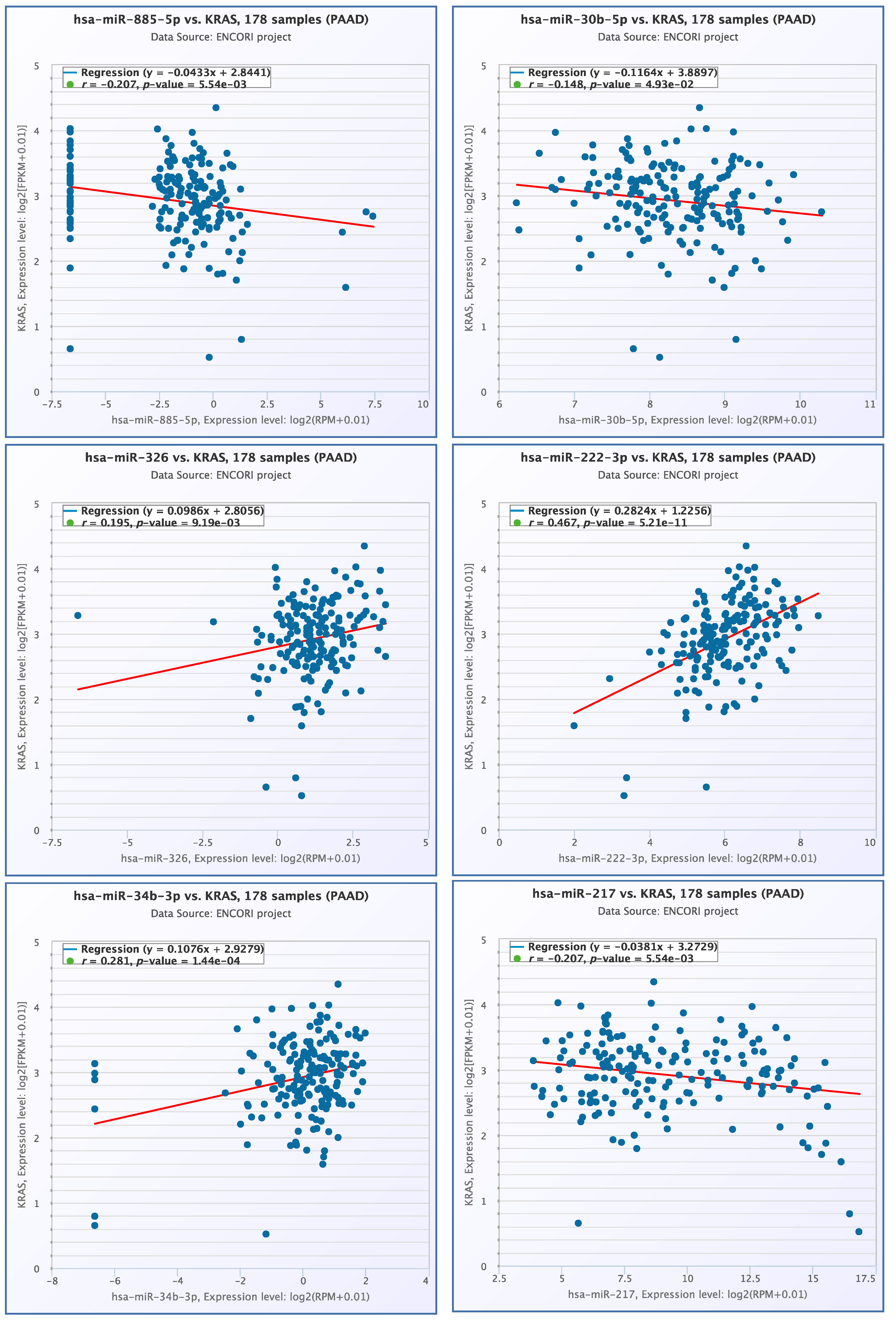
3.4. miRNA- KRAS mRNA Co-Expression Analysis
3.5. Molecular Interaction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klein, A.P. Pancreatic Cancer Epidemiology: Understanding the Role of Lifestyle and Inherited Risk Factors. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Kolbeinsson, H.M.; Chandana, S.; Wright, G.P.; Chung, M. Pancreatic Cancer: A Review of Current Treatment and Novel Therapies. J. Investig. Surg. 2023, 36, 2129884. [Google Scholar] [CrossRef] [PubMed]
- Global Cancer Observatory. International Agency for Research on Cancer. Available online: https://gco.iarc.fr/ (accessed on 22 September 2023).
- Halbrook, C.J.; Lyssiotis, C.A.; Pasca Di Magliano, M.; Maitra, A. Pancreatic Cancer: Advances and Challenges. Cell 2023, 186, 1729–1754. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Liu, W. Pancreatic Cancer: A Review of Risk Factors, Diagnosis, and Treatment. Technol. Cancer Res. Treat. 2020, 19, 1533033820962117. [Google Scholar] [CrossRef] [PubMed]
- Luo, J. KRAS Mutation in Pancreatic Cancer. Semin. Oncol. 2021, 48, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, H.; Liao, X.; Tsai, H. KRAS Mutation: The Booster of Pancreatic Ductal Adenocarcinoma Transformation and Progression. Front. Cell Dev. Biol. 2023, 11, 1147676. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.; Burkhart, D.L.; Haigis, K.M. Classification of KRAS-Activating Mutations and the Implications for Therapeutic Intervention. Cancer Discov. 2022, 12, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chi, L.; Yu, F.; Dai, H.; Gao, C.; Si, X.; Wang, Z.; Liu, L.; Zheng, J.; Shan, L.; et al. Annual Review of KRAS Inhibitors in 2022. Eur. J. Med. Chem. 2023, 249, 115124. [Google Scholar] [CrossRef] [PubMed]
- Rathod, L.S.; Dabhade, P.S.; Mokale, S.N. Recent Progress in Targeting KRAS Mutant Cancers with Covalent G12C-Specific Inhibitors. Drug Discov. Today 2023, 28, 103557. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Vander Heiden, M.G.; McCormick, F. The Metabolic Landscape of RAS-Driven Cancers from Biology to Therapy. Nat. Cancer 2021, 2, 271–283. [Google Scholar] [CrossRef]
- Pupo, E.; Avanzato, D.; Middonti, E.; Bussolino, F.; Lanzetti, L. KRAS-Driven Metabolic Rewiring Reveals Novel Actionable Targets in Cancer. Front. Oncol. 2019, 9, 848. [Google Scholar] [CrossRef] [PubMed]
- Punekar, S.R.; Velcheti, V.; Neel, B.G.; Wong, K.-K. The Current State of the Art and Future Trends in RAS-Targeted Cancer Therapies. Nat. Rev. Clin. Oncol. 2022, 19, 637–655. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Bortoletto, A.S.; Parchem, R.J. KRAS Hijacks the miRNA Regulatory Pathway in Cancer. Cancer Res. 2023, 83, 1563–1572. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Chen, Y.; Zhou, X. The Roles of microRNAs in Epigenetic Regulation. Curr. Opin. Chem. Biol. 2019, 51, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Iwakawa, H.; Tomari, Y. Life of RISC: Formation, Action, and Degradation of RNA-Induced Silencing Complex. Mol. Cell 2022, 82, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Shui, B.; La Rocca, G.; Ventura, A.; Haigis, K.M. Interplay between K-RAS and miRNAs. Trends Cancer 2022, 8, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Cuzziol, C.I.; Marzochi, L.L.; Possebon, V.S.; Kawasaki-Oyama, R.S.; Mattos, M.F.; Junior, V.S.; Ferreira, L.A.M.; Pavarino, É.C.; Castanhole-Nunes, M.M.U.; Goloni-Bertollo, E.M. Regulation of VEGFA, KRAS, and NFE2L2 Oncogenes by MicroRNAs in Head and Neck Cancer. Int. J. Mol. Sci. 2022, 23, 7483. [Google Scholar] [CrossRef] [PubMed]
- Oncogenic KRAS Promotes Pancreatic Ductal Adenocarcinoma (PDAC) through Post-Transcriptionally Regulated KRAS-Induced Granules (KGs). Available online: https://www.researchsquare.com (accessed on 22 September 2023).
- Shui, B.; Beyett, T.S.; Chen, Z.; Li, X.; La Rocca, G.; Gazlay, W.M.; Eck, M.J.; Lau, K.S.; Ventura, A.; Haigis, K.M. Oncogenic K-Ras Suppresses Global miRNA Function. Mol. Cell 2023, 83, 2509–2523.e13. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Wang, Y.; Ling, Y.; Zhou, C.; Wang, H.; Teschendorff, A.E.; Zhao, Y.; Zhao, H.; He, Y.; Zhang, G.; et al. dbDEMC 3.0: Functional Exploration of Differentially Expressed miRNAs in Cancers of Human and Model Organisms. Genom. Proteom. Bioinform. 2022, 20, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Munding, J.B.; Adai, A.T.; Maghnouj, A.; Urbanik, A.; Zöllner, H.; Liffers, S.T.; Chromik, A.M.; Uhl, W.; Szafranska-Schwarzbach, A.E.; Tannapfel, A.; et al. Global microRNA Expression Profiling of Microdissected Tissues Identifies miR-135b as a Novel Biomarker for Pancreatic Ductal Adenocarcinoma. Int. J. Cancer 2012, 131, E86–E95. [Google Scholar] [CrossRef] [PubMed]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The Biochemical Basis of microRNA Targeting Efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An Online Database for Prediction of Functional microRNA Targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; Ziebarth, J.D.; Cui, Y. PolymiRTS Database 3.0: Linking Polymorphisms in microRNAs and Their Target Sites with Human Diseases and Biological Pathways. Nucleic Acids Res. 2014, 42, D86–D91. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.W.; Chen, Y.; Chen, S.; Wang, X. OncomiR: An Online Resource for Exploring Pan-Cancer microRNA Dysregulation. Bioinformatics 2018, 34, 713–715. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and Protein-RNA Interaction Networks from Large-Scale CLIP-Seq Data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [PubMed]
- de Cubas, A.A.; Leandro-García, L.J.; Schiavi, F.; Mancikova, V.; Comino-Méndez, I.; Inglada-Pérez, L.; Perez-Martinez, M.; Ibarz, N.; Ximénez-Embún, P.; López-Jiménez, E.; et al. Integrative Analysis of miRNA and mRNA Expression Profiles in Pheochromocytoma and Paraganglioma Identifies Genotype-Specific Markers and Potentially Regulated Pathways. Endocr. Relat. Cancer 2013, 20, 477–493. [Google Scholar] [CrossRef] [PubMed]
- Ferretti, E.; De Smaele, E.; Po, A.; Di Marcotullio, L.; Tosi, E.; Espinola, M.S.B.; Di Rocco, C.; Riccardi, R.; Giangaspero, F.; Farcomeni, A.; et al. MicroRNA Profiling in Human Medulloblastoma. Int. J. Cancer 2009, 124, 568–577. [Google Scholar] [CrossRef] [PubMed]
- Fassan, M.; Volinia, S.; Palatini, J.; Pizzi, M.; Baffa, R.; De Bernard, M.; Battaglia, G.; Parente, P.; Croce, C.M.; Zaninotto, G.; et al. MicroRNA Expression Profiling in Human Barrett’s Carcinogenesis. Int. J. Cancer 2011, 129, 1661–1670. [Google Scholar] [CrossRef] [PubMed]
- Arndt, G.M.; Dossey, L.; Cullen, L.M.; Lai, A.; Druker, R.; Eisbacher, M.; Zhang, C.; Tran, N.; Fan, H.; Retzlaff, K.; et al. Characterization of Global microRNA Expression Reveals Oncogenic Potential of miR-145 in Metastatic Colorectal Cancer. BMC Cancer 2009, 9, 374. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Wang, Y.-W.; Wang, Y.-Y.; Song, Y.; Zhu, J.; Si, P.-C.; Ma, R. Identification of microRNA Biomarkers in the Blood of Breast Cancer Patients Based on microRNA Profiling. Gene 2017, 619, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, S.K.; Yendamuri, S.; Kannisto, E.; Kucharczuk, J.C.; Singhal, S.; Vachani, A. MicroRNA Expression Profiles of Whole Blood in Lung Adenocarcinoma. PLoS ONE 2012, 7, e46045. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, A.; Jung, M.; Mollenkopf, H.-J.; Wagner, I.; Stephan, C.; Jentzmik, F.; Miller, K.; Lein, M.; Kristiansen, G.; Jung, K. Diagnostic and Prognostic Implications of microRNA Profiling in Prostate Carcinoma. Int. J. Cancer 2010, 126, 1166–1176. [Google Scholar] [CrossRef] [PubMed]
- Lionetti, M.; Biasiolo, M.; Agnelli, L.; Todoerti, K.; Mosca, L.; Fabris, S.; Sales, G.; Deliliers, G.L.; Bicciato, S.; Lombardi, L.; et al. Identification of microRNA Expression Patterns and Definition of a microRNA/mRNA Regulatory Network in Distinct Molecular Groups of Multiple Myeloma. Blood 2009, 114, e20–e26. [Google Scholar] [CrossRef] [PubMed]
- Plieskatt, J.L.; Rinaldi, G.; Feng, Y.; Peng, J.; Yonglitthipagon, P.; Easley, S.; Laha, T.; Pairojkul, C.; Bhudhisawasdi, V.; Sripa, B.; et al. Distinct miRNA Signatures Associate with Subtypes of Cholangiocarcinoma from Infection with the Tumourigenic Liver Fluke Opisthorchis Viverrini. J. Hepatol. 2014, 61, 850–858. [Google Scholar] [CrossRef]
- Manso, R.; Martínez-Magunacelaya, N.; Eraña-Tomás, I.; Monsalvez, V.; Rodríguez-Peralto, J.L.; Ortiz-Romero, P.-L.; Santonja, C.; Cristóbal, I.; Piris, M.A.; Rodríguez-Pinilla, S.M. Mycosis Fungoides Progression Could Be Regulated by microRNAs. PLoS ONE 2018, 13, e0198477. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ye, Y.; Chang, D.W.; Lin, S.-H.; Huang, M.; Tannir, N.M.; Matin, S.; Karam, J.A.; Wood, C.G.; Chen, Z.-N.; et al. Global and Targeted miRNA Expression Profiling in Clear Cell Renal Cell Carcinoma Tissues Potentially Links miR-155-5p and miR-210-3p to Both Tumorigenesis and Recurrence. Am. J. Pathol. 2018, 188, 2487–2496. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.J.; Papoulas, M.; Graubardt, N.; Ovdat, E.; Loewenstein, S.; Kania-Almog, J.; Pasmanik-Chor, M.; Brazowski, E.; Cagnano, E.; Nachmany, I.; et al. Micro-RNA Expression Patterns Predict Metastatic Spread in Solid Pseudopapillary Neoplasms of the Pancreas. Front. Oncol. 2020, 10, 328. [Google Scholar] [CrossRef] [PubMed]
- Reis, P.P.; Drigo, S.A.; Carvalho, R.F.; Lopez Lapa, R.M.; Felix, T.F.; Patel, D.; Cheng, D.; Pintilie, M.; Liu, G.; Tsao, M.-S. Circulating miR-16-5p, miR-92a-3p, and miR-451a in Plasma from Lung Cancer Patients: Potential Application in Early Detection and a Regulatory Role in Tumorigenesis Pathways. Cancers 2020, 12, 2071. [Google Scholar] [CrossRef]
- Kim, M.; Kogan, N.; Slack, F.J. Cis-Acting Elements in Its 3′ UTR Mediate Post-Transcriptional Regulation of KRAS. Oncotarget 2016, 7, 11770–11784. [Google Scholar] [CrossRef] [PubMed]
- Sell, M.C.; Ramlogan-Steel, C.A.; Steel, J.C.; Dhungel, B.P. MicroRNAs in Cancer Metastasis: Biological and Therapeutic Implications. Expert. Rev. Mol. Med. 2023, 25, e14. [Google Scholar] [CrossRef] [PubMed]
- Chhichholiya, Y.; Singh, H.V.; Vashistha, R.; Singh, S.; Munshi, A. Deciphering the Role of KRAS Gene in Oncogenesis: Focus on Signaling Pathways, Genetic Alterations in 3’UTR, KRAS Specific miRNAs and Therapeutic Interventions. Crit. Rev. Oncol./Hematol. 2024, 194, 104250. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Xiao, X.; Xing, X.; Li, X.; Xia, T.; Long, H. KRAS and VEGF Gene 3’-UTR Single Nucleotide Polymorphisms Predicted Susceptibility in Colorectal Cancer. PLoS ONE 2017, 12, e0174140. [Google Scholar] [CrossRef] [PubMed]
- Insodaite, R.; Smalinskiene, A.; Liutkevicius, V.; Ulozas, V.; Poceviciute, R.; Bielevicius, A.; Kucinskas, L. Associations of Polymorphisms Localized in the 3′UTR Regions of the KRAS, NRAS, MAPK1 Genes with Laryngeal Squamous Cell Carcinoma. Genes 2021, 12, 1679. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Lu, F.; Li, X.; Zhou, W.; Li, S.; Jiang, Y.; Wu, H.; Wang, J. Association between KRAS Gene Polymorphisms and Genetic Susceptibility to Breast Cancer in a Chinese Population. J. Clin. Lab. Anal. 2022, 37, e24806. [Google Scholar] [CrossRef] [PubMed]
- Gallegos-Arreola, M.P.; Garibaldi-Ríos, A.F.; Cruz-Sánchez, J.I.; Figuera, L.E.; Ronquillo-Carreón, C.A.; Rosales-Reynoso, M.A.; Gómez-Meda, B.C.; Carrillo-Dávila, I.A.; Puebla-Pérez, A.M.; Montoya-Fuentes, H.; et al. Association of the Rs8720 and Rs12587 KRAS Gene Variants with Colorectal Cancer in a Mexican Population and Their Analysis In Silico. Cells 2023, 12, 1941. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Zhuo, Z.; Hua, R.-X.; Fu, K.; Jia, W.; Zhu, J.; Zhang, J.; Cheng, J.; Zhou, H.; Xia, H.; et al. Association of KRAS and NRAS Gene Polymorphisms with Wilms Tumor Risk: A Four-Center Case-Control Study. Aging 2019, 11, 1551–1563. [Google Scholar] [CrossRef] [PubMed]
- Ulusan, M.; Sen, S.; Yilmazer, R.; Dalay, N.; Demokan, S. The Let-7 microRNA Binding Site Variant in KRAS as a Predictive Biomarker for Head and Neck Cancer Patients with Lymph Node Metastasis. Pathol. Res. Pr. 2022, 239, 154147. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, X.; Song, Q. MicroRNA 885-5p Inhibits Hepatocellular Carcinoma Metastasis by Repressing AEG1. Onco Targets Ther. 2020, 13, 981–988. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Cui, H.; Zeng, S.; Li, L. miR-885-5p Inhibits Invasion and Metastasis in Gastric Cancer by Targeting Malic Enzyme 1. DNA Cell Biol. 2021, 40, 694–705. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, G.; Huang, J.; Chai, L.; Liu, X.; Dai, Q.; Yang, Z. KRAS SNPs Are Related to Colorectal Cancer Susceptibility and Survival in Chinese People. Biomark. Med. 2020, 14, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Jia, C.; Wang, J.; Shi, W.; Wang, X.; Song, Y.; Peng, C. Exosomal Hsa-miR-151a-3p and Hsa-miR-877-5p Are Potential Novel Biomarkers for Predicting Bone Metastasis in Lung Cancer. Aging 2023, 15, 14864–14888. [Google Scholar] [CrossRef] [PubMed]
- Xiong, D.; Dang, Y.; Lin, P.; Wen, D.; He, R.; Luo, D.; Feng, Z.; Chen, G. A circRNA–miRNA–mRNA Network Identification for Exploring Underlying Pathogenesis and Therapy Strategy of Hepatocellular Carcinoma. J. Transl. Med. 2018, 16, 220. [Google Scholar] [CrossRef]
- Gallegos-Arreola, M.P.; Zúñiga-González, G.M.; Gómez-Mariscal, K.; Rosales-Reynoso, M.A.; Luis, L.; Puebla-Pérez, A.M.; Pineda-Razo, T. Association of Rs712 Polymorphism in a Let-7 microRNA-Binding Site of KRAS Gene with Colorectal Cancer in a Mexican Population. Iran. J. Basic. Med. Sci. 2019, 22, 324–327. [Google Scholar] [CrossRef]
- Al-Haddad, R.H.; Jasim, H.M.; Saleh, W.A. Association between KRAS Rs 712 G>T Polymorphism with the Incidence of Breast Tumors in Iraqi Women. Iraqi J. Biotechnol. 2019, 18, 55–61. [Google Scholar]
- Huang, X.; Yang, Y.; Guo, Y.; Cao, Z.L.; Cui, Z.W.; Hu, T.C.; Gao, L.B. Association of a Let-7 KRAS Rs712 Polymorphism with the Risk of Breast Cancer. Genet. Mol. Res. 2015, 14, 16913–16920. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Qiu, R.; He, X.; Hu, Z.; Zheng, F.; Huang, X.; Xie, X.; Chen, F.; Ou, H.; Lin, G. miR-647 Inhibits Hepatocellular Carcinoma Cell Progression by Targeting Protein Tyrosine Phosphatase Receptor Type F. Bioengineered 2022, 13, 1090–1102. [Google Scholar] [CrossRef] [PubMed]
- Qin, K.; Tian, G.; Chen, G.; Zhou, D.; Tang, K. miR-647 Inhibits Glioma Cell Proliferation, Colony Formation and Invasion by Regulating HOXA9. J. Gene Med. 2020, 22, e3153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, M.; Wang, G.; Tian, Y.; He, X. Tumor Promoter Role of miR-647 in Gastric Cancer via Repression of TP73. Mol. Med. Rep. 2018, 18, 3744–3750. [Google Scholar] [CrossRef]
- Block, I.; Burton, M.; Sørensen, K.P.; Andersen, L.; Larsen, M.J.; Bak, M.; Cold, S.; Thomassen, M.; Tan, Q.; Kruse, T.A. Association of miR-548c-5p, miR-7-5p, miR-210-3p, miR-128-3p with Recurrence in Systemically Untreated Breast Cancer. Oncotarget 2018, 9, 9030–9042. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, X.; Wang, G.; Wang, L.; Lin, Y.; Sun, F. Hsa-miR-513b-5p Suppresses Cell Proliferation and Promotes P53 Expression by Targeting IRF2 in Testicular Embryonal Carcinoma Cells. Gene 2017, 626, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Jian, M.; Qi, H.; Mao, W.-Z. MicroRNA 495 Inhibits Proliferation and Metastasis and Promotes Apoptosis by Targeting Twist1 in Gastric Cancer Cells. Oncol. Res. 2019, 27, 389. [Google Scholar] [CrossRef] [PubMed]
- Eun, J.W.; Kim, H.S.; Shen, Q.; Yang, H.D.; Kim, S.Y.; Yoon, J.H.; Park, W.S.; Lee, J.Y.; Nam, S.W. MicroRNA-495-3p Functions as a Tumor Suppressor by Regulating Multiple Epigenetic Modifiers in Gastric Carcinogenesis. J. Pathol. 2018, 244, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Jiang, L.; Guo, C. The Micro RNA Hsa-miR-377-3p Inhibits Tumor Growth in Malignant Melanoma. RSC Adv. 2019, 9, 19057–19064. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yu, P.; Luo, J.; Sun, Z.; Yu, J.; Wang, J. Transcriptomic and microRNA Expression Profiles Identify Biomarkers for Predicting Neo-Chemoradiotherapy Response in Esophageal Squamous Cell Carcinomas (ESCC). Front. Pharmacol. 2021, 12, 626972. [Google Scholar] [CrossRef] [PubMed]
- Gallegos-Arreola, M.P.; García Verdín, P.M.; Magaña-Torres, M.T.; Figuera, L.E.; Zúñiga-González, G.M.; Rosales-Reynoso, M.A.; Gómez-Meda, B.C.; Puebla-Pérez, A.M. Association between Rs61764370, Rs9266, and Rs140080026 Polymorphisms of the KRAS Gene and Breast Cancer Risk in a Mexican Population. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 6454–6464. [Google Scholar] [CrossRef] [PubMed]
- Farokhzad, N.; Hosseini, S.M.; Edalat, H.; Sadeghi, M. Association of Rs61764370 Polymorphism within Let-7 microRNA-Binding Site with Lung Cancer in Iranian Population. Afr. Health Sci. 2020, 20, 1299–1303. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Huang, C.; Li, S.; Yang, C.; Xi, Y.; Wang, L.; Zhang, F.; Fu, Y.; Li, D. Hsa-miR-326 Targets CCND1 and Inhibits Non-Small Cell Lung Cancer Development. Oncotarget 2016, 7, 8341–8359. [Google Scholar] [CrossRef]
- Luo, H.; Zhong, F.; Jing, X.; Lin, H.; Li, Y. miRNA Profiling of Human Nasopharyngeal Carcinoma Cell Lines HONE1 and CNE2 after X-Ray Therapy. Adv. Clin. Exp. Med. 2022, 31, 671–687. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Hong, M.; Zhang, Y.; Wang, J.; Li, L.; Zhu, H.; Sheng, Y.; Wu, W.-S.; Zhang, Z. miR-30e-5p Regulates Leukemia Stem Cell Self-Renewal through the Cyb561/ROS Signaling Pathway. Haematologica 2023, 109, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Laudato, S.; Patil, N.; Abba, M.L.; Leupold, J.H.; Benner, A.; Gaiser, T.; Marx, A.; Allgayer, H. P53-Induced miR-30e-5p Inhibits Colorectal Cancer Invasion and Metastasis by Targeting ITGA6 and ITGB1. Int. J. Cancer 2017, 141, 1879–1890. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Chao, F.; Wang, S.; Song, Z.; Zhuo, Z.; Zhang, J.; Xu, G.; Chen, G. CTHRC1 Affects Malignant Tumor Cell Behavior and Is Regulated by miR-30e-5p in Human Prostate Cancer. Biochem. Biophys. Res. Commun. 2020, 525, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Kwon, A.Y.; Jeong, J.Y.; Park, H.; Hwang, S.; Kim, G.; Kang, H.; Heo, J.H.; Lee, H.J.; Kim, T.H.; An, H.J. miR-22-3p and miR-30e-5p Are Associated with Prognosis in Cervical Squamous Cell Carcinoma. Int. J. Mol. Sci. 2022, 23, 5623. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Cai, J.; Wang, L.; Jiang, L.; Huang, J.; Hu, R.; Ding, F. MicroRNA-30e-5p Suppresses Non-Small Cell Lung Cancer Tumorigenesis by Regulating USP22-Mediated Sirt1/JAK/STAT3 Signaling. Exp. Cell Res. 2018, 362, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Wang, J.; Huang, Z.; Zhang, H.; Wang, X.; Li, Q.; Feng, W. LncRNA OIP5-AS1 Promotes the Autophagy-Related Imatinib Resistance in Chronic Myeloid Leukemia Cells by Regulating miR-30e-5p/ATG12 Axis. Technol. Cancer Res. Treat. 2021, 20, 15330338211052150. [Google Scholar] [CrossRef] [PubMed]
- Qiao, F.; Zhang, K.; Gong, P.; Wang, L.; Hu, J.; Lu, S.; Fan, H. Decreased miR-30b-5p Expression by DNMT1 Methylation Regulation Involved in Gastric Cancer Metastasis. Mol. Biol. Rep. 2014, 41, 5693–5700. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Rao, S.; Cao, R.; Xiao, S.; Cui, X.; Ye, L. miR-30a-5p Suppresses Lung Squamous Cell Carcinoma via ATG5—Mediated Autophagy. Aging 2021, 13, 17462–17472. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Zhang, X.; Gao, P.; Cui, X.; Zhu, C.; Qin, X. LncRNA Loc339803 Acts as CeRNA of miR-30a-5p to Promote the Migration and Invasion of Hepatocellular Carcinoma Cells. J. Cancer 2021, 12, 1061–1072. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Bai, F.; Xu, Y.; Chen, Y.; Chen, L. Intensified Beclin-1 Mediated by Low Expression of Mir-30a-5p Promotes Chemoresistance in Human Small Cell Lung Cancer. Cell. Physiol. Biochem. 2017, 43, 1126–1139. [Google Scholar] [CrossRef]
- Zhu, J.; Zeng, Y.; Xu, C.; Qin, H.; Lei, Z.; Shen, D.; Liu, Z.; Huang, J.-A. Expression Profile Analysis of microRNAs and Downregulated miR-486-5p and miR-30a-5p in Non-Small Cell Lung Cancer. Oncol. Rep. 2015, 34, 1779–1786. [Google Scholar] [CrossRef] [PubMed]
- Tepebaşı, M.Y.; Öztürk, Ö. miR-21, miR-221, and miR-222 Upregulation in Lung Cancer Promotes Metastasis by Reducing Oxidative Stress and Apoptosis. Rev. Assoc. Med. Bras. 2023, 69, e20221688. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Hou, L.; Wei, J.; Du, Y.; Zhao, Y.; Deng, X.; Lin, X. Hsa-miR-217 Inhibits the Proliferation, Migration, and Invasion in Non-Small Cell Lung Cancer Cells Via Targeting SIRT1 and P53/KAI1 Signaling. Balk. Med. J. 2020, 37, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Pliakou, E.; Lampropoulou, D.I.; Dovrolis, N.; Chrysikos, D.; Filippou, D.; Papadimitriou, C.; Vezakis, A.; Aravantinos, G.; Gazouli, M. Circulating miRNA Expression Profiles and Machine Learning Models in Association with Response to Irinotecan-Based Treatment in Metastatic Colorectal Cancer. Int. J. Mol. Sci. 2022, 24, 46. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, X.; Maglic, D.; Dill, M.T.; Mojumdar, K.; Ng, P.K.-S.; Jeong, K.J.; Tsang, Y.H.; Moreno, D.; Bhavana, V.H.; et al. Comprehensive Molecular Characterization of the Hippo Signaling Pathway in Cancer. Cell Rep. 2018, 25, 1304–1317.e5. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Huang, H.-Y.; Lin, Z.; Ranieri, M.; Li, S.; Sahu, S.; Liu, Y.; Ban, Y.; Guidry, K.; Hu, H.; et al. Genome-Wide CRISPR Screens Identify Multiple Synthetic Lethal Targets That Enhance KRASG12C Inhibitor Efficacy. Cancer Res. 2023, 83, 4095–4111. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Yao, W.; Ying, H.; Hua, S.; Liewen, A.; Wang, Q.; Zhong, Y.; Wu, C.-J.; Sadanandam, A.; Hu, B.; et al. Yap1 Activation Enables Bypass of Oncogenic Kras Addiction in Pancreatic Cancer. Cell 2014, 158, 185–197. [Google Scholar] [CrossRef] [PubMed]


| miARN | KRAS SNV | Target Score miRDB * | Context+ Score ** |
|---|---|---|---|
| hsa-miR-885-5p | rs8720 | <50 | −0.155 |
| hsa-miR-877-5p | rs712 | 79 | −0.357 |
| hsa-miR-654-3p | rs190084851 | <50 | 0.069 |
| hsa-miR-647 | rs184195260 | <50 | −0.245 |
| hsa-miR-548c-5p | rs180766260 | 74 | 0.133 |
| hsa-miR-513b-5p | rs1141947 | 67 | −0.066 |
| hsa-miR-495-3p | rs184169974 | <50 | 0.060 |
| hsa-miR-377-3p | rs1141947 | <50 | −0.106 |
| hsa-miR-34b-3p | rs61764370 | <50 | 0.122 |
| hsa-miR-326 | rs150334904 | 72 | −0.283 |
| hsa-miR-30e-5p | rs61764373 | 80 | −0.246 |
| hsa-miR-30b-5p | rs61764373 | 80 | −0.246 |
| hsa-miR-30a-5p | rs61764373 | 80 | −0.225 |
| hsa-miR-222-3p | rs189426424 | <50 | −0.371 |
| hsa-miR-217 | rs192263744 | 92 | −0.175 |
| hsa-miR-181d-5p | rs9266 | 66 | −0.113 |
| miRNA | Cancer Type | Tumor Subtype or Cell Line | Design | Platform | Status | Reference |
|---|---|---|---|---|---|---|
| hsa-miR-885-5p | Pheochromocytoma | Tumors with germline mutations in RET | Tissue: Cancer vs. Normal | qRT-PCR | UP | [30] |
| hsa-miR-326 | Brain cancer | Medulloblastoma | Tissue: Cancer vs. Normal | qRT-PCR | DOWN | [31] |
| Esophageal cancer | Barrett’s carcinogenesis | Tissue: Cancer vs. Normal | qRT-PCR | DOWN | [32] | |
| hsa-miR-30b-5p | Colon cancer | Metastatic colorectal cancer | Tissue: Cancer vs. Normal | Northern Blot | DOWN | [33] |
| Brain cancer | Medulloblastoma | Tissue: Cancer vs. Normal | qRT-PCR | DOWN | [31] | |
| Breast cancer | N/A | Tissue: Cancer vs. Normal | qRT-PCR | UP | [34] | |
| hsa-miR-30a-5p | Lung cancer | Lung adenocarcinoma | Blood cells: cancer vs. Normal | qRT-PCR | DOWN | [35] |
| hsa-miR-222-3p | Prostate cancer | N/A | Tissue: Cancer vs. Normal | qRT-PCR | DOWN | [36] |
| Lymphoma | Multiple myeloma (TC4) | Tissue: Subtype1 vs. Subtype2 | qRT-PCR | UP | [37] | |
| Cholangiocarcinoma | N/A | Tissue: Cancer vs. Normal | qPCR | UP | [38] | |
| Lymphoma | Cutaneous T-cell lymphoma | Tissue: Cancer vs. Normal | qRT-PCR | UP | [39] | |
| Kidney cancer | N/A | Tissue: Cancer vs. Normal | qRT-PCR | UP | [40] | |
| hsa-miR-217 | PC | Solid pseudopapillary neoplasm of pancreas | Tissue: Cancer vs. Normal | Real-Time PCR | DOWN | [41] |
| Lung cancer | N/A | Blood | qRT-PCR | UP | [42] |
| miRNA | Type of Association | Log2 Average Expression (Deceased) | Log2 Average Expression (Survivors) | p Value |
|---|---|---|---|---|
| hsa-miR-30a-5p | Mortality | 14.22 | 14.02 | 0.012 |
| hsa-miR-30b-5p | Survival | 8.35 | 8.56 | 0.046 |
| hsa-miR-377-3p | Survival | 1.30 | 2.10 | 0.047 |
| hsa-miR-495-3p | Survival | 3.65 | 4.45 | 0.013 |
| hsa-miR-885-5p | Survival | 0.11 | 2.65 | 0.000412 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garibaldi-Ríos, A.F.; Figuera, L.E.; Zúñiga-González, G.M.; Gómez-Meda, B.C.; García-Verdín, P.M.; Carrillo-Dávila, I.A.; Gutiérrez-Hurtado, I.A.; Torres-Mendoza, B.M.; Gallegos-Arreola, M.P. In Silico Identification of Dysregulated miRNAs Targeting KRAS Gene in Pancreatic Cancer. Diseases 2024, 12, 152. https://doi.org/10.3390/diseases12070152
Garibaldi-Ríos AF, Figuera LE, Zúñiga-González GM, Gómez-Meda BC, García-Verdín PM, Carrillo-Dávila IA, Gutiérrez-Hurtado IA, Torres-Mendoza BM, Gallegos-Arreola MP. In Silico Identification of Dysregulated miRNAs Targeting KRAS Gene in Pancreatic Cancer. Diseases. 2024; 12(7):152. https://doi.org/10.3390/diseases12070152
Chicago/Turabian StyleGaribaldi-Ríos, Asbiel Felipe, Luis E. Figuera, Guillermo Moisés Zúñiga-González, Belinda Claudia Gómez-Meda, Patricia Montserrat García-Verdín, Irving Alejandro Carrillo-Dávila, Itzae Adonai Gutiérrez-Hurtado, Blanca Miriam Torres-Mendoza, and Martha Patricia Gallegos-Arreola. 2024. "In Silico Identification of Dysregulated miRNAs Targeting KRAS Gene in Pancreatic Cancer" Diseases 12, no. 7: 152. https://doi.org/10.3390/diseases12070152
APA StyleGaribaldi-Ríos, A. F., Figuera, L. E., Zúñiga-González, G. M., Gómez-Meda, B. C., García-Verdín, P. M., Carrillo-Dávila, I. A., Gutiérrez-Hurtado, I. A., Torres-Mendoza, B. M., & Gallegos-Arreola, M. P. (2024). In Silico Identification of Dysregulated miRNAs Targeting KRAS Gene in Pancreatic Cancer. Diseases, 12(7), 152. https://doi.org/10.3390/diseases12070152








