Regulation of Gene Expression by m6Am RNA Modification
Abstract
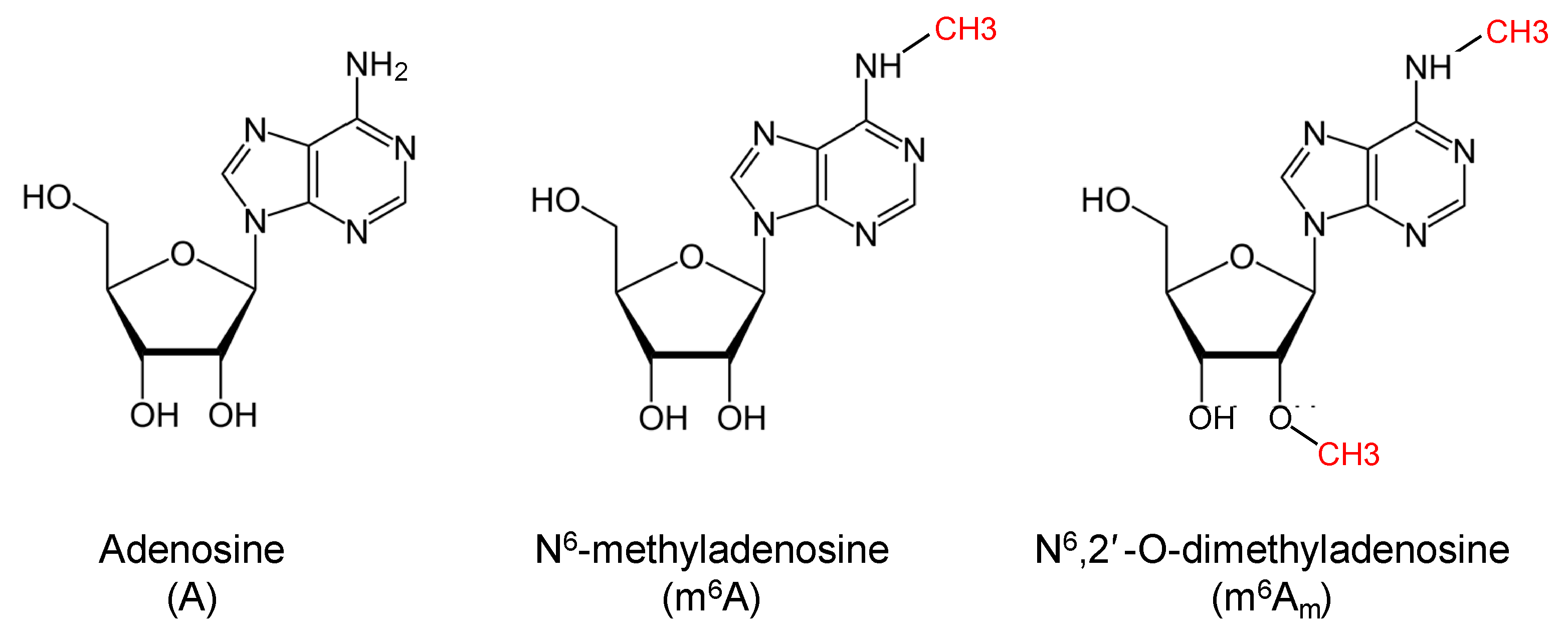
:1. Introduction
2. Methodologies for m6Am Detection
3. Regulators of m6Am Levels
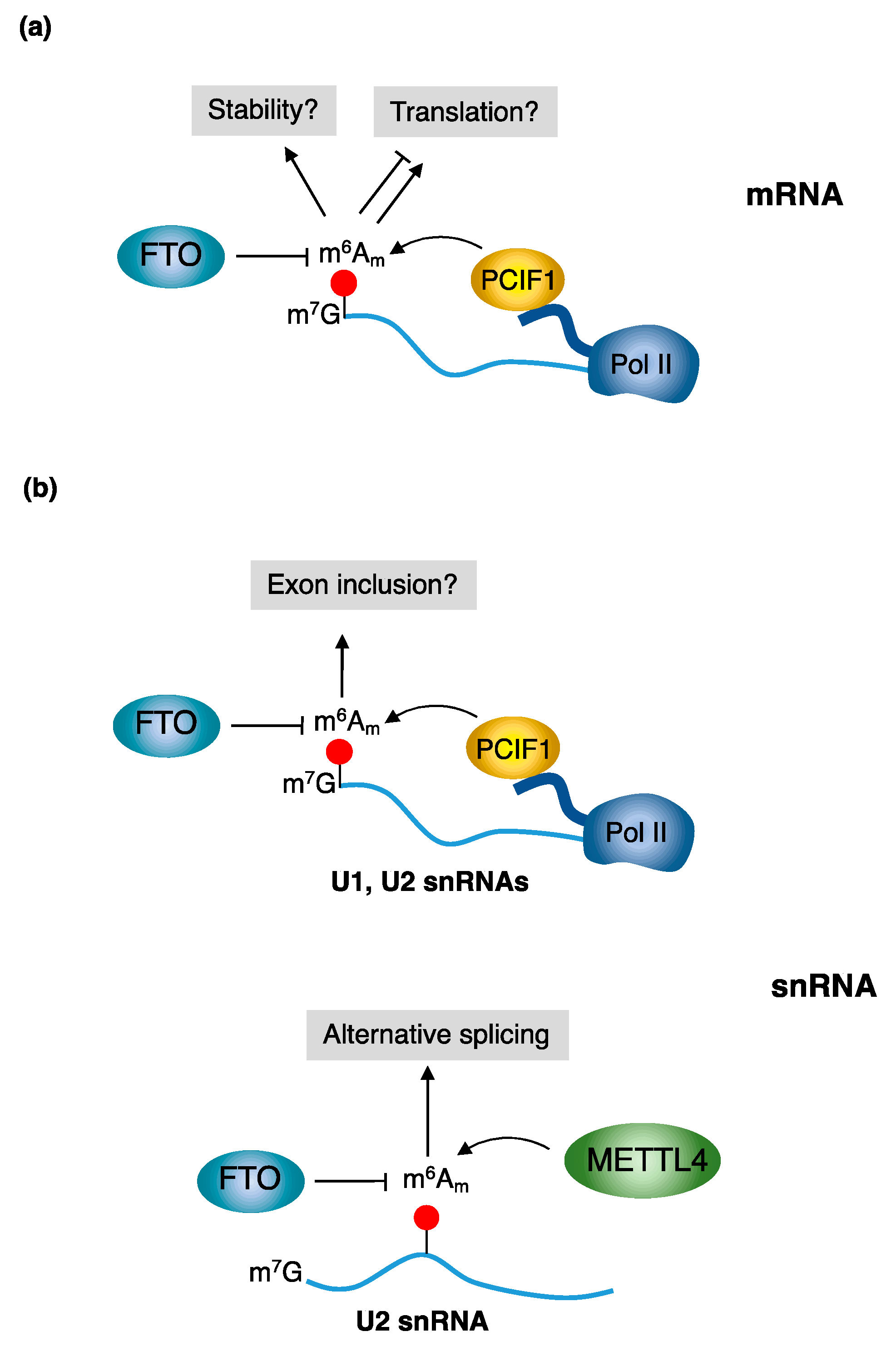
3.1. m6Am Writers: PCIF1 and METTL4
- PCIF1 (CAPAM)
- METTL4
3.2. m6Am Eraser: FTO
- FTO
4. Function of m6Am in mRNA Expression
4.1. The Role of m6Am in mRNA Stability
4.2. The Role of m6Am in mRNA Translation
4.3. The Role of m6Am in Splicing Regulation
5. The Role of m6Am in Cancer
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, A.; Robb, G.; Chan, S. mRNA capping: Biological functions and applications. Nucleic Acids Res. 2016, 44, 7511–7526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, C.; Gershowitz, A.; Moss, B. N6, O-2′-dimethyladenosine a novel methylated ribonucleoside next to the 5’ terminal of animal cell and virus mRNAs. Nature 1975, 257, 251–253. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Alvin Chew, B.L.; Lai, Y.; Dong, H.; Xu, L.; Balamkundu, S.; Cai, W.M.; Cui, L.; Liu, C.F.; Fu, X.Y.; et al. Quantifying the RNA cap epitranscriptome reveals novel caps in cellular and viral RNA. Nucleic Acids Res. 2019, 47, e130. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Li, K.; Cai, J.; Zhang, M.; Zhang, X.; Xiong, X.; Meng, H.; Xu, X.; Huang, Z.; Peng, J.; et al. Landscape and Regulation of m6A and m6Am Methylome across Human and Mouse Tissues. Mol. Cell 2020, 77, 426–440. [Google Scholar] [CrossRef]
- Wetzel, C.; Limbach, P.A. Mass spectrometry of modified RNAs: Recent developments. Analyst 2016, 141, 16–23. [Google Scholar] [CrossRef] [Green Version]
- Kruse, S.; Zhong, S.; Bodi, Z.; Button, J.; Alcocer, M.J.; Hayes, C.J.; Fray, R. A novel synthesis and detection method for cap-associated adenosine modifications in mouse mRNA. Sci. Rep. 2011, 1, 126. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, S.; Mumbach, M.R.; Jovanovic, M.; Wang, T.; Maciag, K.; Bushkin, G.G.; Mertins, P.; Ter-Ovanesyan, D.; Habib, N.; Cacchiarelli, D.; et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014, 8, 284–296. [Google Scholar] [CrossRef] [Green Version]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef]
- Sendinc, E.; Valle-Garcia, D.; Dhall, A.; Chen, H.; Henriques, T.; Navarrete-Perea, J.; Sheng, W.; Gygi, S.P.; Adelman, K.; Shi, Y. PCIF1 Catalyzes m6Am mRNA Methylation to Regulate Gene Expression. Mol. Cell 2019, 75, 620–630.e9. [Google Scholar] [CrossRef]
- Koh, C.W.Q.; Goh, Y.T.; Goh, W.S.S. Atlas of quantitative single-base-resolution N6-methyl-adenine methylomes. Nat. Commun. 2019, 10, 5636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.; Li, K.; Zhang, X.; Liu, J.; Zhang, M.; Meng, H.; Yi, C. m6Am-seq reveals the dynamic m6Am methylation in the human transcriptome. Nat. Commun. 2021, 12, 4778. [Google Scholar] [CrossRef] [PubMed]
- Muthmann, N.; Albers, M.; Rentmeister, A. CAPturAM, a Chemo-Enzymatic Strategy for Selective Enrichment and Detection of Physiological CAPAM-Targets. Angew. Chem. Int. Ed. Engl. 2022, 62, e202211957. [Google Scholar] [CrossRef] [PubMed]
- Keith, J.M.; Ensinger, M.J.; Moss, B. HeLa cell RNA (2′-O-methyladenosine-N6-)-methyltransferase specific for the capped 5′-end of messenger RNA. J. Biol. Chem. 1978, 253, 5033–5039. [Google Scholar] [CrossRef] [PubMed]
- Akichika, S.; Hirano, S.; Shichino, Y.; Suzuki, T.; Nishimasu, H.; Ishitani, R.; Sugita, A.; Hirose, Y.; Iwasaki, S.; Nureki, O.; et al. Cap-specific terminal N6-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 2019, 363, eaav0080-9. [Google Scholar] [CrossRef] [PubMed]
- Boulias, K.; Toczydłowska-Socha, D.; Hawley, B.R.; Liberman, N.; Takashima, K.; Zaccara, S.; Guez, T.; Vasseur, J.-J.; Debart, F.; Aravind, L.; et al. Identification of the m6Am Methyltransferase PCIF1 Reveals the Location and Functions of m6Am in the Transcriptome. Mol. Cell 2019, 75, 631–643. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, M.; Li, K.; Bai, D.; Yi, C. Cap-specific, terminal N6-methylation by a mammalian m6Am methyltransferase. Cell Res. 2019, 29, 80–82. [Google Scholar] [CrossRef] [Green Version]
- Pandey, R.R.; Delfino, E.; Homolka, D.; Roithova, A.; Chen, K.-M.; Li, L.; Franco, G.; Vågbø, C.B.; Taillebourg, E.; Fauvarque, M.-O.; et al. The Mammalian Cap-Specific m6Am RNA Methyltransferase PCIF1 Regulates Transcript Levels in Mouse Tissues. Cell Rep. 2020, 32, 108038. [Google Scholar] [CrossRef]
- Hirose, Y.; Iwamoto, Y.; Sakuraba, K.; Yunokuchi, I.; Harada, F.; Ohkuma, Y. Human phosphorylated CTD-interacting protein, PCIF1, negatively modulates gene expression by RNA polymerase II. Biochem. Biophys. Res. Commun. 2008, 369, 449–455. [Google Scholar] [CrossRef]
- Goh, Y.T.; Koh, C.W.Q.; Sim, D.Y.; Roca, X.; Goh, W.S.S. METTL4 catalyzes m6Am methylation in U2 snRNA to regulate pre-mRNA splicing. Nucleic Acids Res. 2020, 48, 9250–9261. [Google Scholar] [CrossRef]
- Chen, H.; Gu, L.; Orellana, E.A.; Wang, Y.; Guo, J.; Liu, Q.; Wang, L.; Shen, Z.; Wu, H.; Gregory, R.I.; et al. METTL4 is an snRNA m6Am methyltransferase that regulates RNA splicing. Cell Res. 2020, 30, 544–547. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Mo, J.; Chen, H.; Hu, Z.; Wang, B.; Wu, J.; Liang, Z.; Xie, W.; Du, K.; Peng, M.; et al. Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4. Nat. Commun. 2022, 13, 5636. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Wu, T.; Cui, X.; Zhu, P.; Tan, C.; Dou, X.; Hsu, K.W.; Lin, Y.T.; Peng, P.H.; Zhang, L.S.; et al. N6- Deoxyadenosine Methylation in Mammalian Mitochondrial DNA. Mol. Cell 2020, 78, 382–395.e8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Hou, Y.; Wang, Y.; Gao, T.; Ma, Z.; Yang, Y.; Zhang, P.; Yi, F.; Zhan, J.; Zhang, H.; et al. Regulation of Adipocyte Differentiation by METTL4, a 6mA Methylase. Sci. Rep. 2020, 10, 8285. [Google Scholar] [CrossRef]
- Gu, L.; Wang, L.; Chen, H.; Hong, J.; Shen, Z.; Dhall, A.; Lao, T.; Liu, C.; Wang, Z.; Xu, Y.; et al. CG14906 (mettl4) mediates m6A methylation of U2 snRNA in Drosophila. Cell Discov. 2020, 6, 44. [Google Scholar] [CrossRef]
- Wei, J.; Liu, F.; Lu, Z.; Fei, Q.; Ai, Y.; He, P.C.; Shi, H.; Cui, X.; Su, R.; Klungland, A.; et al. Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cell 2018, 71, 973–985. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Wei, L.-H.; Wang, Y.; Xiao, Y.; Liu, J.; Zhang, W.; Yan, N.; Amu, G.; Tang, X.; Zhang, L.; et al. Structural insights into FTO′s catalytic mechanism for the demethylation of multiple RNA substrates. Proc. Natl. Acad. Sci. 2019, 116, 2919–2924. [Google Scholar] [CrossRef] [Green Version]
- Hirayama, M.; Wei, F.-Y.; Chujo, T.; Oki, S.; Yakita, M.; Kobayashi, D.; Araki, N.; Takahashi, N.; Yoshida, R.; Nakayama, H.; et al. FTO Demethylates Cyclin D1 mRNA and Controls Cell-Cycle Progression. Cell Rep. 2020, 31, 107464. [Google Scholar] [CrossRef]
- Mauer, J.; Luo, X.; Blanjoie, A.; Jiao, X.; Grozhik, A.V.; Patil, D.P.; Linder, B.; Pickering, B.F.; Vasseur, J.J.; Chen, Q.; et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 2017, 541, 371–375. [Google Scholar] [CrossRef] [Green Version]
- Sikorski, P.J.; Warminski, M.; Kubacka, D.; Ratajczak, T.; Nowis, D.; Kowalska, J.; Jemielity, J. The identity and methylation status of the first transcribed nucleotide in eukaryotic mRNA 5′ cap modulates protein expression in living cells. Nucleic Acids Res. 2020, 48, 1607–1626. [Google Scholar] [CrossRef]
- Ben-Haim, M.S.; Pinto, Y.; Moshitch-Moshkovitz, S.; Hershkovitz, V.; Kol, N.; Diamant-Levi, T.; Beeri, M.S.; Amariglio, N.; Cohen, H.Y.; Rechavi, G. Dynamic regulation of N6,2′-O-dimethyladenosine (m6Am) in obesity. Nat. Commun. 2021, 12, 7185. [Google Scholar] [CrossRef] [PubMed]
- Drazkowska, K.; Tomecki, R.; Warminski, M.; Baran, N.; Cysewski, D.; Depaix, A.; Kasprzyk, R.; Kowalska, J.; Jemielity, J.; Sikorski, P.J. 2′- O -Methylation of the second transcribed nucleotide within the mRNA 5′ cap impacts the protein production level in a cell-specific manner and contributes to RNA immune evasion. Nucleic Acids Res. 2022, 50, 9051–9071. [Google Scholar] [CrossRef] [PubMed]
- Grudzien-Nogalska, E.; Kiledjian, M. New insights into decapping enzymes and selective mRNA decay. Wiley Interdisciplinary Reviews. RNA 2017, 8, e1379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidya, E.; Duchaine, T.F. Eukaryotic mRNA Decapping Activation. Front. Genet. 2022, 13, 832547. [Google Scholar] [CrossRef]
- Herzog, V.A.; Reichholf, B.; Neumann, T.; Rescheneder, P.; Bhat, P.; Burkard, T.R.; Wlotzka, W.; von Haeseler, A.; Zuber, J.; Ameres, S.L. Thiol-linked alkylation of RNA to assess expression dynamics. Nat. Methods 2017, 14, 1198–1204. [Google Scholar] [CrossRef] [Green Version]
- Mahat, D.B.; Kwak, H.; Booth, G.T.; Jonkers, I.H.; Danko, C.G.; Patel, R.K.; Waters, C.T.; Munson, K.; Core, L.J.; Lis, J.T. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq). Nat. Protoc. 2016, 11, 1455–1476. [Google Scholar] [CrossRef]
- Niedzwiecka, A.; Marcotrigiano, J.; Stepinski, J.; Jankowska-Anyszka, M.; Wyslouch-Cieszynska, A.; Dadlez, M.; Gingras, A.; Mak, P.; Darzynkiewicz, E.; Sonenberg, N.; et al. Biophysical studies of eIF4E cap-binding protein: Recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 2002, 319, 615–635. [Google Scholar] [CrossRef] [PubMed]
- Mauer, J.; Sindelar, M.; Despic, V.; Guez, T.; Hawley, B.R.; Vasseur, J.J.; Rentmeister, A.; Gross, S.S.; Pellizzoni, L.; Debart, F.; et al. FTO controls reversible m6Am RNA methylation during snRNA biogenesis. Nat. Che. Biol. 2019, 15, 340–347. [Google Scholar] [CrossRef]
- Hensel, J.; Duex, J.E.; Owens, C.; Dancik, G.M.; Edwards, M.G.; Frierson, H.F.; Theodorescu, D. Patient Mutation Directed shRNA Screen Uncovers Novel Bladder Tumor Growth Suppressors. Mol. Cancer Res. 2015, 13, 1306–1315. [Google Scholar] [CrossRef] [Green Version]
- Relier, S.; Ripoll, J.; Guillorit, H.; Amalric, A.; Achour, C.; Boissière, F.; Vialaret, J.; Attina, A.; Debart, F.; Choquet, A.; et al. FTO-mediated cytoplasmic m6Am demethylation adjusts stem-like properties in colorectal cancer cell. Nat. Commun. 2021, 12, 1716. [Google Scholar] [CrossRef]
- Zhuo, W.; Sun, M.; Wang, K.; Zhang, L.; Li, K.; Yi, D.; Li, M.; Sun, Q.; Ma, X.; Liu, W.; et al. m6Am methyltransferase PCIF1 is essential for aggressiveness of gastric cancer cells by inhibiting TM9SF1 mRNA translation. Cell Discov. 2022, 8, 48. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Peng, Z.; Luo, Y.; Wang, L.; Yu, P.; Deng, W.; An, Y.; Shi, T.; Ma, D. High-throughput functional screening for autophagy-related genes and identification of TM9SF1 as an autophagosome-inducing gene. Autophagy 2009, 5, 52–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, M.Z.; Zhang, Y.G.; Jin, W.L.; Wang, X.P. A Pan-Cancer Analysis of the Oncogenic and Immunogenic Role of m6Am Methyltransferase PCIF1. Front. Oncol. 2021, 11, 753393. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Zhou, J.; Hu, Z.; Zhang, S.; Wu, Y.; Musunuru, P.P.; Zhang, T.; Yang, L.; Luo, X.; Bai, J.; et al. Effects of the m6Am methyltransferase PCIF1 on cell proliferation and survival in gliomas. Biochim. Biophys. Acta Mol. Basis Dis. 2022, 1868, 166498. [Google Scholar] [CrossRef] [PubMed]
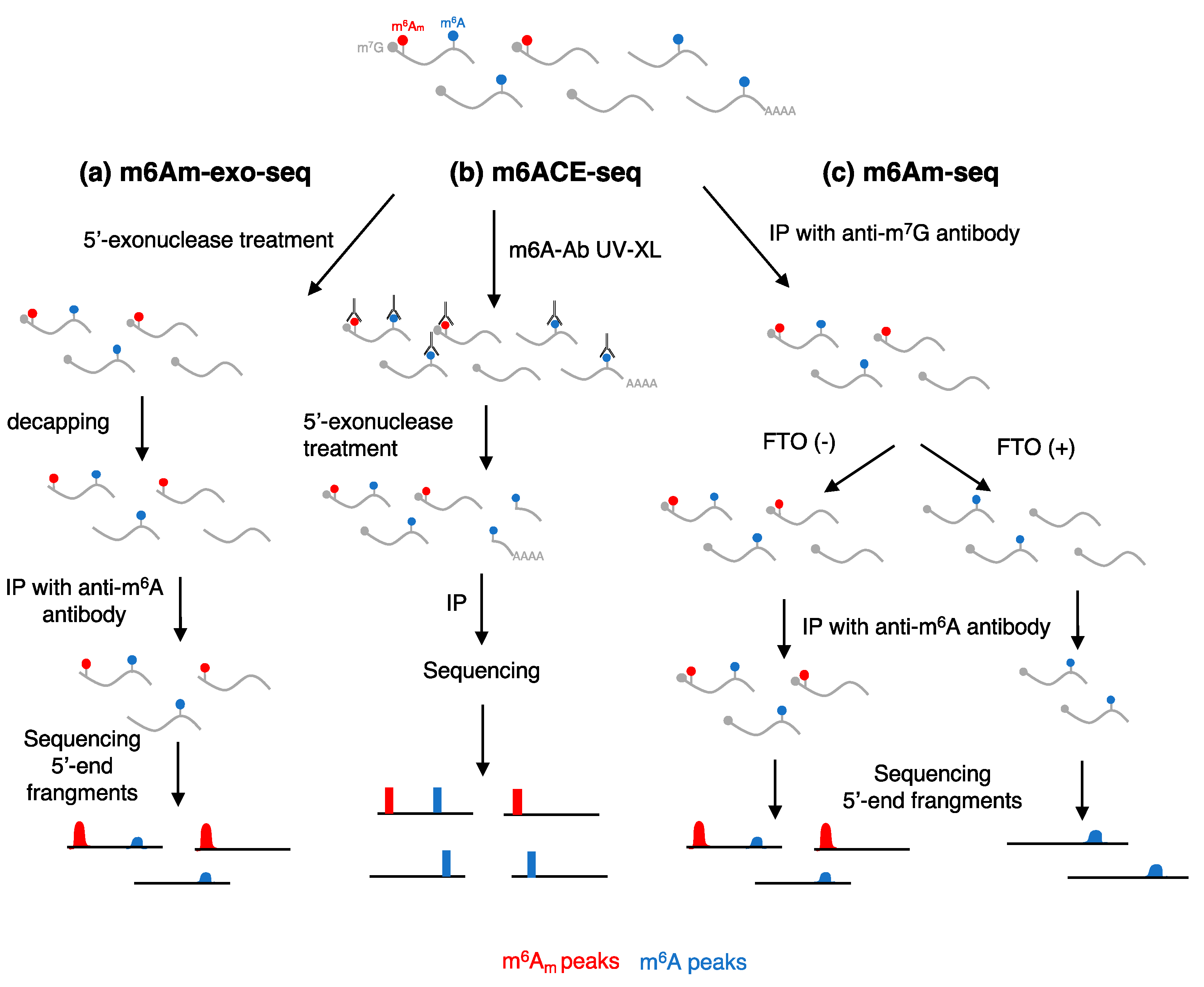
| Method | Principle | Advantages | Limitations |
|---|---|---|---|
| MeRIP-seq (m6A-seq) [8] | Utilizes anti-m6A antibody to enrich m6A- and m6Am-containing fragments | Easy to perform, kit available from different suppliers | Low resolution, required dedicated bioinformatic analysis for m6Am identification, cannot distinguish between cap-m6Am and m6A in 5′-RNA fragments, antibody cross-reactivity |
| miCLIP [9] | Utilizes CLIP with anti-m6A antibody to identify m6A- and m6Am in RNA fragments | Single-nucleotide resolution | Required dedicated bioinformatic analysis for m6Am identification, antibody cross-reactivity |
| m6am-exo-seq [10] | Utilizes 5′ -> 3′ digestion to degrade uncapped RNAs after fragmentation followed by m6A IP | Allows sequencing of cap-m6Am fragments | Cannot distinguish between cap-m6Am and m6A in 5′-RNA fragments, antibody cross-reactivity |
| m6ACE-seq [11] | Utilizes crosslinking of anti-m6A antibody, followed by 5′-> 3′ digestion and m6A-IP to identify m6A- and m6Am in RNA fragments | Single-nucleotide resolution, allows mapping of both m6A and cap-m6Am | Required dedicated bioinformatic analysis, antibody cross-reactivity |
| m6Am-seq [12] | Utilizes anti-m7G-antibody to purify 5′-RNA fragments, followed by digestion with recombinant FTO and m6A IP | Allows mapping of both m6A and cap-m6Am | Cannot distinguish between cap-m6Am and m6A in 5′-RNA fragments, requires recombinant FTO protein, FTO activity is not specific for m6Am and is influenced by sequence and structure, antibody cross-reactivity |
| CAPturAM [13] | Cap-m6Am are enzymatically propargylated by PCIF1, using synthetic AdoMet analog, selectively biotinylated and enriched with magnetic streptavidin-beads | Antibody-independent method | Never applied to transcriptome studies |
| Enzyme/ Experimental Procedure | Molecular Effect of m6Am | Model System |
|---|---|---|
| FTO/KO [29] | Enhanced mRNA stability | HEK293T |
| FTO/OE [29] | Reduced mRNA stability | HEK293T |
| PCIF/KO [10,15,17] | No effect on mRNA stability [10,15,17]/increased translation [10]/reduced translation [15] | HEK293T, MEL624 |
| PCIF1/KO [16] | Reduced stability of low expressed mRNAs/no effect on translation | HEK293T |
| PCIF1/KO [18] | Reduced stability of pseudogenes in testis/no effect on translation | Mouse |
| Transfection of in vitro transcribed RNAs [30] | Positive correlation with mRNA stability | JAWS II |
| Transfection of in vitro transcribed RNAs [30] | No effect on mRNA stability | HeLa, 3T3-L1 |
| Sequencing [31] | Enhanced mRNA stability in fat liver | Mouse |
| Transfection of in vitro transcribed RNAs [32] | Positive effect of m6Am on translation | A549, JAWS II |
| Transfection of in vitro transcribed RNAs [32] | Negative effect of m6Am on translation | 3T3-L1 |
| Transfection of in vitro transcribed RNAs [32] | No effect on translation | THP1 |
| Transfection of in vitro transcribed RNAs [10] | Negative effect of m6Am on translation | HEK293T |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cesaro, B.; Tarullo, M.; Fatica, A. Regulation of Gene Expression by m6Am RNA Modification. Int. J. Mol. Sci. 2023, 24, 2277. https://doi.org/10.3390/ijms24032277
Cesaro B, Tarullo M, Fatica A. Regulation of Gene Expression by m6Am RNA Modification. International Journal of Molecular Sciences. 2023; 24(3):2277. https://doi.org/10.3390/ijms24032277
Chicago/Turabian StyleCesaro, Bianca, Marco Tarullo, and Alessandro Fatica. 2023. "Regulation of Gene Expression by m6Am RNA Modification" International Journal of Molecular Sciences 24, no. 3: 2277. https://doi.org/10.3390/ijms24032277
APA StyleCesaro, B., Tarullo, M., & Fatica, A. (2023). Regulation of Gene Expression by m6Am RNA Modification. International Journal of Molecular Sciences, 24(3), 2277. https://doi.org/10.3390/ijms24032277







