A New Practice to Monitor the Fabrication Process of Fab-Targeting Ligands from Bevacizumab by LC-MS: Preparation and Analytical Characterization
Abstract
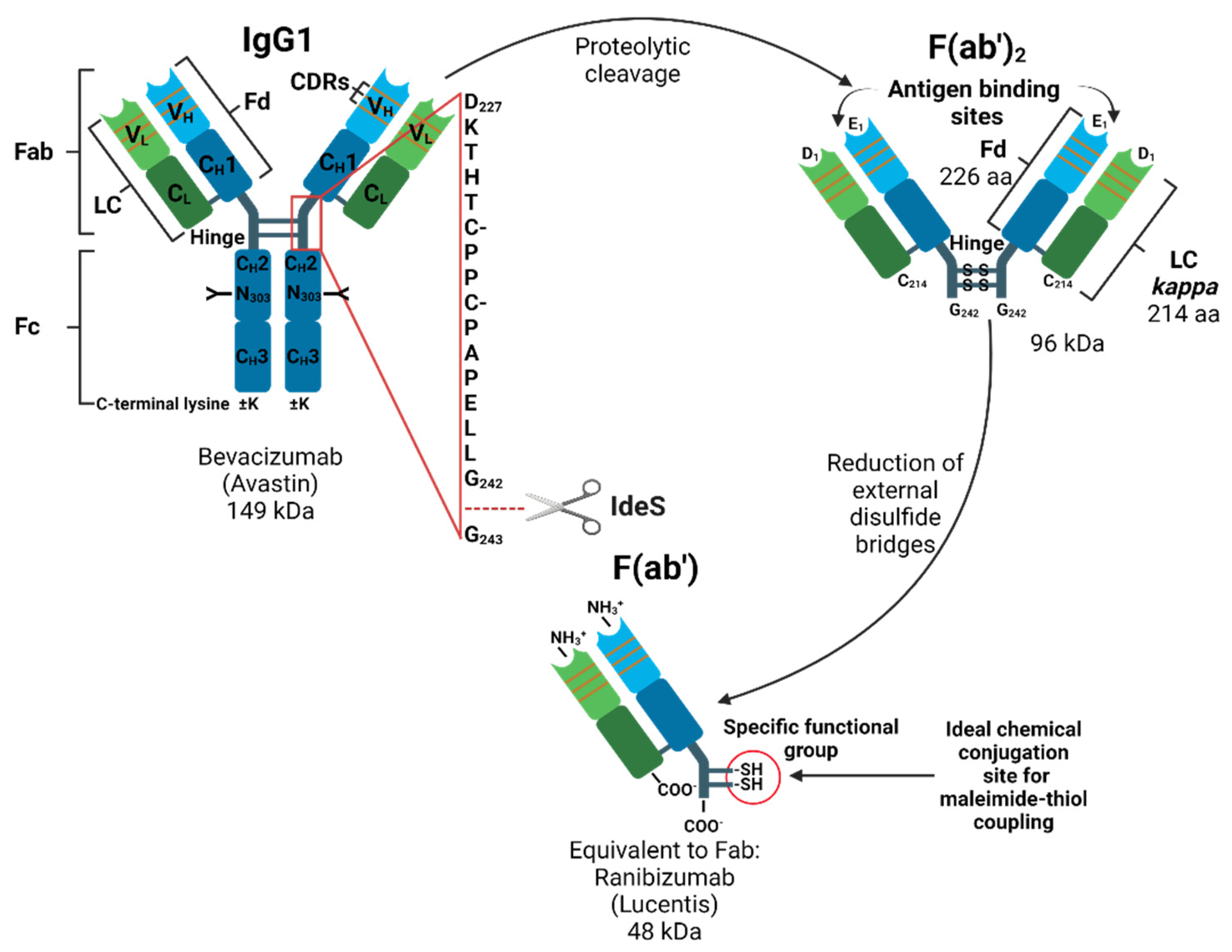
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Enzymatic Digests and Sample Preparation
2.3. Chromatographic System
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ford, C.H.; Newman, C.E.; Johnson, J.R.; Woodhouse, C.S.; Reeder, T.A.; Rowland, G.F.; Simmonds, R.G. Localisation and toxicity study of a vindesine-anti-CEA conjugate in patients with advanced cancer. Br. J. Cancer 1983, 47, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Tsuchikama, K.; An, Z. Antibody-drug conjugates: Recent advances in conjugation and linker chemistries. Protein Cell 2018, 9, 33–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manjappa, A.S.; Chaudhari, K.R.; Venkataraju, M.P.; Dantuluri, P.; Nanda, B.; Sidda, C.; Sawant, K.K.; Murthy, R.S. Antibody derivatization and conjugation strategies: Application in preparation of stealth immunoliposome to target chemotherapeutics to tumor. J. Control. Release 2011, 150, 2–22. [Google Scholar] [CrossRef]
- Birrer, M.J.; Moore, K.N.; Betella, I.; Bates, R.C. Antibody-drug conjugate-based therapeutics: State of the science. J. Natl. Cancer Inst. 2019, 111, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Senter, P.D.; Sievers, E.L. The discovery and development of brentuximab vedotin for use in relapsed Hodgkin lymphoma and systemic anaplastic large cell lymphoma. Nat. Biotechnol. 2012, 30, 631–637. [Google Scholar] [CrossRef]
- Kedmi, R.; Veiga, N.; Ramishetti, S.; Goldsmith, M.; Rosenblum, D.; Dammes, N.; Hazan-Halevy, I.; Nahary, L.; Leviatan-Ben-Arye, S.; Harlev, M.; et al. A modular platform for targeted RNAi therapeutics. Nat. Nanotechnol. 2018, 13, 214–219. [Google Scholar] [CrossRef]
- Lippold, S.; Nicolardi, S.; Wuhrer, M.; Falck, D. Proteoform-resolved FcRIIIa binding assay for Fab glycosylated monoclonal antibodies achieved by affinity chromatography mass spectrometry of Fc moieties. Front. Chem. 2019, 7, 698. [Google Scholar] [CrossRef]
- Presta, L.G.; Chen, H.; O’Connor, S.J.; Chisholm, V.; Meng, Y.G.; Krummen, L.; Winkler, M.; Ferrara, N. Humanization of an anti-vascular endothelial growth factor monoclonal antibody for the therapy of solid tumors and other disorders. Cancer Res. 1997, 57, 4593–4599. [Google Scholar]
- Seo, N.; Polozova, A.; Zhang, M.; Yates, Z.; Cao, S.; Li, H.; Kuhns, S.; Maher, G.; McBride, H.J.; Liu, J. Analytical and functional similarity of Amgen biosimilar ABP 215 to bevacizumab. MAbs 2018, 10, 678–691. [Google Scholar] [CrossRef] [Green Version]
- Kabat, E.A.; Wu, T.T.; Perry, H.; Gottesman, K.; Foeller, C. Sequences of Proteins of Immunological Interest, 5th ed.; NIH Publication No. 91-3242; NIH: Bethesda, MD, USA, 1991. [Google Scholar]
- Steinbrook, R. The price of sight--ranibizumab, bevacizumab, and the treatment of macular degeneration. N. Engl. J. Med. 2006, 355, 1409–1412. [Google Scholar] [CrossRef]
- Plyukhova, A.A.; Budzinskaya, M.V.; Starostin, K.M.; Rejdak, R.; Bucolo, C.; Reibaldi, M.; Toro, M.D. Comparative safety of bevacizumab, ranibizumab, and aflibercept for treatment of neovascular age-related macular degeneration (AMD): A systematic review and network meta-analysis of direct comparative studies. J. Clin. Med. 2020, 9, 1522. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhang, J.; Zhang, Y.; Ma, G.; Su, Z. Specific conjugation of the hinge region for homogeneous preparation of antibody fragment-drug conjugate: A case study for doxorubicin-PEG-anti-CD20 Fab’ synthesis. Bioconjug. Chem. 2016, 27, 238–246. [Google Scholar] [CrossRef] [PubMed]
- D’Atri, V.; Fekete, S.; Stoll, D.; Lauber, M.; Beck, A.; Guillarme, D. Characterization of an antibody-drug conjugate by hydrophilic interaction chromatography coupled to mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2018, 1080, 37–41. [Google Scholar] [CrossRef] [PubMed]
- D’Atri, V.; Goyon, A.; Bobaly, B.; Beck, A.; Fekete, S.; Guillarme, D. Protocols for the analytical characterization of therapeutic monoclonal antibodies. III—Denaturing chromatographic techniques hyphenated to mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2018, 1096, 95–106. [Google Scholar] [CrossRef]
- Goyon, A.; Excoffier, M.; Janin-Bussat, M.C.; Bobaly, B.; Fekete, S.; Guillarme, D.; Beck, A. Determination of isoelectric points and relative charge variants of 23 therapeutic monoclonal antibodies. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2017, 1065–1066, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Goyon, A.; D’Atri, V.; Colas, O.; Fekete, S.; Beck, A.; Guillarme, D. Characterization of 30 therapeutic antibodies and related products by size exclusion chromatography: Feasibility assessment for future mass spectrometry hyphenation. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2017, 1065–1066, 35–43. [Google Scholar] [CrossRef]
- Waibl, F.; Fernandez-Quintero, M.L.; Kamenik, A.S.; Kraml, J.; Hofer, F.; Kettenberger, H.; Georges, G.; Liedl, K.R. Conformational ensembles of antibodies determine their hydrophobicity. Biophys. J. 2021, 120, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Alenazi, N.A.; Hussein, M.A.; Alamry, K.A.; Asiri, A.M. Modified polyether-sulfone membrane: A mini review. Des. Monomers Polym. 2017, 20, 532–546. [Google Scholar] [CrossRef] [Green Version]
- Rodrigo, G.; Gruvegard, M.; Van Alstine, J.M. Antibody fragments and their purification by protein L affinity chromatography. Antibodies 2015, 4, 259–277. [Google Scholar] [CrossRef] [Green Version]
- Malpiedi, L.P.; Diaz, C.A.; Nerli, B.B.; Pessoa, A. Single-chain antibody fragments: Purification methodologies. Process Biochem. 2013, 48, 1242–1251. [Google Scholar] [CrossRef]
- Cruz, A.R.; den Boer, M.A.; Strasser, J.; Zwarthoff, S.A.; Beurskens, F.J.; de Haas, C.J.C.; Aerts, P.C.; Wang, G.B.; de Jong, R.N.; Bagnoli, F.; et al. Staphylococcal protein A inhibits complement activation by interfering with IgG hexamer formation. Proc. Natl. Acad. Sci. USA 2021, 118, e2016772118. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L.; Ultsch, M.H.; de Vos, A.M.; Wells, J.A. Convergent solutions to binding at a protein-protein interface. Science 2000, 287, 1279–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Branco, R.J.; Dias, A.M.; Roque, A.C. Understanding the molecular recognition between antibody fragments and protein A biomimetic ligand. J. Chromatogr. A 2012, 1244, 106–115. [Google Scholar] [CrossRef] [PubMed]
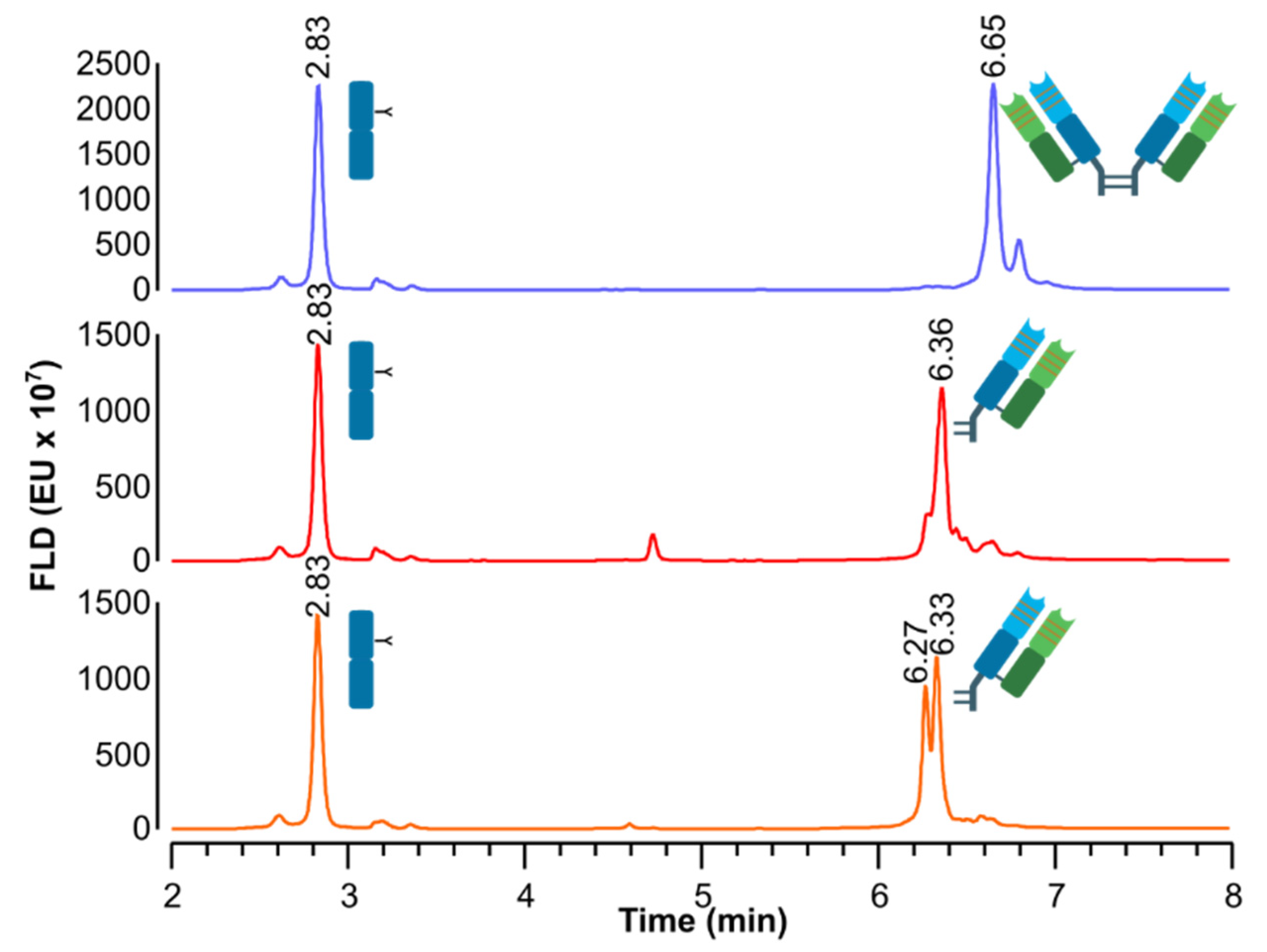


| Samples | tr (Min) | Assignment | Theoretical Masses (Da) 1 | Experimental Masses (Da) | Δm (Da) | ppm (Error) |
|---|---|---|---|---|---|---|
| IdeS-digested bevacizumab | 2.88 | Fc/2–K G0F | 25,232.28 | 25,230.90 | 1.38 | 55 |
| Fc/2–K G1F | 25,394.42 | 25,393.78 | 0.64 | 25 | ||
| 6.70 | F(ab’)2 | 98,770.35 | 98,770.67 | −0.32 | −3 | |
| IdeS-digested bevacizumab, reduced by DTT | 2.89 | Fc/2–K G0F | 25,232.28 | 25,231.83 | 0.45 | 18 |
| Fc/2–K G1F | 25,394.42 | 25,393.37 | 1.05 | 41 | ||
| 6.40 | Fab (Cys. linked) | 49,385.18 | 49,384.89 | 0.29 | 6 | |
| IdeS-digested bevacizumab, reduced by β-MEA | 2.87 | Fc/2–K G0F | 25,232.28 | 25,231.12 | 1.16 | 46 |
| Fc/2–K G1F | 25,394.42 | 25,393.26 | 1.16 | 46 | ||
| 6.32 | Fab (Cys. linked) | 49,385.18 | 49,383.43 | 1.75 | 35 | |
| 6.37 | Fab (Cys. linked) | 49,385.18 | 49,382.80 | 2.38 | 48 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marquet, F.; D’Atri, V.; Guillarme, D.; Borchard, G. A New Practice to Monitor the Fabrication Process of Fab-Targeting Ligands from Bevacizumab by LC-MS: Preparation and Analytical Characterization. Sci. Pharm. 2022, 90, 5. https://doi.org/10.3390/scipharm90010005
Marquet F, D’Atri V, Guillarme D, Borchard G. A New Practice to Monitor the Fabrication Process of Fab-Targeting Ligands from Bevacizumab by LC-MS: Preparation and Analytical Characterization. Scientia Pharmaceutica. 2022; 90(1):5. https://doi.org/10.3390/scipharm90010005
Chicago/Turabian StyleMarquet, Franck, Valentina D’Atri, Davy Guillarme, and Gerrit Borchard. 2022. "A New Practice to Monitor the Fabrication Process of Fab-Targeting Ligands from Bevacizumab by LC-MS: Preparation and Analytical Characterization" Scientia Pharmaceutica 90, no. 1: 5. https://doi.org/10.3390/scipharm90010005
APA StyleMarquet, F., D’Atri, V., Guillarme, D., & Borchard, G. (2022). A New Practice to Monitor the Fabrication Process of Fab-Targeting Ligands from Bevacizumab by LC-MS: Preparation and Analytical Characterization. Scientia Pharmaceutica, 90(1), 5. https://doi.org/10.3390/scipharm90010005







