A Unifying Framework for Analysis of Spatial-Temporal Event Sequence Similarity and Its Applications
Abstract
:1. Introduction
2. Materials and Methods
2.1. Eventization and Spatiotemporal Event Sequences (STES)
2.2. Matrix Representation of STES
2.3. Development of Similarity Measures for Spatiotemporal Event Sequences

2.3.1. Similarity Measures between Event Sequences without Considering Event Magnitude
2.3.2. Similarity Measures between Event Sequences Considering Event Magnitude
- —global similarity between event sequences ,
- —the event levels of two corresponding co-occurring events in and at timestamp , inherited from original measurements,
- —the relative event levels of two corresponding co-occurring events in and at timestamp , respectively:
- —the total number of co-occurring timestamps,
- —absolute value of difference between relative event levels of two corresponding co-occurring events in and at time stamp ,
- —cardinality of the union of two event sequences ,
- n—the number of ordinal attribute-based event levels.
3. Results and Discussion
3.1. Implementation Examples
3.2. Performance Evaluation
3.2.1. Execution Speed for a Binary Event Matrix
3.2.2. Accuracy Evaluation with Synthetic Datasets Using 1-NN Classifier
3.3. Application Example
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Software Availability
References
- Bollobas, B.; Das, G.; Gunopulos, D.; Mannila, H. Time-series similarity problems and well-separated geometric sets. In Proceedings of the Thirteenth Annual Symposium on Computational Geometry, Nice, France, 4–6 June 1997; pp. 454–456. [Google Scholar]
- Fu, T.-C. A review on time series data mining. Eng. Appl. Artif. Intell. 2011, 24, 164–181. [Google Scholar] [CrossRef]
- Du, F.; Shneiderman, B.; Plaisant, C.; Malik, S.; Perer, A. Coping with volume and variety in temporal event sequences: Strategies for sharpening analytic focus. IEEE Trans. Vis. Comput. Graph. 2016, 23, 1636–1649. [Google Scholar] [CrossRef] [PubMed]
- Shurkhovetskyy, G.; Andrienko, N.; Andrienko, G.; Fuchs, G. Data abstraction for visualizing large time series. Comput. Graph. Forum 2018, 37, 125–144. [Google Scholar] [CrossRef]
- Yeh, C.-C.M.; Zhu, Y.; Ulanova, L.; Begum, N.; Ding, Y.; Dau, H.A.; Zimmerman, Z.; Silva, D.F.; Mueen, A.; Keogh, E. Time series joins, motifs, discords and shapelets: A unifying view that exploits the matrix profile. Data Min. Knowl. Discov. 2018, 32, 83–123. [Google Scholar] [CrossRef]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [Green Version]
- Maurya, M.R.; Rengaswamy, R.; Venkatasubramanian, V. Fault diagnosis using dynamic trend analysis: A review and recent developments. Eng. Appl. Artif. Intell. 2007, 20, 133–146. [Google Scholar] [CrossRef]
- Tao, C.; Wongsuphasawat, K.; Clark, K.; Plaisant, C.; Shneiderman, B.; Chute, C.G. Towards event sequence representation, reasoning and visualization for EHR data. In Proceedings of the 2nd ACM SIGHIT International Health Informatics Symposium, Miami, FL, USA, 28–30 January 2012; ACM: New York, NY, USA, 2012; pp. 801–806. [Google Scholar]
- Stehle, S.; Peuquet, D.J. Analyzing spatio-temporal patterns and their evolution via sequence alignment. Spat. Cogn. Comput. 2015, 15, 68–85. [Google Scholar] [CrossRef]
- Prinzie, A.; Van den Poel, D. Modeling complex longitudinal consumer behavior with Dynamic Bayesian networks: An Acquisition Pattern Analysis application. J. Intell. Inf. Syst. 2011, 36, 283–304. [Google Scholar] [CrossRef]
- Yang, J.; McAuley, J.; Leskovec, J.; LePendu, P.; Shah, N. Finding progression stages in time-evolving event sequences. In Proceedings of the 23rd International Conference on World Wide Web, Seoul, Korea, 7–11 April 2014; ACM: New York, NY, USA, 2014; pp. 783–794. [Google Scholar]
- Hamming, R.W. Error detecting and error correcting codes. Bell Syst. Tech. J. 1950, 29, 147–160. [Google Scholar] [CrossRef]
- Levenshtein, V.I. Binary codes capable of correcting deletions, insertions, and reversals. Sov. Phys. Dokl. 1966, 10, 707–710. [Google Scholar]
- Jacobs, B.E.; Walczak, C.A. A generalized query-by-example data manipulation language based on database logic. IEEE Trans. Softw. Eng. 1983, SE-9, 40–57. [Google Scholar] [CrossRef]
- André-Jönsson, H.; Badal, D.Z. Using signature files for querying time-series data. In Proceedings of the European Symposium on Principles of Data Mining and Knowledge Discovery, Trondheim, Norway, 24–27 June 1997; Springer: Berlin/Heidelberg, Germany, 1997; pp. 211–220. [Google Scholar]
- Mannila, H.; Moen, P. Similarity between event types in sequences. In Proceedings of the International Conference on Data Warehousing and Knowledge Discovery, Florence, Italy, 30 August–1 September 1999; Springer: Berlin/Heidelberg, Germany, 1999; pp. 271–280. [Google Scholar]
- Mannila, H.; Ronkainen, P. Similarity of event sequences. In Proceedings of the TIME’97: 4th International Workshop on Temporal Representation and Reasoning, Dayton Beach, FL, USA, 10–11 May 1997; IEEE: New York, NY, USA, 1997; pp. 136–139. [Google Scholar]
- Wongsuphasawat, K.; Plaisant, C.; Taieb-Maimon, M.; Shneiderman, B. Querying event sequences by exact match or similarity search: Design and empirical evaluation. Interact. Comput. 2012, 24, 55–68. [Google Scholar] [CrossRef] [Green Version]
- Chung, N.C.; Miasojedow, B.; Startek, M.; Gambin, A. Jaccard/Tanimoto similarity test and estimation methods for biological presence-absence data. BMC Bioinform. 2019, 20, 644. [Google Scholar] [CrossRef] [PubMed]
- Vorontsov, I.E.; Kulakovskiy, I.V.; Makeev, V.J. Jaccard index based similarity measure to compare transcription factor binding site models. Algorithms Mol. Biol. 2013, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Luu, V.-T.; Forestier, G.; Weber, J.; Bourgeois, P.; Djelil, F.; Muller, P.-A. A review of alignment based similarity measures for web usage mining. Artif. Intell. Rev. 2020, 53, 1529–1551. [Google Scholar] [CrossRef]
- Obweger, H.; Suntinger, M.; Schiefer, J.; Raidl, G. Similarity searching in sequences of complex events. In Proceedings of the 2010 Fourth International Conference on Research Challenges in Information Science (RCIS), Nice, France, 19–21 May 2010; IEEE: New York, NY, USA, 2010; pp. 631–640. [Google Scholar]
- Andrienko, G.; Andrienko, N.; Mladenov, M.; Mock, M.; Poelitz, C. Extracting events from spatial time series. In Proceedings of the 2010 14th International Conference Information Visualisation, London, UK, 26–29 July 2010; pp. 48–53. [Google Scholar]
- Mirbagheri, S.M.; Hamilton, H.J. Similarity Matching of Temporal Event-Interval Sequences. In Proceedings of the Canadian Conference on Artificial Intelligence, Online, 13–15 May 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 420–425. [Google Scholar]
- Jassby, A.D.; Powell, T.M. Detecting changes in ecological time series. Ecology 1990, 71, 2044–2052. [Google Scholar] [CrossRef]
- Rude, A.; Beard, K. High-Level Event Detection in Spatially Distributed Time Series; Springer: Berlin/Heidelberg, Germany, 2012; pp. 160–172. [Google Scholar]
- Abadi, D.; Madden, S.; Lindner, W. Sensor Network Integration with Streaming Database Systems. In Data Stream Management; Springer: Berlin/Heidelberg, Germany, 2016; pp. 409–428. [Google Scholar]
- Hogenboom, F.; Frasincar, F.; Kaymak, U.; De Jong, F.; Caron, E. A survey of event extraction methods from text for decision support systems. Decis. Support Syst. 2016, 85, 12–22. [Google Scholar] [CrossRef]
- Wang, T.-Y.; Yang, M.-H.; Wu, J.-Y. Distributed Detection of Dynamic Event Regions in Sensor Networks With a Gibbs Field Distribution and Gaussian Corrupted Measurements. IEEE Trans. Commun. 2016, 64, 3932–3945. [Google Scholar] [CrossRef]
- Shahar, Y. A framework for knowledge-based temporal abstraction. Artif. Intell. 1997, 90, 79–133. [Google Scholar] [CrossRef] [Green Version]
- Yin, J.; Hu, D.H.; Yang, Q. Spatio-Temporal Event Detection Using Dynamic Conditional Random Fields. In Proceedings of the 21st International Jont Conference on Artifical intelligence, Pasadena, CA, USA, 11–17 July 2009; pp. 1321–1327. [Google Scholar]
- Guralnik, V.; Srivastava, J. Event detection from time series data. In Proceedings of the Fifth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Diego, CA, USA, 15–18 August 1999; ACM: New York, NY, USA, 1999; pp. 33–42. [Google Scholar]
- Jaccard, P. Étude comparative de la distribution florale dans une portion des Alpes et des Jura. Bull. Soc. Vaud. Sci. Nat. 1901, 37, 547–579. [Google Scholar]
- Bershad, B.; Draves, R.P.; Forin, A. Using microbenchmarks to evaluate system performance. In Proceedings of the Third Workshop on Workstation Operating Systems, Key Biscayne, FL, USA, 23–24 April 1992; IEEE: New York, NY, USA, 1992; pp. 148–153. [Google Scholar]
- Peterson, M.R.; Doom, T.E.; Raymer, M.L. Ga-facilitated knn classifier optimization with varying similarity measures. In Proceedings of the 2005 IEEE Congress on Evolutionary Computation, Edinburgh, UK, 2–5 September 2005; IEEE: New York, NY, USA, 2005; pp. 2514–2521. [Google Scholar]
- Prasath, V.; Alfeilat, H.A.A.; Lasassmeh, O.; Hassanat, A. Distance and similarity measures effect on the performance of K-nearest neighbor classifier-a review. arXiv 2017, arXiv:1708.04321. [Google Scholar]
- Wang, X.; Mueen, A.; Ding, H.; Trajcevski, G.; Scheuermann, P.; Keogh, E. Experimental comparison of representation methods and distance measures for time series data. Data Min. Knowl. Discov. 2013, 26, 275–309. [Google Scholar] [CrossRef] [Green Version]
- Ros, F.; Guillaume, S. A hierarchical clustering algorithm and an improvement of the single linkage criterion to deal with noise. Expert Syst. Appl. 2019, 128, 96–108. [Google Scholar] [CrossRef]






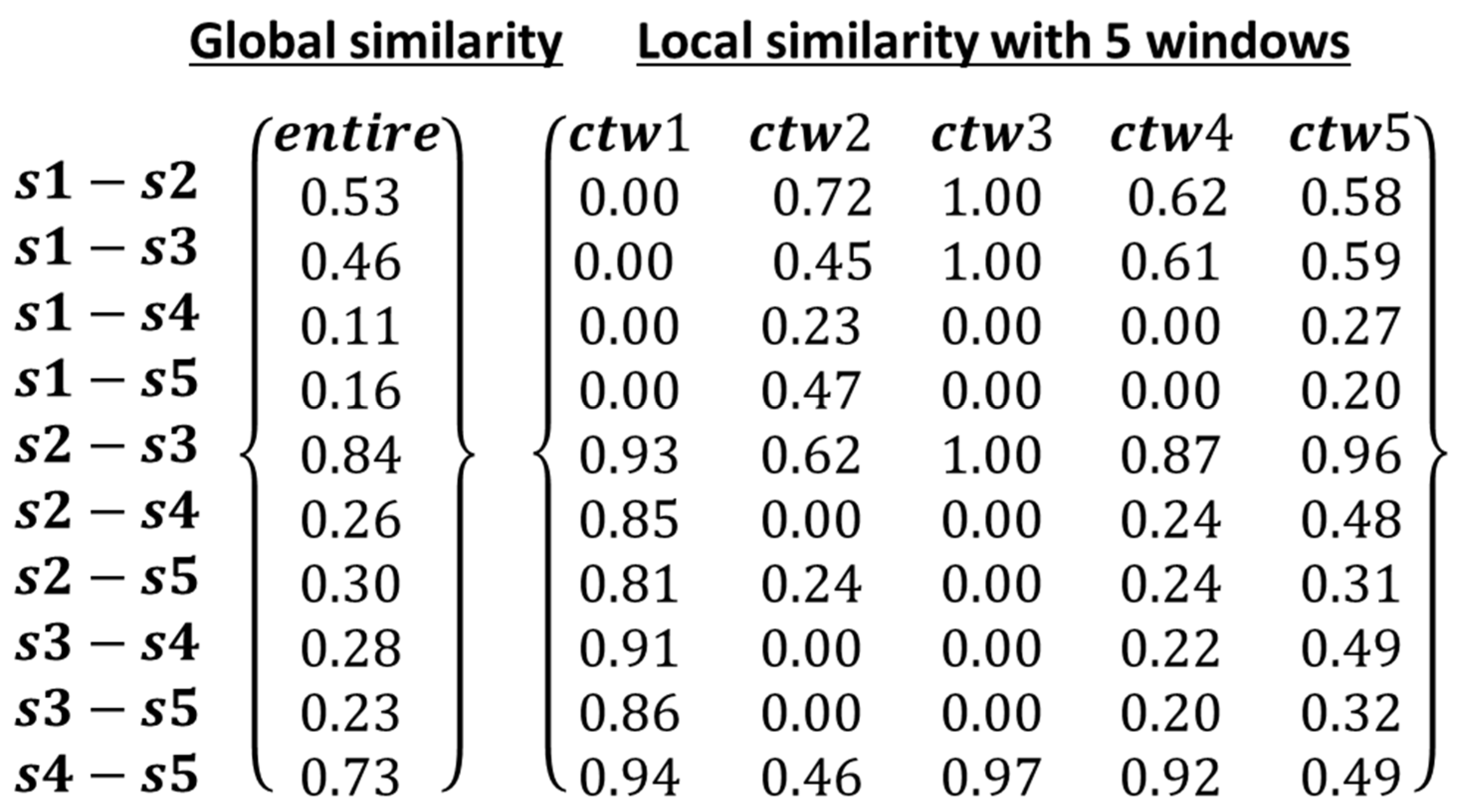



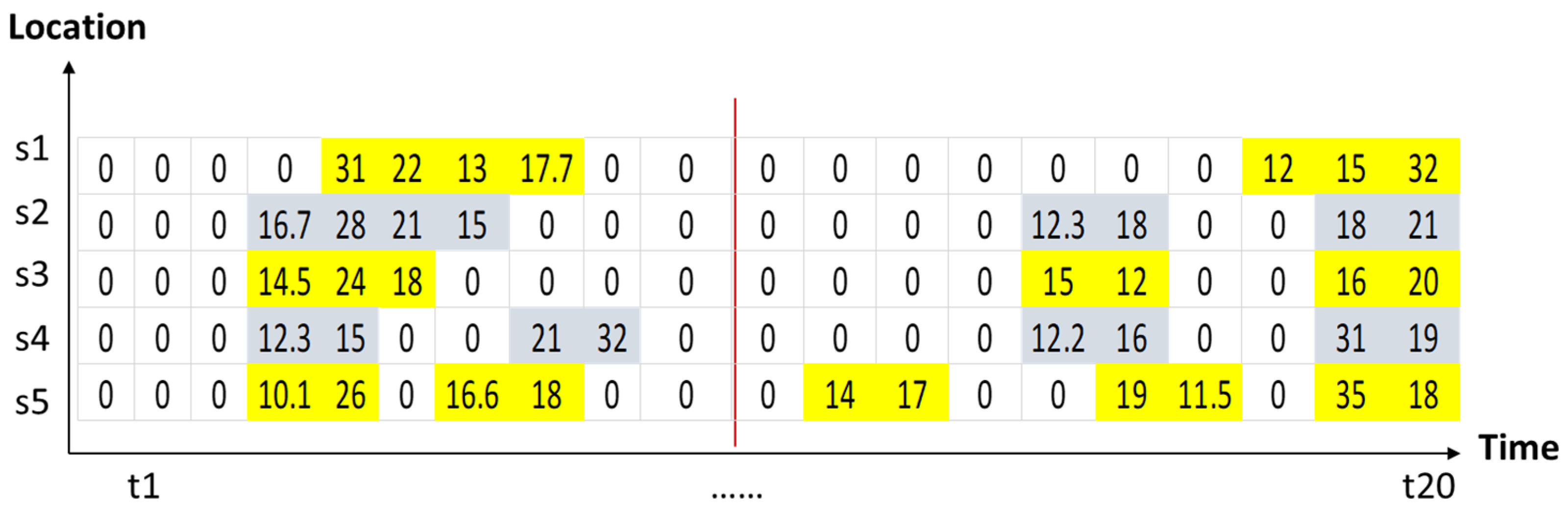

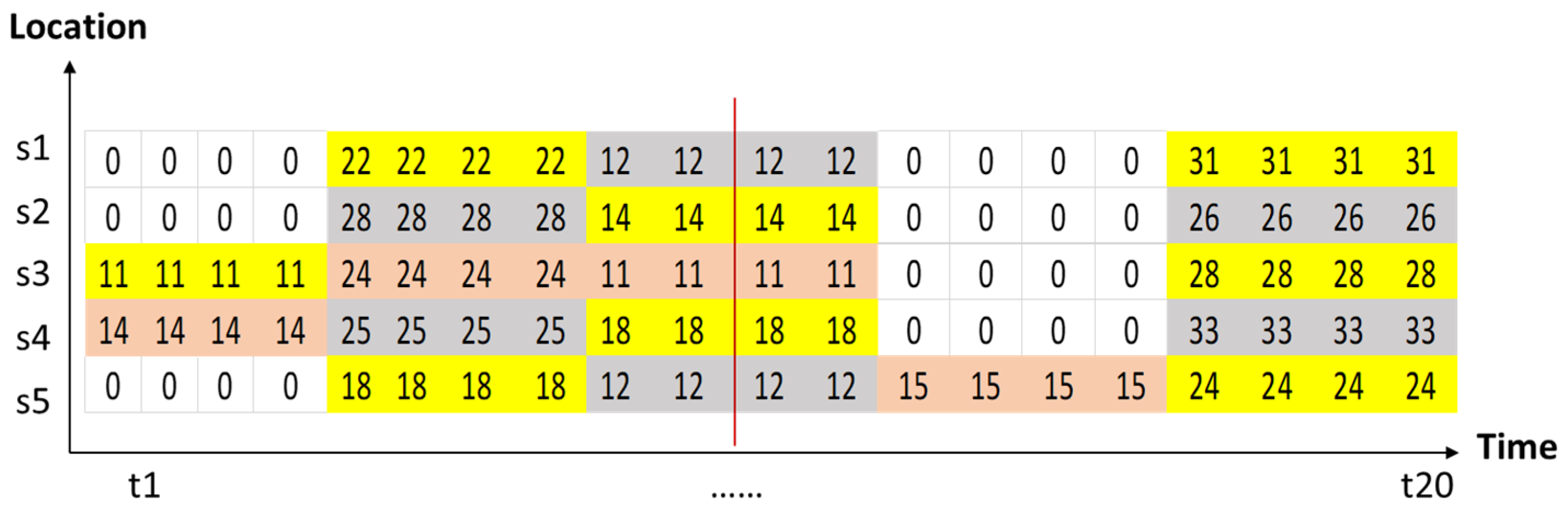







| t1 | t2 | t3 | t4 | t5 | t6 | t7 | t8 | t9 | t10 | t11 | t12 | t13 | t14 | t15 | t16 | t17 | t18 | t19 | t20 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| s1 | 0.22 | 0.35 | 1.20 | 0.56 | 3.10 | 2.20 | 1.30 | 1.77 | 0.30 | 0.00 | 1.00 | 0.55 | 2.10 | 0.50 | 1.55 | 0.80 | 0.20 | 1.20 | 1.50 | 2.20 |
| s2 | 0.25 | 2.50 | 0.40 | 1.67 | 2.80 | 2.10 | 1.50 | 0.60 | 0.20 | 0.00 | 1.00 | 0.44 | 2.00 | 0.33 | 1.23 | 1.80 | 0.10 | 0.10 | 1.80 | 2.10 |
| s3 | 0.28 | 2.10 | 0.45 | 1.45 | 2.40 | 1.80 | 0.44 | 0.80 | 0.10 | 0.00 | 1.00 | 0.70 | 1.50 | 0.80 | 1.50 | 1.20 | 0.00 | 0.00 | 1.60 | 2.00 |
| s4 | 0.31 | 1.70 | 0.50 | 1.23 | 0.50 | 0.60 | 0.55 | 2.10 | 0.20 | 0.00 | 0.00 | 1.50 | 0.50 | 2.10 | 0.22 | 1.60 | 0.10 | 0.22 | 0.10 | 1.90 |
| s5 | 0.34 | 1.60 | 0.55 | 1.01 | 0.60 | 0.67 | 1.66 | 1.80 | 0.10 | 0.00 | 0.00 | 1.40 | 0.70 | 2.50 | 0.52 | 1.90 | 1.15 | 0.30 | 0.50 | 1.80 |
| t1 | t2 | t3 | t4 | t5 | t6 | t7 | t8 | t9 | t10 | t11 | t12 | t13 | t14 | t15 | t16 | t17 | t18 | t19 | t20 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| s1 | 0 | 0 | 10.2 | 5.6 | 31 | 22 | 13 | 17.7 | 3 | 0 | 10 | 5.5 | 21 | 5 | 15.5 | 8 | 2 | 12 | 15 | 32 |
| s2 | 2.5 | 5 | 4 | 16.7 | 28 | 21 | 15 | 6 | 2 | 0 | 10 | 4.4 | 20 | 3.3 | 12.3 | 18 | 1 | 1 | 18 | 21 |
| s3 | 0 | 1 | 4.5 | 14.5 | 24 | 18 | 4.4 | 8 | 0 | 0 | 10 | 7 | 15 | 8 | 15 | 12 | 0 | 0 | 16 | 20 |
| s4 | 3.1 | 7 | 5 | 12.3 | 15 | 6 | 5.5 | 21 | 32 | 0 | 0 | 15 | 5 | 1 | 12.2 | 16 | 1 | 2.2 | 31 | 19 |
| s5 | 3.4 | 6 | 5.5 | 10.1 | 26 | 6.7 | 16.6 | 18 | 0 | 0 | 0 | 14 | 17 | 5 | 5.2 | 19 | 11.5 | 3 | 35 | 18 |
| t1 | t2 | t3 | t4 | t5 | t6 | t7 | t8 | t9 | t10 | t11 | t12 | t13 | t14 | t15 | t16 | t17 | t18 | t19 | t20 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| s1 | 9.8 | 9.8 | 9.8 | 9.8 | 22 | 22 | 22 | 22 | 12 | 12 | 12 | 12 | 8.8 | 8.8 | 8.8 | 8.8 | 31 | 31 | 31 | 31 |
| s2 | 9.1 | 9.1 | 9.1 | 9.1 | 28 | 28 | 28 | 28 | 14 | 14 | 14 | 14 | 5 | 5 | 5 | 5 | 26 | 26 | 26 | 26 |
| s3 | 11 | 11 | 11 | 11 | 24 | 24 | 24 | 24 | 11 | 11 | 11 | 11 | 7 | 7 | 7 | 7 | 28 | 28 | 28 | 28 |
| s4 | 14 | 14 | 14 | 14 | 25 | 25 | 25 | 25 | 18 | 18 | 18 | 18 | 9 | 9 | 9 | 9 | 33 | 33 | 33 | 33 |
| s5 | 8 | 8 | 8 | 8 | 18 | 18 | 18 | 18 | 12 | 12 | 12 | 12 | 15 | 15 | 15 | 15 | 24 | 24 | 24 | 24 |
| Algorithm | Min | lq | Mean | Median | uq | Max | n_eval |
|---|---|---|---|---|---|---|---|
| STES.sim1 | 503 | 549 | 676 | 587 | 657 | 2328 | 100 |
| EditD Dynamic | 4904 | 5250 | 5942 | 5474 | 6319 | 12,467 | 100 |
| EditD_Rstringdist | 2064 | 2280 | 2591 | 2408 | 2625 | 5501 | 100 |
| Jaccard_Rstringdist | 1863 | 2021 | 2651 | 2167 | 2556 | 8504 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, F.; Beard, K. A Unifying Framework for Analysis of Spatial-Temporal Event Sequence Similarity and Its Applications. ISPRS Int. J. Geo-Inf. 2021, 10, 594. https://doi.org/10.3390/ijgi10090594
Xu F, Beard K. A Unifying Framework for Analysis of Spatial-Temporal Event Sequence Similarity and Its Applications. ISPRS International Journal of Geo-Information. 2021; 10(9):594. https://doi.org/10.3390/ijgi10090594
Chicago/Turabian StyleXu, Fuyu, and Kate Beard. 2021. "A Unifying Framework for Analysis of Spatial-Temporal Event Sequence Similarity and Its Applications" ISPRS International Journal of Geo-Information 10, no. 9: 594. https://doi.org/10.3390/ijgi10090594
APA StyleXu, F., & Beard, K. (2021). A Unifying Framework for Analysis of Spatial-Temporal Event Sequence Similarity and Its Applications. ISPRS International Journal of Geo-Information, 10(9), 594. https://doi.org/10.3390/ijgi10090594





