Variables Selection for Aboveground Biomass Estimations Using Satellite Data: A Comparison between Relative Importance Approach and Stepwise Akaike’s Information Criterion
Abstract
:1. Introduction
2. Materials and Methods
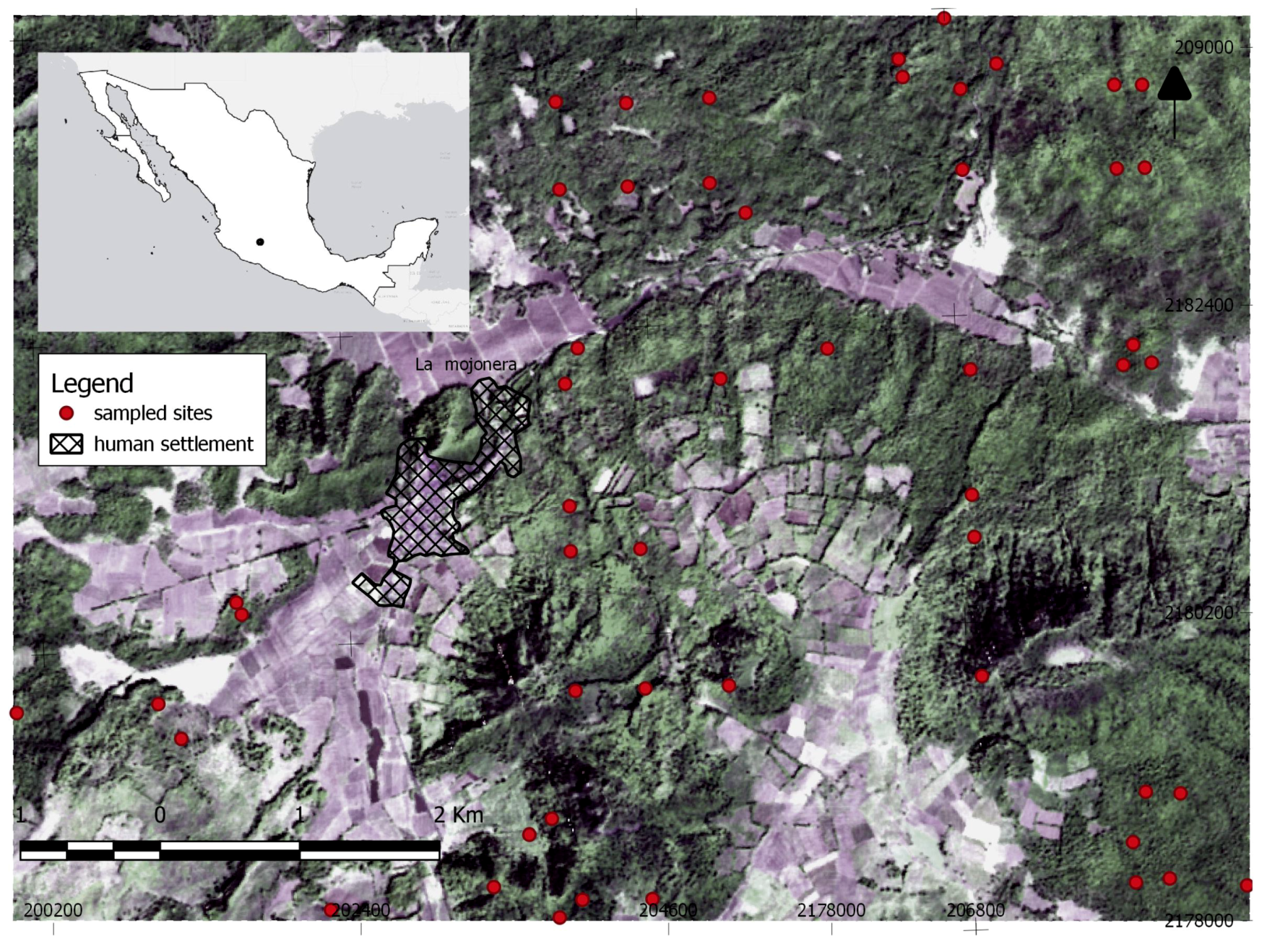
2.1. The Study Area
2.2. Fieldwork Data
2.3. Satellite Images
2.4. Independent Variables
2.5. Statistical Methods
2.5.1. Correlation Analysis and Model Assumptions
2.5.2. Variable Selection Methods
2.5.3. Validation of the Regression Models
3. Results
3.1. State and Structure of the Forest
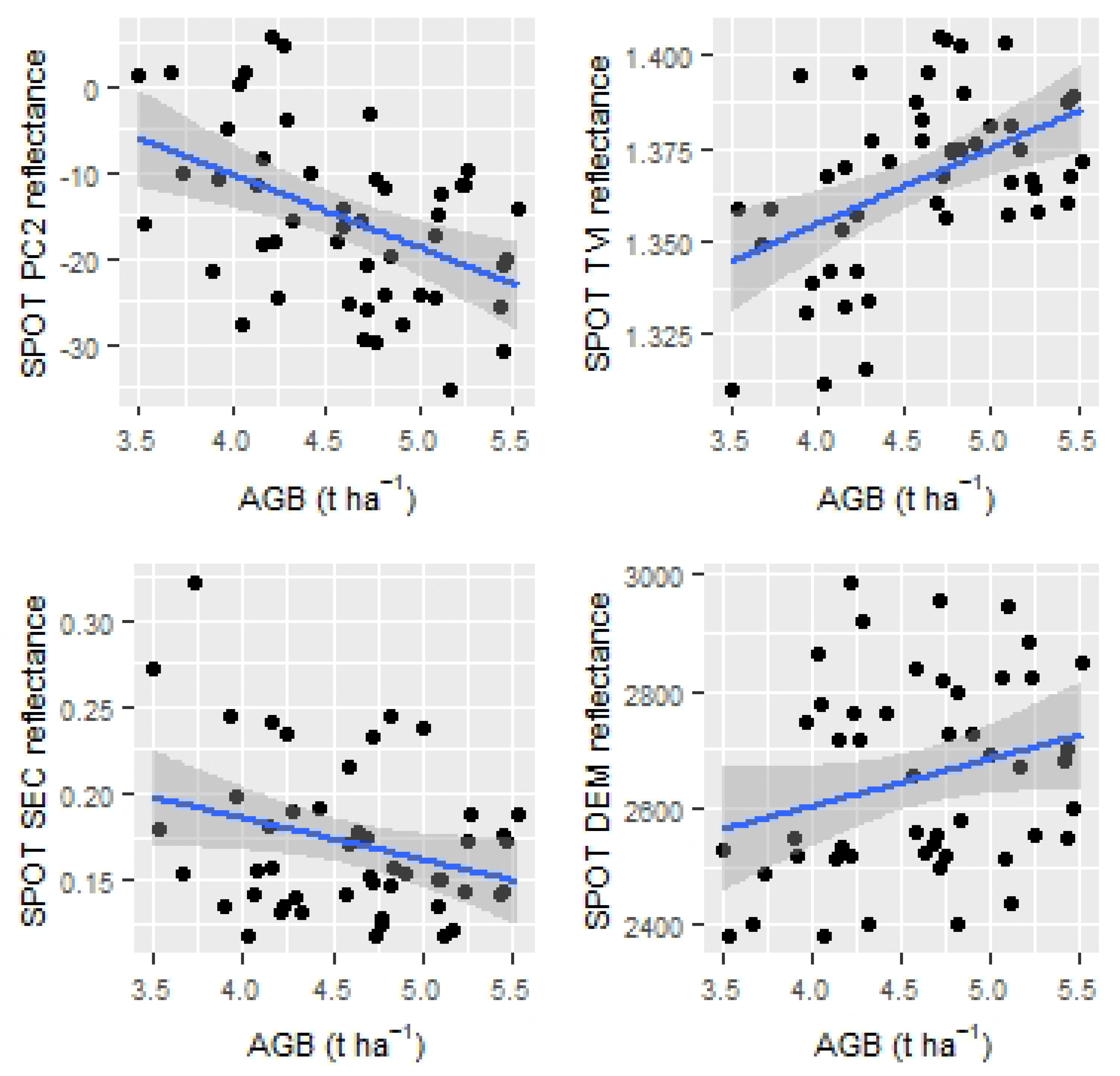
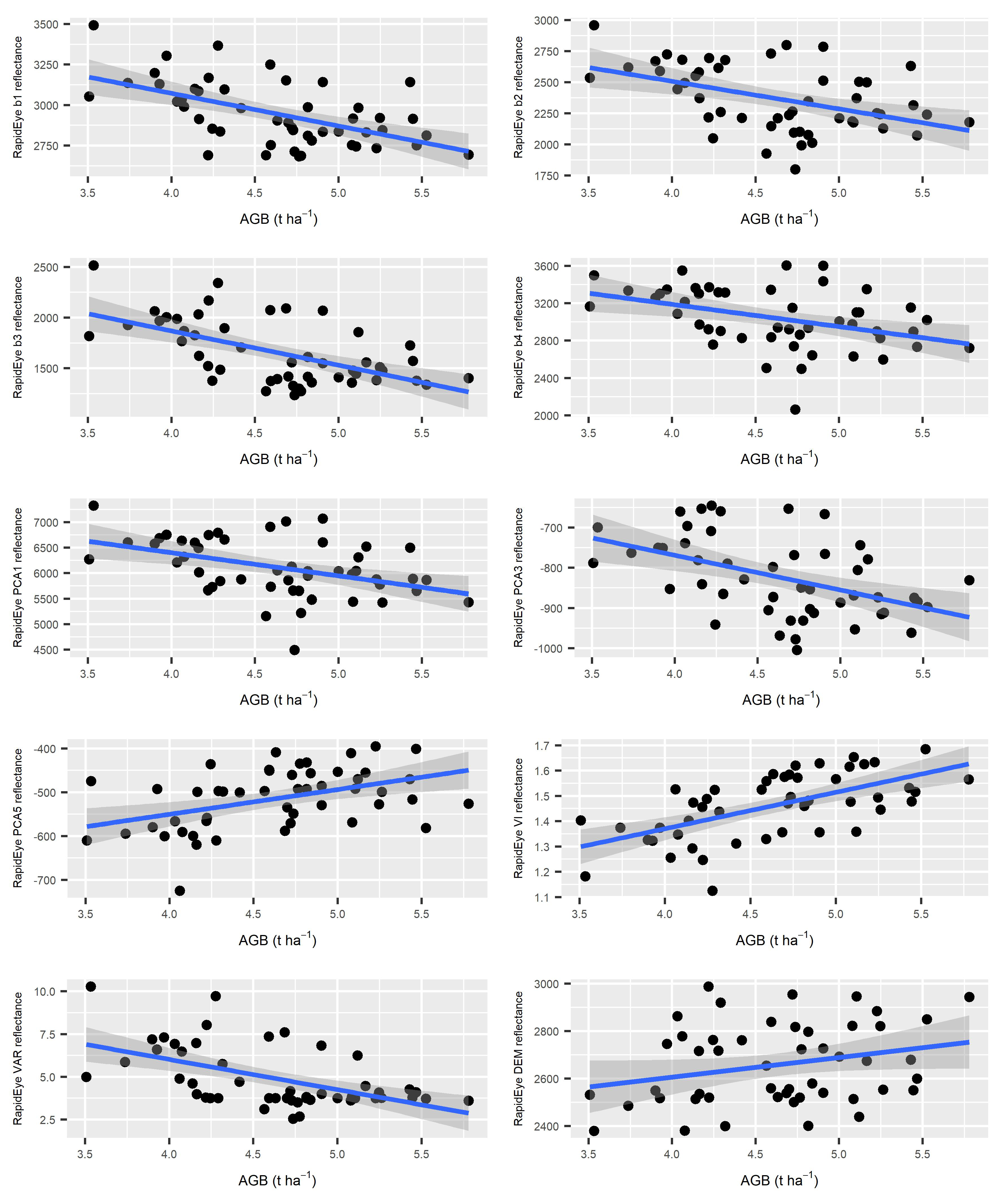
3.2. Correlation Analysis
3.3. Estimation of the AGB
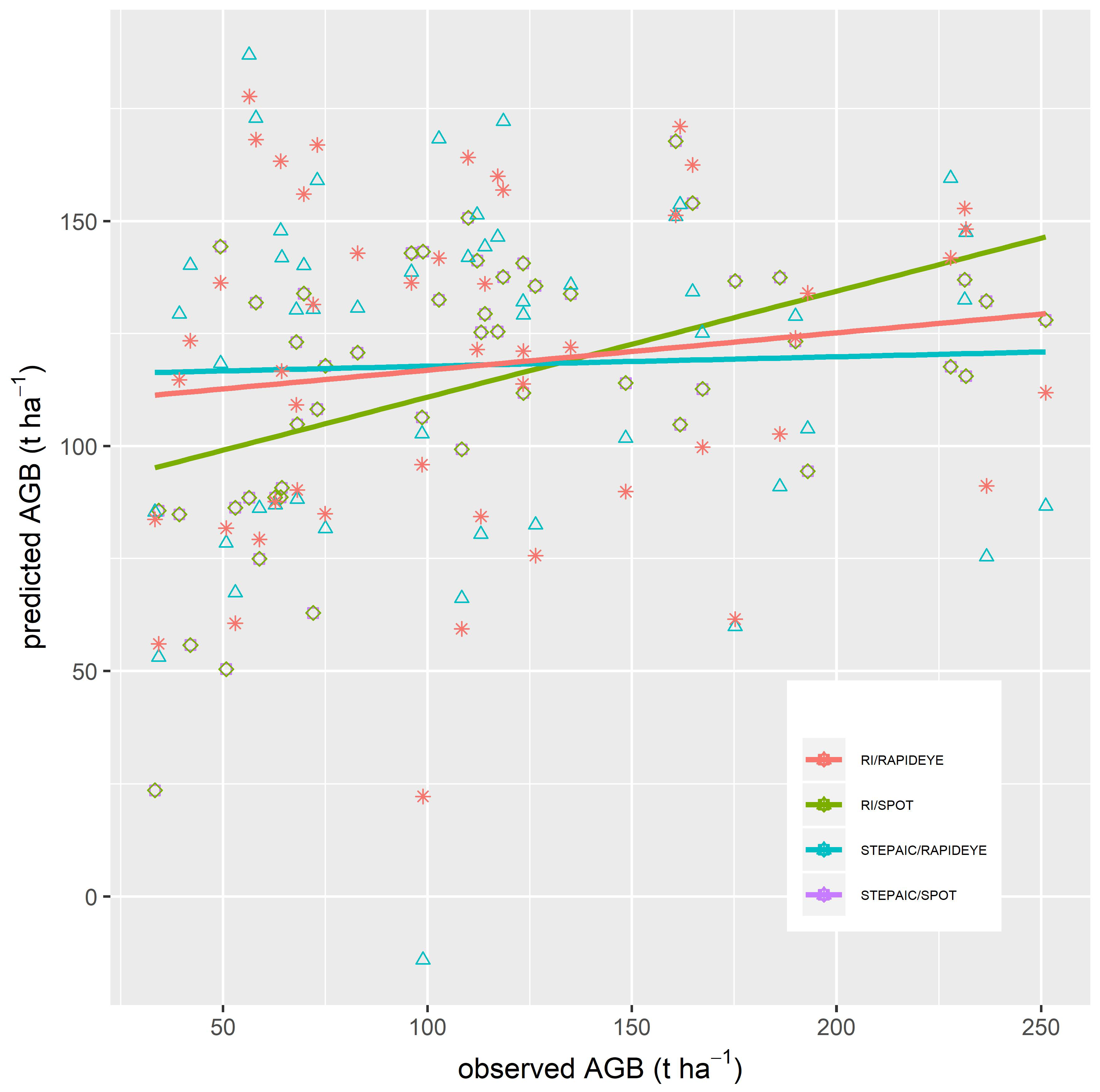
3.4. Validation of the Regression Models
4. Discussion
4.1. Correlation Analysis
4.2. Estimation of the AGB
4.3. Validation of the Regression Models
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Spectral Bands |
|---|
| Green band |
| Red band |
| NIR band |
| SWIR band |
| Principal Components |
| Principal component 1 (PC1) |
| Principal component 2 (PC2) |
| Principal component 3 (PC3) |
| Principal component 4 (PC4) |
| Vegetation indices |
| Normalized difference vegetation index (NDVI ) |
| NIR/red reflectance ratio index (RATIO) |
| NIR/green reflectance ratio index (GR) |
| Green–red reflectance ratio index (VI) |
| Ratio vegetation index (RVI) |
| Brightness index (Brightness) |
| Soil adjusted vegetation index (SAVI) |
| Transformed vegetation index (TVI) |
| Corrected transformed vegetation index (CTVI) |
| Thiam’s transformed vegetation index (TTVI) |
| Normalized ratio vegetation index (NRVI) |
| Digital Elevation Model |
| Altitude |
| Slope |
| Orientation |
| Solar Radiance |
| 7 × 7 GLCM(grey-level co-occurrence matrix) textures per band |
| Mean (green band, red band, NIR band, and SWIR band) |
| Variance (VAR) (green band, red band, NIR band, and SWIR band) |
| Correlation (green band, red band, NIR band, and SWIR band) |
| Dissimilarity (green band, red band, NIR band, and SWIR band) |
| Entropy (green band, red band, NIR band, and SWIR band) |
| Second moment (SEC) (green band, red band, NIR band, and SWIR band) |
| Contrast (green band, red band, NIR band, and SWIR band) |
| Homogeneity (green band, red band, NIR band, and SWIR band) |
| Spectral Bands |
|---|
| Blue band |
| Green band |
| Red band |
| Red edge band |
| NIR band |
| Principal Components |
| Principal component 1 (PC1) |
| Principal component 2 (PC2) |
| Principal component 3 (PC3) |
| Principal component 4 (PC4) |
| Principal component 5 (PC5) |
| Vegetation indices |
| Normalized difference vegetation index (NDVI) |
| NIR/red reflectance ratio index (RATIO) |
| NIR/green reflectance ratio index (GR) |
| Green–red reflectance ratio index (VI) |
| Ratio vegetation index (RVI) |
| Brightness index (Brightness) |
| Soil adjusted vegetation index (SAVI) |
| Transformed vegetation index (TVI) |
| Corrected transformed vegetation index (CTVI) |
| Thiam’s transformed vegetation index (TTVI) |
| Normalized ratio vegetation index (NRVI) |
| Normalized difference red edge (NDRE) |
| Ratio index (RI) |
| Digital Elevation Model |
| Altitude |
| Slope |
| Orientation |
| Solar Radiance |
| 3 × 3 GLCM(grey-level co-occurrence matrix) textures per band |
| Mean (blue band, green band, red band, NIR band, and red edge band) |
| Variance (VAR) (blue band, green band, red band, NIR band, and red edge band) |
| Correlation (blue band, green band, red band, NIR band, and red edge band) |
| Dissimilarity (blue band, green band, red band, NIR band, and red edge band) |
| Entropy (blue band, green band, red band, NIR band, and red edge band) |
| Second moment (SEC) (blue band, green band, red band, NIR band, and red edge band) |
| Contrast (blue band, green band, red band, NIR band, and red edge band) |
| Homogeneity (blue band, green band, red band, NIR band, and red edge band) |
References
- Dixon, R.K.; Brown, S.; Houghton, R.A.R.; Solomon, A.M.; Trexler, M.C.; Wisniewski, J. Carbon pools and flux of global forest ecosystems. Science 1994, 263, 185–190. [Google Scholar] [CrossRef]
- Houghton, R.A.; Hall, F.; Goetz, S.J. Importance of biomass in the global carbon cycle. J. Geophys. Res. Biogeosci. 2009, 114, 1–13. [Google Scholar] [CrossRef]
- Toan, T.L.; Quegan, S.; Davidson, M. The BIOMASS mission: Mapping global forest biomass to better understand the terrestrial carbon cycle. Remote Sens. Environ. 2011, 115, 2850–2860. [Google Scholar] [CrossRef] [Green Version]
- Marta-Pedroso, C.; Laporta, L.; Proença, V.; Azevedo, J.C.; Domingos, T. Changes in the ecosystem services provided by forests and their economic valuation: A review. For. Landsc. Glob. Chang. Chall. Res. Manag. 2014, 3, 107–137. [Google Scholar] [CrossRef]
- Timothy, D.; Onisimo, M.; Riyad, I. Quantifying aboveground biomass in African environments: A review of the trade-offs between sensor estimation accuracy and costs. Trop. Ecol. 2016, 57, 393–405. [Google Scholar]
- Lu, D.; Chen, Q.; Wang, G.; Liu, L.; Li, G.; Moran, E. A survey of remote sensing-based aboveground biomass estimation methods in forest ecosystems. Int. J. Digit. Earth 2014, 1–43. [Google Scholar] [CrossRef]
- Yavasli, D.D. Recent approaches in above ground biomass estimation methods. Aegean Geogr. J. 2013, 21, 39–49. [Google Scholar]
- Kayitakire, F.; Hamel, C.; Defourny, P. Retrieving forest structure variables based on image texture analysis and IKONOS-2 imagery. Remote Sens. Environ. 2006, 102, 390–401. [Google Scholar] [CrossRef]
- Castillo-Santiago, M.A.; Ricker, M.; de Jong, B.H.J. Estimation of tropical forest structure from SPOT-5 satellite images. Int. J. Remote Sens. 2010, 31, 2767–2782. [Google Scholar] [CrossRef]
- Li, W.; Niu, Z.; Liang, X.; Li, Z.; Huang, N.; Gao, S.; Wang, C.; Muhammad, S. Geostatistical modeling using LiDAR-derived prior knowledge with SPOT-6 data to estimate temperate forest canopy cover and above-ground biomass via stratified random sampling. Int. J. Appl. Earth Obs. Geoinf. 2015, 41, 88–98. [Google Scholar] [CrossRef]
- Aguirre-Salado, C.A.; Valdez-Lazalde, J.R.; Ángeles-Pérez, G.; de los Santos-Posadas, H.M.; Haapanen, R.; Aguirre-Salado, A.I. Mapeo de carbono arbóreo aéreo en bosques manejados de pino Patula en Hidalgo, México. Agrociencia 2009, 43, 209–220. [Google Scholar]
- Du, L.; Zhou, T.; Zou, Z.; Zhao, X.; Huang, K.; Wu, H. Mapping forest biomass using remote sensing and national forest inventory in China. Forests 2014, 5, 1267–1283. [Google Scholar] [CrossRef]
- Sousa, A.M.O.; Cristina, A.; Mesquita, P.; Marques, J.R. Biomass estimation with high resolution satellite images: A case study of Quercus rotundifolia. ISPRS J. Photogramm. Remote Sens. 2015, 101, 69–79. [Google Scholar] [CrossRef] [Green Version]
- Wani, A.A.; Joshi, P.K.; Singh, O. Estimating biomass and carbon mitigation of temperate coniferous forests using spectral modeling and field inventory data. Ecol. Inform. 2015, 25, 63–70. [Google Scholar] [CrossRef]
- Zhu, X.; Liu, D. Improving forest aboveground biomass estimation using seasonal Landsat NDVI time-series. ISPRS J. Photogramm. Remote Sens. 2015, 102, 222–231. [Google Scholar] [CrossRef]
- Wallner, A.; Elatawneh, A.; Schneider, T.; Knoke, T. Estimation of forest structural information using RapidEye satellite data. Forestry 2014, 88, 96–107. [Google Scholar] [CrossRef] [Green Version]
- Latifi, H.; Fassnacht, F.E.; Hartig, F.; Berger, C.; Hernández, J.; Corvalán, P.; Koch, B. Stratified aboveground forest biomass estimation by remote sensing data. Int. J. Appl. Earth Obs. Geoinf. 2015, 38, 229–241. [Google Scholar] [CrossRef]
- Muukkonen, P.; Heiskanen, J. Estimating biomass for boreal forests using ASTER satellite data combined with standwise forest inventory data. Remote Sens. Environ. 2005, 99, 434–447. [Google Scholar] [CrossRef]
- Wulder, M.A.; White, J.C.; Fournier, R.A.; Luther, J.E.; Magnussen, S. Spatially Explicit Large Area Biomass Estimation: Three Approaches Using Forest Inventory and Remotely Sensed Imagery in a GIS. Sensors 2008, 8, 529–560. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Sha, Z.; Yu, M.; Bai, Y.; Zhang, L. A comparison of two models with Landsat data for estimating above ground grassland biomass in Inner Mongolia, China. Ecol. Model. 2009, 220, 1810–1818. [Google Scholar] [CrossRef]
- Tian, X.; Su, Z.; Chen, E.; Li, Z.; van der Tol, C.; Guo, J.; He, Q. Estimation of forest above-ground biomass using multi-parameter remote sensing data over a cold and arid area. Int. J. Appl. Earth Obs. Geoinf. 2012, 14, 160–168. [Google Scholar] [CrossRef]
- Lu, D. The potential and challenge of remote sensing-based biomass estimation. Int. J. Remote Sens. 2006, 27, 1297–1328. [Google Scholar] [CrossRef]
- Whittingham, M.J.; Stephens, P.A.; Bradbury, R.B.; Freckleton, R.P. Why do we still use stepwise modelling in ecology and behaviour? J. Anim. Ecol. 2006, 75, 1182–1189. [Google Scholar] [CrossRef] [PubMed]
- Kronseder, K.; Ballhorn, U.; Böhmc, V.; Siegert, F. Above ground biomass estimation across forest types at different degradation levels in central kalimantan using lidar data. Int. J. Appl. Earth Obs. Geoinf. 2012, 18, 37–48. [Google Scholar] [CrossRef]
- Vaglio Laurin, G.; Chen, Q.; Lindsell, J.A.; Coomes, D.A.; Frate, F.D.; Guerriero, L.; Pirotti, F.; Valentini, R. Above ground biomass estimation in an African tropical forest with lidar and hyperspectral data. ISPRS J. Photogramm. Remote Sens. 2014, 89, 49–58. [Google Scholar] [CrossRef]
- Duan, Z.G.; Zhao, D.; Zeng, Y.; Zhao, Y.J.; Wu, B.F.; Zhu, J.J. Estimation of the forest aboveground biomass at regional scale based on remote sensing. Geomat. Inf. Sci. Wuhan Univ. 2015, 40, 1400–1407. [Google Scholar]
- Zhao, P.; Lu, D.; Wang, G.; Wu, C.; Huang, Y.; Yu, S. Examining spectral reflectance saturation in landsat imagery and corresponding solutions to improve forest aboveground biomass estimation. Remote Sens. 2016, 8, 469. [Google Scholar] [CrossRef]
- Lukacs, P.M.; Burnham, K.P.; Anderson, D.R. Model selection bias and Freedman’s paradox. Ann. Inst. Stat. Math. 2010, 62, 117–125. [Google Scholar] [CrossRef]
- Ratner, B. Variable selection methods in regression: Ignorable problem, outing notable solution. J. Target. Meas. Anal. Mark. 2010, 18, 65–75. [Google Scholar] [CrossRef]
- Sileshi, G.W. A critical review of forest biomass estimation models, common mistakes and corrective measures. For. Ecol. Manag. 2014, 329, 237–254. [Google Scholar] [CrossRef]
- Yamashita, T.; Yamashita, K.; Kamimura, R. A Stepwise AIC Method for Variable Selection in Linear Regression. Commun. Stat. Theory Methods 2007, 36, 2395–2403. [Google Scholar] [CrossRef]
- Johnson, J.W.; LeBreton, J.M. History and use of relative importance indices in organizational research. Organ. Res. Methods 2004, 7, 238–257. [Google Scholar] [CrossRef]
- Johnson, J.W. A Heuristic Method for Estimating the Relative Weight of Predictor Variables in Multiple Regression. Multivar. Behav. Res. 2000, 35, 1–19. [Google Scholar] [CrossRef]
- Budescu, D.V. Dominance analysis: A new approach to the problem of relative importance of predictors in multiple regression. Psychol. Bull. 1993, 114, 542–551. [Google Scholar] [CrossRef]
- Lindeman, R.H.; Merenda, P.F.; Gold, R.Z. Introduction to Bivariate and Multivariate Analysis; Scott, Foresman: Glenview, IL, USA, 1980; Volume 4. [Google Scholar]
- Chevan, A.; Sutherland, M. Hierarchical Partitioning. Am. Stat. 1991, 45, 90–96. [Google Scholar] [CrossRef]
- Feldman, B. Relative Importance and Value. SSRN 2005. [Google Scholar] [CrossRef]
- Grömping, U. Relative importance for linear regression in R: The package relaimpo. J. Stat. Softw. 2006, 17, 139–147. [Google Scholar] [CrossRef]
- Grömping, U. Estimators of Relative Importance in Linear Regression Based on Variance Decomposition. Am. Stat. 2007, 61, 139–147. [Google Scholar] [CrossRef]
- Lipovetsky, S.; Conklin, W.M. Predictor relative importance and matching regression parameters. J. Appl. Stat. 2015, 42, 1017–1031. [Google Scholar] [CrossRef]
- Chi, H.; Sun, G.; Huang, J.; Guo, Z.; Ni, W.; Fu, A. National Forest Aboveground Biomass Mapping from ICESat/GLAS Data and MODIS Imagery in China. Remote Sens. 2015, 7, 5534–5564. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.; Chen, Q.; Wang, G.; Moran, E.; Batistella, M.; Zhang, M.; Vaglio Laurin, G.; Saah, D. Aboveground Forest Biomass Estimation with Landsat and LiDAR Data and Uncertainty Analysis of the Estimates. Int. J. For. Res. 2012, 2012, 1–16. [Google Scholar] [CrossRef]
- Foody, G.M. Remote sensing of tropical forest environments: Towards the monitoring of environmental resources for sustainable development. Int. J. Remote Sens. 2003, 24, 4035–4046. [Google Scholar] [CrossRef]
- Návar, J. Allometric equations for tree species and carbon stocks for forests of northwestern Mexico. For. Ecol. Manag. 2009, 257, 427–434. [Google Scholar] [CrossRef]
- Aguilar, R.; Ghilardi, A.; Vega, E.; Skutsch, M.; Oyama, K. Sprouting productivity and allometric relationships of two oak species managed for traditional charcoal making in central Mexico. Biomass Bioenergy 2012, 36, 192–207. [Google Scholar] [CrossRef]
- Acosta, M.; Carrillo, F.; Gómez, R.G. Biomass and Carbon Assessment in Two Tree Species in a Cloudy Forest. Revista Mexicana de Ciencias Agrícolas 2011, 2, 529–543. [Google Scholar]
- Norberto Vigil, N. Estimación De Biomasa Y Contenido De Carbono en Cupressus Lindleyi Klotzsch Ex Endl. en El Campo Forestal Experimental “Las Cruces”. Ph.D. Thesis, Universidad Autónoma Chapingo, Texcoco, Mexico, 2010. [Google Scholar]
- Neubert, M.; Meinel, G. Atmospheric and Terrain Correction of IKONOS Imagery Using ATCOR3. In Proceedings of the ISPRS Hannover Workshop 2005: High-Resolution Earth Imaging for Geospatial Information, Hannover, Germany, 17–20 May 2005; Volume 3. [Google Scholar]
- Zvoleff, A. Calculate Textures from Grey-Level Co-Occurrence Matrices (GLCMs) in R. 2015. Available online: https://rdrr.io/cran/glcm/ (accessed on 20 May 2019).
- Lu, D.; Batistella, M. Exploring TM image texture and its relationships with biomass estimation in Rondônia, Brazilian Amazon. Acta Amaz. 2005, 35, 249–257. [Google Scholar] [CrossRef]
- Ozdemir, I.; Karnieli, A. Predicting forest structural parameters using the image texture derived from worldview-2 multispectral imagery in a dryland forest, Israel. Int. J. Appl. Earth Obs. Geoinf. 2011, 13, 701–710. [Google Scholar] [CrossRef]
- Acevedo, J.A.; Londoño, R.A.; Hernandez, G. Análisis de textura en imágenes de satélite en el ámbito de la biodiversidad y la estructura en un bosque de los Andes Colombianos. Revista Gestión y Ambiente 2008, 11, 137–146. [Google Scholar]
- Eckert, S. Improved forest biomass and carbon estimations using texture measures from worldView-2 satellite data. Remote Sens. 2012, 4, 810–829. [Google Scholar] [CrossRef]
- Ramsey, P.H. Critical Values for Spearman’s Rank Order Correlation. J. Educ. Stat. 2008, 14, 245–253. [Google Scholar] [CrossRef]
- Rouse, J.W. Monitoring the Vernal Advancement and Retrogradation (Green Wave Effect) of Natural Vegetation; NASA: Washington, DC, USA, 1973.
- Schneider, T.; Tian, J.; Elatawneh, A.; Rappl, A.; Reinartz, P. Tracing Structural Changes of a Complex Forest by a multiple systems approach. In Proceedings of the 1st European Association of Remote Sensing Laboratories Workshop on Temporal Analysis of Satellite Images, Mykonos, Greece, 23–25 May 2012; pp. 159–165. [Google Scholar]
- Jordan, C.F. Derivation of Leaf-Area Index from Quality of Light on the Forest Floor. Ecol. Soc. Am. 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Lyon, J.G.; Yuan, D.; Lunetta, R.S.; Elvidge, C.D. A Change Detection Experiment Using Vegetation Indices. Photogramm. Eng. Remote Sens. 1998, 64, 143–150. [Google Scholar]
- Kanemasu, E.T. Seasonal Canopy Reflectance Patterns of Wheat, Sorghum, and Soybean. Remote Sens. Environ. 1974, 1974. 3, 43–47. [Google Scholar] [CrossRef]
- Definiens. Developer XD 2.0.4. Reference Book; Definiens AG: Munchen, Germany, 2012. [Google Scholar]
- Barnes, E.M.; Clarke, T.R.; Richards, S.E.; Colaizzi, P.D.; Haberland, J.; Kostrzewski, M.; Waller, P.; Choi, C.; Riley, E.; Thompson, T.; et al. Coincident detection of crop water stress, nitrogen status and canopy density using ground based multispectral data. In Proceedings of the Fifth International Conference on Precision Agriculture, Bloomington, IN, USA, 16–19 July 2000; pp. 16–19. [Google Scholar]
- Huete, A.R. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Deering, D.W.; Rouse, J.W.; Haas, R.H.; Schell, J.A. Measuring forage production of grazing units from landsat MSS data. In Proceedings of the 10th International Symposium of Remote Sensing of Environment, Ann Arbor, MI, USA, 6–10 October 1975; pp. 1169–1178. [Google Scholar]
- Perry, C.R.; Lautenschlager, L.F. Funcional Equivalence of Spectral Vegetation Indices. Remote Sens. Environ. 1984, 14, 169–182. [Google Scholar] [CrossRef]
- Thiam, A.K.; Khoudiedji, A. Geographic Information Systems and Remote Sensing Methods for Assessing and Monitoring Land Degradation in the Sahel Region: The Case of Southern Mauritania; Clark University: Worcester, MA, USA, 1998; 490p, ISBN 9780591716474. [Google Scholar]
- Richardson, A.J.; Wiegand, C.L. Distinguishing vegetation from soil background information. Photogrammetric Engineering and Remote Sensing. Photogramm. Eng. Remote Sens. 1977, 43, 1541–1552. [Google Scholar]
- Baret, F.; Guyot, G. Potentials and limits of vegetation indices for LAI and APAR assessment. Remote Sens. Environ. 1991, 35, 161–173. [Google Scholar] [CrossRef]
- Li, G. Robust regression. In Exploring Data Tables, Trends, and Shapes; Wiley: New York, NY, USA, 1985. [Google Scholar]
- Rousseeuw, P.J.; Leroy, A.M. Robust Regression and Outlier Detection; John Wiley & Sons: Hoboken, NJ, USA, 2005; Volume 589. [Google Scholar]
- Grömping, U. Relative Importance of Regressors in Linear Models. Package ‘Relaimpo’. Version 2.2. 2013. Available online: https://www.scribd.com/document/321113503/2013-Package-relaimpo-pdf (accessed on 20 May 2019).
- R Core Team. R: A Language and Environment for Statistical Computing; The R Project for Statistical Computing; R Core Team: Vienna, Austria, 2015. [Google Scholar]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Caballero, F.F. Información en Análisis Factorial. Aspectos Teóricos y Computacionales. Ph.D. Thesis, University of Granada, Granada, Spain, 2011. [Google Scholar]
- Bozdogan, H. Model selection and Akaike’s Information Criterion (AIC): The general theory and its analytical extensions. Psychometrika 1987, 52, 345–370. [Google Scholar] [CrossRef]
- Walsh, C.; Nally, R.M. Hierarchical Partitioning;R Project For Statistical Computing. 2003. Available online: https://cran.r-project.org/web/packages/hier.part/hier.part.pdf (accessed on 1 February 2019).
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning; Springer: New York, NY, USA, 2000; Volume 7, p. 618. [Google Scholar]
- Witten, I.; Frank, E.; Hall, M.; Pal, C. Data Mining: Practical machine learning tools and techniques; Morgan Kaufmann: Burlington, MA, USA, 2016. [Google Scholar]
- Kizha, A.R.; Han, H.S. Predicting aboveground biomass in second growth coast redwood: Comparing localized with generic allometric models. Forests 2016, 7, 96. [Google Scholar] [CrossRef]
- Refaeilzadeh, P.; Tang, L.; Liu, H.; Ross, K.A. Cross-Validation. Encycl. Database Syst. 2009, 532–538. [Google Scholar] [CrossRef]
- Yao, X.; Wang, S.; Liu, M.; Fu, B.; Lü, Y.; Sun, F. Comparison of Four Spatial Interpolation Methods for Estimating Soil Moisture in a Complex Terrain Catchment. PLoS ONE 2013, 8, e54660. [Google Scholar] [CrossRef] [PubMed]
- Statistics Solutions. Table of Critical Values: Pearson Correlation. 2018. Available online: https://www.statisticssolutions.com/table-of-critical-values-pearson-correlation/ (accessed on 1 February 2019).
- Ojoyi, M.; Mutanga, O.; Odindi, J.; Abdel-Rahman, E.M. Application of topo-edaphic factors and remotely sensed vegetation indices to enhance biomass estimation in a heterogeneous landscape in the Eastern Arc Mountains of Tanzania. Geocarto Int. 2015, 1–21. [Google Scholar] [CrossRef]
- Sheen, S.W. A Principal Component Analysis of Vegetation Characteristics in North America A Principal Component Analysis of Vegetation Characteristics in North America. North 2004, 17, 173–182. [Google Scholar]
- Estornell, J.; Martí-Gavilá, J.M.; Sebastiá, M.T.; Mengual, J. Principal component analysis applied to remote sensing. Model. Sci. Educ. Learn. 2013, 6, 83–89. [Google Scholar] [CrossRef] [Green Version]
- Campbell, J.B. Introduction to Remote Sensing; Guilford Press: New York, NY, USA, 2002. [Google Scholar]
- Aguirre-Salado, C.A.; Treviño-Garza, E.J.; Aguirre-Calderón, O.A.; Jiménez-Pérez, J.; González-Tagle, M.A.; Valdez-Lazalde, J.R.; Miranda-Aragón, L.; Aguirre-Salado, A.I. Construction of aboveground biomass models with remote sensing technology in the intertropical zone in Mexico. J. Geogr. Sci. 2012, 22, 669–680. [Google Scholar] [CrossRef]
- Munoz-Ruiz, M.A.; Valdez-Lazalde, J.R.; de los Santos-Posadas, H.M.; Angeles-Perez, G.; Monterroso-Rivas, A.I. Inventory and Mapping of Temperate Forest in Hidalgo, Mexico through Spot and Field Data. Agrociencia 2014, 48, 847–862. [Google Scholar]
- Asner, G.P.; Warner, A.S. Canopy shadow in IKONOS satellite observations of tropical forests and savannas. Remote Sens. Environ. 2003, 87, 521–533. [Google Scholar] [CrossRef]
- Hero, J.M.; Castley, J.G.; Butler, S.A.; Lollback, G.W. Biomass estimation within an Australian eucalypt forest: Meso-scale spatial arrangement and the influence of sampling intensity. For. Ecol. Manag. 2013, 310, 547–554. [Google Scholar] [CrossRef]
- Ene, L.T.; Næsset, E.; Gobakken, T.; Gregoire, T.G.; Ståhl, G.; Nelson, R. Assessing the accuracy of regional LiDAR-based biomass estimation using a simulation approach. Remote Sens. Environ. 2012, 123, 579–592. [Google Scholar] [CrossRef]
- Peña, D. Análisis de datos multivariantes; McGraw-Hill España: Madrid, Spain, 2002; p. 515. [Google Scholar]

| Common Name | Genus | Allometric Equation | DBH Range (cm) | |
|---|---|---|---|---|
| Pine | Pinus | Y = 0.1229DBH | 5.7–57.4 | 0.91 [44] |
| Oak | Quercus | Y = 0.0890DBH | 7.6–62.5 | 0.95 [44] |
| Oak with sprouts | Quercus | Y = 0.0342DBH | 3.8–26.6 | 0.94 [45] |
| Tepamo | Alnus | Y = 0.1649DBH | 10.0–40.0 | 0.97 [46] |
| Cedar | Cupressus | Y = 0.5266DBH | 5.0–50 | 0.93 [47] |
| Madroño and other genus | Arbutus | Y = 0.0890DBH | 7.6–62.5 | 0.94 [44] |
| Normalized difference vegetation (NDVI) | [55] | |
| Ratio index (RI) | [56] | |
| NIR/red reflectance ratio index (RATIO) | [57] | |
| NIR/green reflectance ratio index (GR) | [58] | |
| green–red reflectance ratio index (VI) | [59] | |
| Brightness | [60] | |
| Normalized difference red edge (NDRE) | [61] | |
| Soil adjusted vegetation index (SAVI) | [62] | |
| Transformed vegetation index (TVI) | [63] | |
| Corrected transformed vegetation index (CTVI) | [64] | |
| Thiam’s transformed vegetation index (TTVI) | [65] | |
| Ratio vegetation index (RVI) | [66] | |
| Normalized ratio vegetation index (NRVI) | [67] |
| Parameters | Average | Standard Deviation | Standard Error (RMS) | Min | Max |
|---|---|---|---|---|---|
| AGB (ton ha−1 ) | 116 | 66 | 9 | 24 | 323 |
| basal area ( m2 ha−1 ) | 19 | 9 | 1 | 4 | 42 |
| tree height (m) | 15 | 6 | 0 | 4 | 32 |
| diameter of breast height (DBH) (cm) | 22 | 7 | 1 | 11 | 48 |
| woodland density (trees ha−1) | 700 | 433 | 57 | 100 | 1940 |
| stumps density (stumps ha−1 ) | 163 | 134 | 17 | 0 | 480 |
| regeneration height (cm) | 30 | 20 | 2 | 0 | 70 |
| Variables | SPOT-5 | RapidEye |
|---|---|---|
| Blue band | −0.489 ** | |
| Green band | −0.393 ** | |
| Red band | −0.511 ** | |
| Red edge | −0.323 * | |
| Principal component 1 (PC1) | −0.354 ** | |
| Principal component 2 (PC2) | −0.320 * | |
| Principal component 3 (PC3) | −0.423 ** | |
| Principal component 5 (PC5) | 0.423 ** | |
| Green–red vegetation index (VI) | 0.535 ** | |
| Transformed vegetation index (TVI) | 0.276 * | |
| 3 × 3 variance red texture (VAR) | −0.486 ** | |
| 7 × 7 second moment NIR (SEC) | −0.247 * | |
| Altitude (DEM) | 0.275 * | 0.275 * |
| Variables | SPOT-5 Image | RapidEye Image | ||
|---|---|---|---|---|
| STEPWISE-AIC | RI | STEPWISE-AIC | RI | |
| Red band | 0.005 (0.003) | |||
| Red edge | 0.012 * (0.006) | |||
| PC1 | -0.008 * (0.004) | |||
| PC3 | -0.008 * (0.004) | |||
| PC5 | 0.011 ** (0.005) | 0.002 (0.001) | ||
| TVI | 11.032 *** (2.768) | 11.032 *** (2.768) | ||
| VI | 2.195 *** (0.523) | |||
| SEC | −3.075 ** (1.484) | −3.075 ** (1.484) | ||
| DEM | 0.001 * (0.0004) | 0.001 * (0.0004) | ||
| Constant | −11.966 *** | −11.966 *** | 7.645 *** | 2.365 ** |
| Observations | 49 | 49 | 49 | 49 |
| 0.359 | 0.359 | 0.437 | 0.420 | |
| Adjusted | 0.317 | 0.317 | 0.317 | 0.394 |
| Standard residual error (SRE) | 0.451 (df = 45) | 0.451 (df = 45) | 0.431 (df = 43) | 0.423 (df = 46) |
| F statistics | 8.414 *** (df = 3; 45) | 8.414 *** (df = 3; 45) | 6.668 (df = 5; 43) | 16.632 *** (df = 2; 46) |
| RapidEye Image | ||||
|---|---|---|---|---|
| Iteration | STEPWISE-AIC | RI | ||
| RMSE | MAPE | RMSE | MAPE | |
| 1 | 55.77 | 40.97 | 55.60 | 38.08 |
| 2 | 58.89 | 44.61 | 58.39 | 40.56 |
| 3 | 68.53 | 44.25 | 61.78 | 41.61 |
| 4 | 57.75 | 39.11 | 58.74 | 39.54 |
| 5 | 57.89 | 41.93 | 56.96 | 38.19 |
| 6 | 64.21 | 46.18 | 53.41 | 34.47 |
| 7 | 54.39 | 39.33 | 57.49 | 39.65 |
| 8 | 56.21 | 38.18 | 53.73 | 35.77 |
| 9 | 54.92 | 38.18 | 53.50 | 35.59 |
| 10 | 55.37 | 39.69 | 54.06 | 38.71 |
| SPOT-5 image | ||||
| Iteration | STEPWISE-AIC | RI | ||
| RMSE | MAPE | RMSE | MAPE | |
| 1 | 55.79 | 39.51 | 55.79 | 39.51 |
| 2 | 56.46 | 40.24 | 56.46 | 40.24 |
| 3 | 61.20 | 43.21 | 61.20 | 43.21 |
| 4 | 59.40 | 41.77 | 59.40 | 41.77 |
| 5 | 58.25 | 40.89 | 58.25 | 40.89 |
| 6 | 56.85 | 38.38 | 56.85 | 38.38 |
| 7 | 55.66 | 38.04 | 55.66 | 38.04 |
| 8 | 59.15 | 39.81 | 59.15 | 39.81 |
| 9 | 56.07 | 39.17 | 56.07 | 39.17 |
| 10 | 57.67 | 40.12 | 57.67 | 40.12 |
| Iteration | STEPWISE-AIC | MAPE | Iteration | RI | MAPE |
|---|---|---|---|---|---|
| Fold 1 | |||||
| 1 | AGB ∼ PC2 | 50.59 | 1 | AGB ∼ PC2 + DEM | 44.32 |
| 2 | AGB ∼ TVI + DEM | 30.36 | 2 | AGB ∼ TVI + DEM | 30.36 |
| 3 | AGB ∼ TVI + SEC + DEM | 53.84 | 3 | AGB ∼ TVI + SEC + DEM | 53.84 |
| 4 | AGB ∼ PC2 + SEC + DEM | 45.12 | 4 | AGB ∼ PC2 + SEC + DEM | 45.12 |
| 5 | AGB ∼ TVI + DEM | 38.65 | 5 | AGB ∼ TVI + SEC + DEM | 32.6 |
| 6 | AGB ∼ TVI + SEC | 45.48 | 6 | AGB ∼ TVI + SEC | 45.48 |
| 7 | AGB ∼ TVI + DEM | 52.62 | 7 | AGB ∼ TVI + SEC + DEM | 45.61 |
| 8 | AGB ∼ PC2 | 46.66 | 8 | AGB ∼ PC2 + SEC | 41.87 |
| 9 | AGB ∼ TVI + DEM | 45.28 | 9 | AGB ∼ TVI + DEM | 45.28 |
| 10 | AGB ∼ PC2 + DEM | 37.97 | 10 | AGB ∼ PC2 + SEC + DEM | 36.36 |
| Fold 2 | |||||
| 1 | AGB ∼ TVI + SEC | 43.79 | 1 | AGB ∼ TVI + SEC | 43.79 |
| 2 | AGB ∼ TVI + SEC + DEM | 29.44 | 2 | AGB ∼ TVI + SEC + DEM | 29.44 |
| 3 | AGB ∼ PC2 + DEM | 55.61 | 3 | AGB ∼ PC2 + SEC+ DEM | 54.56 |
| 4 | AGB ∼ TVI + DEM | 52.79 | 4 | AGB ∼ TVI + SEC + DEM | 49.52 |
| 5 | AGB ∼ TVI + SEC + DEM | 54.42 | 5 | AGB ∼ TVI + SEC + DEM | 54.42 |
| AGB ∼ PC2 + DEM | 35.43 | 6 | AGB ∼ PC2 + DEM | 35.43 | |
| 7 | AGB ∼ PC2 + SEC + DEM | 43.03 | 7 | AGB ∼ PC2 + SEC + DEM | 43.03 |
| 8 | AGB ∼ PC2 + TVI + SEC + DEM | 53.51 | 8 | AGB ∼ PC2 + SEC + DEM | 54.27 |
| 9 | AGB ∼ PC2 + SEC + DEM | 47.57 | 9 | AGB ∼ PC2 + SEC + DEM | 47.57 |
| 10 | AGB ∼ PC2 + SEC + DEM | 48.34 | 10 | AGB ∼ PC2 + SEC +DEM | 48.34 |
| Fold 3 | |||||
| 1 | AGB ∼ TVI + SEC + DEM | 42.54 | 1 | AGB ∼ PC2 + SEC + DEM | 41.17 |
| 2 | AGB ∼ PC2 + TVI + SEC + DEM | 62.36 | 2 | AGB ∼ PC2 + SEC + DEM | 57.94 |
| 3 | AGB ∼ TVI + SEC | 31.94 | 3 | AGB ∼ TVI + SEC | 31.94 |
| 4 | AGB ∼ PC2 + SEC | 40.86 | 4 | AGB ∼ PC2 + SEC | 40.86 |
| 5 | AGB ∼ PC2 + DEM | 43.33 | 5 | AGB ∼ PC2 + SEC + DEM | 41.59 |
| 6 | AGB ∼ TVI + SEC + DEM | 49.65 | 6 | AGB ∼ TVI + SEC + DEM | 49.65 |
| 7 | AGB ∼ TVI + SEC + DEM | 38.84 | 7 | AGB ∼ TVI + SEC + DEM | 38.84 |
| 8 | AGB ∼ TVI + DEM | 31.33 | 8 | AGB ∼ TVI + DEM | 31.33 |
| 9 | AGB ∼ TVI + SEC | 49.82 | 9 | AGB ∼ TVI + SEC | 49.82 |
| 10 | AGB ∼ TVI + SEC + DEM | 51.47 | 10 | AGB ∼ TVI + SEC + DEM | 51.47 |
| Iteration | STEPWISE-AIC | MAPE | Iteration | RI | MAPE |
|---|---|---|---|---|---|
| Fold 1 | |||||
| 1 | AGB ∼ b3 + b4 + PC1 + PC3 + PC5 + VAR + DEM | 62.04 | 1 | AGB ∼ VI + b3 + PC1 + PC5 | 52.93 |
| 2 | AGB ∼ b4 + PC1 + PC5 | 39.14 | 2 | AGB ∼ b4 + PC3 + PC5 | 39.14 |
| 3 | AGB ∼ b3 + b4 + PC1 + PC3 + PC5 + VI + VAR | 59.46 | 3 | AGB ∼ b3 + PC5 | 56.63 |
| 4 | AGB ∼ b3 + b4 + PC1 + PC3 + PC5 + VAR + DEM | 38.12 | 4 | AGB ∼ VI + b4 + VAR + DEM | 32.07 |
| 5 | AGB ∼ b4 + PC1 + PC3 + PC5 | 38.61 | 5 | AGB ∼ VI + b4 | 42.35 |
| 6 | AGB ∼ b3 + PC1 + PC5 + VI | 55.96 | 6 | AGB ∼ VI | 41.97 |
| 7 | AGB ∼ b3 + b4 + PC1 + PC3 + PC5 | 34.26 | 7 | AGB ∼ VI + PC5 | 39.65 |
| 8 | AGB ∼ b3 + b4 + PC1 + PC3 + PC5 | 39.43 | 8 | AGB ∼ VI + b3 + PC1 + PC5 | 44.03 |
| 9 | AGB ∼ b4 + PC1 + PC3 + PC5 | 45.23 | 9 | AGB ∼ VI + PC5 | 44.38 |
| 10 | AGB ∼ b4 + PC1 + PC3 + PC5 + VAR | 42.45 | 10 | AGB ∼ VI | 33.47 |
| Fold 2 | |||||
| 1 | AGB ∼ b4 + PC1 + PC5 | 44.43 | 1 | AGB ∼ b3 + PC5 | 43.52 |
| 2 | AGB ∼ b3 + b4 + PC1 + PC3 + PC5 | 59.52 | 2 | AGB ∼ VI | 48.01 |
| 3 | AGB ∼ b3 + b4 + VI | 35.47 | 3 | AGB ∼ VI + DEM | 30.14 |
| 4 | AGB ∼ b3 + b4 + PC1 + PC3 + PC5 + VI | 51.70 | 4 | AGB ∼ VI + PC1 + PC5 | 55.98 |
| 5 | AGB ∼ b3 + b4 + PC1 + PC3 + PC5 + VI | 54.64 | 5 | AGB ∼ b3 + b4 + PC1 + PC3 + PC5 + VI | 54.64 |
| 6 | AGB ∼ b3 + b4 + PC1 + PC3 + PC5 + DEM | 40.26 | 6 | AGB ∼ VI + PC5 + DEM | 34.56 |
| 7 | AGB ∼ b3 + b4 + PC1 + PC3 + PC5 + VAR + DEM | 39.28 | 7 | AGB ∼ VI + PC5 + DEM | 33.49 |
| 8 | AGB ∼ b3 + b4 + PC1 + PC3 + PC5 + VI + VAR + DEM | 55.14 | 8 | AGB ∼ VI | 32.31 |
| 9 | AGB ∼ b3 + b4 + PC1 + PC3 + PC5 | 35.48 | 9 | AGB ∼ b3 + b4 + PC1 + PC3 + PC5 | 35.48 |
| 10 | AGB ∼ b4 + PC1 + PC3 + VI | 50.12 | 10 | AGB ∼ VI + b3 + PC1 + PC5 | 52.95 |
| Fold 3 | |||||
| 1 | AGB ∼ b3 + b4 + PC1 + PC3 + PC5 | 31.86 | 1 | AGB ∼ VI + PC5 | 31.2 |
| 2 | AGB ∼ b3 + b4 + PC1 + PC3 + PC5 + VAR | 41.57 | 2 | AGB ∼ VI | 30.73 |
| 3 | AGB ∼ b4 + PC1 + PC5 | 52.02 | 3 | AGB ∼ PC5 + b3 + PC1 | 52.82 |
| 4 | AGB ∼ b4 + PC1 + PC3 + VI | 37.41 | 4 | AGB ∼ VI + PC5 | 35.07 |
| 5 | AGB ∼ b4 + PC1 + PC3 + PC5 + VI | 60.39 | 5 | AGB ∼ VI + b4 + PC1 + PC3 | 46.68 |
| 6 | AGB ∼ b3 + b4 + PC1 | 49.76 | 6 | AGB ∼ b3 + VAR | 38.68 |
| 7 | AGB ∼ VI | 49.09 | 7 | AGB ∼ VI | 49.09 |
| 8 | AGB ∼ b3 + PC1 | 44.29 | 8 | AGB ∼ b3 | 41.60 |
| 9 | AGB ∼ VI | 38.59 | 9 | AGB ∼ VI | 38.59 |
| 10 | AGB ∼ b3 + b4 + PC1 + PC3 + PC5 + VI + DEM | 64.54 | 10 | AGB ∼ b3 + b4 + PC1 + PC3 + PC5 + DEM | 53.80 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adame-Campos, R.L.; Ghilardi, A.; Gao, Y.; Paneque-Gálvez, J.; Mas, J.-F. Variables Selection for Aboveground Biomass Estimations Using Satellite Data: A Comparison between Relative Importance Approach and Stepwise Akaike’s Information Criterion. ISPRS Int. J. Geo-Inf. 2019, 8, 245. https://doi.org/10.3390/ijgi8060245
Adame-Campos RL, Ghilardi A, Gao Y, Paneque-Gálvez J, Mas J-F. Variables Selection for Aboveground Biomass Estimations Using Satellite Data: A Comparison between Relative Importance Approach and Stepwise Akaike’s Information Criterion. ISPRS International Journal of Geo-Information. 2019; 8(6):245. https://doi.org/10.3390/ijgi8060245
Chicago/Turabian StyleAdame-Campos, Rita Libertad, Adrian Ghilardi, Yan Gao, Jaime Paneque-Gálvez, and Jean-François Mas. 2019. "Variables Selection for Aboveground Biomass Estimations Using Satellite Data: A Comparison between Relative Importance Approach and Stepwise Akaike’s Information Criterion" ISPRS International Journal of Geo-Information 8, no. 6: 245. https://doi.org/10.3390/ijgi8060245






